Abstract
Class imbalance is a phenomenon of asymmetry that degrades the performance of traditional classification algorithms such as the Support Vector Machine (SVM) and Extreme Learning Machine (ELM). Various modifications of SVM and ELM have been proposed to handle the class imbalance problem, which focus on different aspects to resolve the class imbalance. The Universum Support Vector Machine (USVM) incorporates the prior information in the classification model by adding Universum data to the training data to handle the class imbalance problem. Various other modifications of SVM have been proposed which use Universum data in the classification model generation. Moreover, the existing ELM-based classification models intended to handle class imbalance do not consider the prior information about the data distribution for training. An ELM-based classification model creates two symmetry planes, one for each class. The Universum-based ELM classification model tries to create a third plane between the two symmetric planes using Universum data. This paper proposes a novel hybrid framework called Reduced-Kernel Weighted Extreme Learning Machine Using Universum Data in Feature Space (RKWELM-UFS) to handle the classification of binary class-imbalanced problems. The proposed RKWELM-UFS combines the Universum learning method with a Reduced-Kernelized Weighted Extreme Learning Machine (RKWELM) for the first time to inherit the advantages of both techniques. To generate efficient Universum samples in the feature space, this work uses the kernel trick. The performance of the proposed method is evaluated using 44 benchmark binary class-imbalanced datasets. The proposed method is compared with 10 state-of-the-art classifiers using AUC and G-mean. The statistical t-test and Wilcoxon signed-rank test are used to quantify the performance enhancement of the proposed RKWELM-UFS compared to other evaluated classifiers.
1. Introduction
The performance of a classification problem is affected by various data complexity measures such as class imbalance, class overlapping, length of the decision boundary, small disjuncts of classes, etc. In the classification domain, most of the real-world problems are class imbalanced. Examples of such problems are cancer detection [1,2], fault detection [3], intrusion detection system [4], software test optimization [5], speech quality assessment [6], pressure prediction [7], etc. In a problem when the number of samples in one class outnumbers the numbers of samples in some other class, it is considered as a class imbalanced/asymmetric problem. The class with a greater number of instances is the majority class and the class with fewer instances is the minority class. In real-world problems, usually, the minority class instances have more importance than the majority class.
Traditional classifiers such as the support vector machine (SVM), Naive Bayes, decision tree, and extreme learning machine (ELM) are biased towards the correct classification of majority class data. Various approaches have been proposed to handle such class-imbalanced classification problems, which can be classified as data sampling, algorithmic and hybrid methods [8].
In classification, the idea of using additional data along with the original training data has been used widely for better training of the model. The virtual example method, oversampling method, noise injection method, and Universum data creation method are some examples that use additional data. The oversampling method generates additional data in the majority class to balance the data distribution in the classes. In the virtual example and noise injection methods, labeled synthetic data are created that may not come from the same distribution as the original data. Universum data creation methods allow the classifier to encode prior knowledge by representing meaningful concepts in the same domain as the problem at hand as stated in [9]. In Universum learning-based classification models, the Universum data are added to the training data to enhance performance. Universum data are data that do not belong to any of the target classes. The two main factors which affect the performance of Universum data are the number of Universum data created and the method used for the creation of Universum data. Different methods have been used for the creation of Universum; among those, the two most common methods widely used are the use of examples from other classes and random averaging [9].
Several methods have been proposed that use Universum data in the training of SVM based classifiers to handle the class imbalance problem, such as the Universum Support Vector Machine (USVM) [9], Twin support vector machine with Universum data (TUSVM) [10], and Cost-Sensitive Universum-SVM (CS-USVM) [11]. A Universum support vector machine-based model for EEG signal classification has been proposed in [12]. A nonparallel support vector machine for a classification problem with Universum learning has been proposed in [13]. An improved non-parallel Universum support vector machine and its safe sample screening rules are proposed in [14]. Tencer et al. [15] used Universum data with other classifiers such as fuzzy models to demonstrate its usefulness in combination with fuzzy models. Recently, a Multiple Universum Empirical Kernel Learning (MUEKL) [16] classifier has been proposed to handle class imbalance by combining the Universum learning with Multiple Empirical Kernel Learning (MEKL).
Extreme Learning Machine (ELM) [17] is a single hidden-layer feed-forward neural network designed for regression and classification with fast speed and better generalization performance, but it cannot handle the classification of class-imbalanced problems effectively. Various ELM based models have been proposed to handle the classification of class imbalance problems, such as Weighted Extreme Learning Machine (WELM) [18], Class-Specific Cost Regulation Extreme Learning Machine (CCR-ELM) [19], Class-Specific Kernelized Extreme Learning Machine (CSKELM) [20], Reduced-Kernelized Weighted Extreme Learning Machine (RKWELM) [21], UnderBagging-based Kernelized Weighted Extreme Learning Machine (UBKWELM) [22], and UnderBagging-based Reduced-Kernelized Weighted Extreme Learning Machine (UBRKWELM) [21]. The proposed work is motivated by the idea that none of the existing ELM-based models for classification encode prior knowledge in the training model using Universum data.
This work proposes a novel hybrid classification model called Reduced-Kernel Weighted Extreme Learning Machine using Universum data in Feature Space (RKWELM-UFS) which incorporates the Universum data in the RKWELM model. The contributions of the proposed approach are listed below.
- This work is the first attempt that utilized the Universum data in a Reduced-Kernelized Weighted Extreme Learning Machine (RKWELM)-based classification model to handle the class imbalance problem.
- The Weighted Kernelized Synthetic Minority Oversampling Technique (WKSMOTE) [23] is an oversampling-based classification method in which the synthetic samples are created in the feature space of the Support Vector Machine (SVM). Inspired by WKSMOTE, the proposed work creates the Universum samples in the feature space.
- The proposed method uses the kernel trick to create the Universum samples in the feature space between randomly selected instances of the majority and minority classes.
- In a classification problem, the samples located near the decision boundary contribute more to better training. The creation of Universum samples in feature space ensures that the Universum samples lie near the decision boundary.
The rest of the paper is structured as follows. In the related work section, Universum learning, class imbalance learning, ELM classifier, and its variants are discussed in detail. The proposed work section provides a detailed explanation of the proposed RKWELM-UFS classifier. The experimental setup and result analysis section provide the specification of the dataset used in the experiments, parameter settings of the proposed algorithm, the evaluation metrics used for performance evaluation, and the experimental results obtained in form of various tables and figures. The last section provides the concluding remarks and future research directions.
2. Related Work
The following section provides the literature related to Universum learning, class imbalance learning, and some of the existing ELM-based models to handle class imbalance learning.
2.1. Universum Learning
The idea of using Universum data is close to the idea of using the prior knowledge in Bayesian classifiers [9]. However, there is a conceptual difference between the two approaches, i.e., the prior knowledge is knowledge about decision rules used in Bayesian inference, while the Universum is knowledge about the admissible collection of examples. Similarly to the Bayesian prior probability, the Universum data encode prior information.
It has been observed by various researchers [9,15,24] that the effect of Universum is dependent on the quality of Universum samples created. A safe sample screening rule for Universum support vector machines, in which the non-contributed data can be identified and safely eliminated before the training process, can obtain the same solution as solving the original problem is proposed in [25]. An improved version of the non-parallel Universum support vector machine and its safe sample screening rule is proposed in [14]. It is suggested in [24] that not all the Universum samples are helpful for effective classification, so they proposed selecting the informative Universum samples for semi-supervised learning, which is a method used to identify informative samples among the Universum samples. An empirical study on the Universum support vector machine (USVM), which describes some practical conditions for evaluating the effectiveness of random averaging for the creation of Universum data, is performed in [26].
2.2. Class Imbalance Learning
The classification performance of traditional classifiers degrades when there is an imbalance in the ratio of the majority and minority class data. Different approaches have been used in classification to deal with the problem of class imbalance. Table 1 provides the categorization of the proposed methods and other methods used in this work for comparison. Table 1 also provides the strategy and basic ideas used in the respective methods. The broad categories of these approaches are discussed in the following subsections.

Table 1.
Categorization and comparison of the proposed method and other methods in comparison used to handle classification of class imbalance problems.
2.2.1. Data Level Approach
The data-level methods are based on balancing the ratio of data to convert an imbalanced classification problem into a balanced classification problem. These methods can be seen as data pre-processing methods because they try to handle the class imbalance present in the data before the classification model generation. The data-level approaches can be broadly categorized as under-sampling, oversampling and, hybrid sampling methods.
The under-sampling methods remove some of the data (i.e., the majority samples) to decrease the imbalance ratio of a training dataset. These methods may suffer from data loss, as some of the important samples may be removed. The efficiency of an under-sampling method lies in its ability to select the right samples which can be removed from the dataset. The under-sampling methods reduce the time complexity of a given class-imbalanced classification problem. A combined weighted multi-objective optimizer for instance reduction in a two-class imbalanced data problem is proposed in [27]. Clustering-Based Under-Sampling (CBUS) [28] uses clustering of majority class data for the under-sampling. Fast Clustering-Based Under-Sampling (FCBUS) [29] is a modified version of CBUS which clusters the minority class data for under-sampling to reduce the time complexity of CBUS.
The oversampling method adds some additional data (in the minority class) to decrease the imbalance ratio of the training dataset. The additional samples are obtained by creating synthetic minority class samples or replicating the existing minority class samples. These methods can lead to over-fitting problems in model generation. The oversampling methods increase the time complexity of a given class imbalance classification problem. The synthetic minority oversampling technique (SMOTE) [30] is a popular oversampling method, widely used to handle class imbalance, in which synthetic minority samples are created. Several variants of SMOTE have been proposed to further enhance the performance of class imbalance dataset classification, such as Borderline SMOTE, Borderline SMOTE1, Borderline SMOTE2, Safe- Level-SMOTE, MSMOTE [31], and CSMOTE [32]. The hybrid sampling methods such as SCUT [16] try to reduce the class imbalance by using both oversampling and under-sampling.
2.2.2. Algorithmic Approach
There are some approaches in which the classification algorithm is able to handle class imbalance problems, such as cost-sensitive and one-class learning approaches. The cost-sensitive methods assign a different cost to the misclassification of different classes. In an imbalance problem, generally, the misclassification cost of minority class samples is higher than the misclassification cost of majority class samples. The efficiency of any cost-sensitive method lies in the selection of misclassification costs for different classes. Multiple Random Empirical Kernel Learning (MREKL) [33] is a cost-sensitive classification model which emphasizes the importance of samples located in overlapping regions of positive and negative classes and ignores the effects of noisy samples to achieve better performance in class imbalance problems. Weighted Extreme Learning Machine (WELM) [18] is a weighted version of Extreme Learning Machine (ELM) [17] that minimizes the weighted error by incorporating a weight matrix in the optimization problem of ELM. Class-Specific Extreme Learning Machine (CSELM) [34] is a variant of WELM which replaces the weight matrix with two constant weight values for each class. Class-Specific Kernel Extreme Learning Machine (CSKELM) [20] is the modification of CSELM which uses the Gaussian kernel function to map the input data to feature space. Class-Specific Cost-Regulation Extreme Learning Machine (CCR-KELM) [19] is the variant of KELM which uses different regularization perimeters for the classes.
The one-class learning approach is also called single-class learning. In these methods, the classifier learns only one class as the target class. In this approach generally, the minority class is considered as the target class. Multi-Kernel Support Vector Data Description with boundary information proposes a novel method called MKL-SVDD [35] by introducing Multi- Kernel Learning (MKL) into the traditional Support Vector Data Description (SVDD) based on the boundary information to form one-class learning.
2.2.3. Hybrid Approach
In a hybrid approach, multiple classification approaches are combined to handle a class imbalance problem. Some hybrid techniques combine ensemble techniques with data sampling methods such as over-sampling or under-sampling to handle class imbalance problems. RUSBoost [36] is a hybrid technique that combines random under-sampling with boosting to create an ensemble of classifiers. UBKELM [22] and UBRKELM [21] are two hybrid classification models that combine underbagging with KELM and RKELM respectively. BPSO-AdaBoost-KNN [37] is a method that implements BPSO as the feature selection algorithm and then designs an AdaBoost-KNN classifier to convert the traditional weak classifier into a strong classifier. UBoost: Boosting with the Universum [38] is a technique that combined the Universum sample creation with a boosting framework. An Adaptive-Boosting (AdaBoost) algorithm [39] uses multiple iterations to learn multiple classifiers in a serial manner to generate a single strong learner.
Some hybrid techniques combine cost-sensitive approaches with ensemble techniques such as Ensemble of Weighted Extreme Learning Machine (EWELM) [40] and Boosting Weighted Extreme Learning machine (BWELM) [41]. In EWELM, the weight of each component classifier in the ensemble is optimized by using a differential evolution algorithm. BWELM is a modified AdaBoost framework that combines multiple Weighted ELM-based classifiers in a boosting manner. The main idea of BWELM is to find the better weights in each base classifier.
2.3. Extreme Learning Machine (ELM) and Its Variants to Handle Class Imbalance Learning
ELM [17,42] is a generalized single hidden-layer feed-forward neural network, which provides good generalization performance and disposes of the iterative time-consuming training process. It uses the Moore–Penrose pseudoinverse for computing the weights between the hidden and the output layer which make it fast. For a given classification dataset with N training samples , where is the input feature vector and is the output label vector. Here, the vector/matrix transpose is denoted by superscript T. During the training time, these weights are randomly generated and are not changed further. The hidden neurons bias matrix is denoted by , where is the bias of the jth hidden neuron. In ELM, for a given training/testing sample, i.e., , the hidden layer output is calculated as follows:
Here, is the activation function of the hidden neurons. In ELM, for a binary classification problem, the decision function, i.e., for a sample is given as:
where is the output weight matrix. The hidden layer output matrix H can be written as follows:
ELM minimizes the training error and the norm of the output weights as:
In the original implementation of ELM [17], the minimal norm least-square method instead of the standard optimization method was used to find β.
where is the Moore–Penrose generalized inverse of matrix H. In [17,42] the orthogonal projection method is used to calculate , which can be used in two cases.
When is nonsingular then,
When is singular then
In ELM the constrained optimization-based problem for classification with multiple output nodes was formulated as follows:
Subjected to:
The output layer weights can be obtained using two solutions
Case 1. Where the Number of Training Samples is Not Huge:
Case 2. Where the Number of Training Samples is Huge:
2.3.1. Weighted Extreme Learning Machine (WELM)
Conventional ELM does not account for good generalization performance while dealing with the class-imbalance learning problems. Weighted Extreme Learning Machine (WELM) [18] is a cost-sensitive version of ELM which was proposed for handling the class-imbalanced learning problem effectively. In cost-sensitive learning methods, the different cost is assigned to the misclassification of different class samples. In WELM, two generalized weighting schemes were proposed. These generalized weighting schemes assign weights to the training samples as per their class distribution. In WELM [18], the following optimization problem is formulated:
Subjected to:
Here, C is the regularization parameter and is a diagonal matrix whose diagonal elements are the weights assigned to the training samples. The two weighting schemes proposed by WELM are:
Weighting scheme W1:
Here, and is the total number of samples belonging to kth class.
Weighting scheme W2:
Here, represents the average number of samples for all classes. Weight is assigned to the ith samples. Samples belonging to the minority class will be assigned weights equal to, in both the weighting schemes. The second weighting scheme assigns a lesser weight to the majority class samples compared to the first weighting scheme. The two variants of WELM are sigmoid node-based WELM and Gaussian kernel-based WELM, which are described as follows.
- Sigmoid node-based Weighted Extreme Learning Machine
The Sigmoid node-based WELM uses random input weights and Sigmoid activation function i.e., G(.), to find the hidden layer output matrix H given in Equation (3). The solution of the optimization problem of WELM as given in [18] is reproduced below:
The two solutions are given for two cases. The first solution is given for the case when the number of training samples is smaller than the number of selected hidden layer neurons. The second solution is given for the case where the number of selected hidden layer neurons is smaller than the number of training samples.
- Gaussian kernel-based Weighted Extreme Learning Machine (KWELM)
In KELM [42], the kernel matrix of the hidden layer is represented as follows:
The Gaussian kernel-based WELM maps the input data to the feature space as follows:
Here, represents the kernel width parameter, represents the ith sample and represents the jth centroid; represents the distance of the jth centroid to the ith input sample. The number of Gaussian kernel functions i.e., the centroids used in [32] was equal to the number of training samples. On applying Mercer’s condition, the kernel matrix of KELM [42] can be represented as given below:
The output of KWELM is determined in [18] which is represented as follow:
Compared to the Sigmoid node-based WELM, KWELM has better classification performance, as stated in [18].
2.3.2. Reduced Kernel Weighted Extreme Learning Machine (RKWELM)
Reduced-Kernel Extreme Learning Machine (RKELM) [43] is a fast and accurate kernel-based supervised algorithm for classification. Unlike Support Vector Machine (SVM) or Least-Square SVM (LS-SVM), which identify the support vectors or weight vectors iteratively, the RKELM randomly selects a subset of the available data samples as centroids or mapping samples. The weighted version of RKELM i.e., Reduced-Kernel Weighted Extreme Learning Machine (RKWELM) is proposed in [21] for class imbalance learning. In RKWELM, a reduced number of kernels are selected, which act as the centroids. The number of Gaussian kernel functions used in RKWELM is denoted as where . The kernel matrix of the hidden layer can be reproduced as given by the following equation.
Here, represents the ith sample and represents the jth centroid and . In the case when the output of RKWELM can be given by the following equation.
The final output of RKWELM, as given in [43], is computed as:
2.3.3. UnderBagging-Based Kernel Extreme Learning Machine (UBKELM)
UnderBagging-Based Kernel Extreme Learning Machine (UBKELM) [22] is an ensemble of KELM. UBKELM creates several balanced training subsets by random under-sampling of the majority class samples. K is the number of balanced subsets that are created by selecting M number of majority samples and all the minority samples in each subset, where M is the number of minority samples in the training dataset and K is the ceiling value of the imbalance ratio of the training dataset. In the subset creation, the majority samples are selected using the random under-sampling method. There are two variants of UBKELM, i.e., UnderBagging-Based Kernel Extreme Learning Machine-Max Voting (UBKELM-MV) and UnderBagging-Based Kernel Extreme Learning Machine-Soft Voting (UBKELM-SV) in which the ultimate outcome of the ensemble is computed by majority voting and soft voting respectively.
2.3.4. UnderBagging-Based Reduced-Kernelized Weighted Extreme Learning Machine
UnderBagging-based Reduced-Kernelized Weighted Extreme Learning Machine (UBRKELM) [21] is an ensemble of Reduced Kernelized Weighted Extreme Learning Machine (RKWELM). The UBRKELM creates several balanced training subsets and learns multiple classification models with these balanced training subsets using RKWELM as the classification algorithm. K is the number of balanced subsets that are created by selecting M number of majority samples and all the minority samples in each subset, where M is the number of minority samples in the training dataset and K is the ceiling value of the imbalance ratio of the training dataset. In UBRKELM the reduced number of kernel functions is used as centroids to learn an RKELM model. Two variants of UBRKWELM are proposed, UBRKWELM-MV and UBRKWELM-SV, in which the final outcome of the ensemble is computed by majority voting and soft voting respectively.
3. Proposed Method
This work proposes a novel Reduced-Kernel Weighted Extreme Learning Machine using Universum data in Feature Space (RKWELM-UFS) to handle the class imbalance classification problem. In the proposed work, the Universum data along with the original training data is provided to the classifier for training purposes, to improve its learning capability. The proposed method creates Universum samples in the feature space because the mapping of input data from the input space to the feature space is not conformal.
The following subsections describe the process of creation of the Universum samples in the input space, the process of creation of the Universum samples in the feature space, the proposed RKWELM-UFS classifier, and the computational complexity of the proposed RKWELM-UFS classification model. Algorithm 1 provides the pseudo-code of the proposed RKWELM-UFS.
3.1. Generation of Universum Samples in the Input Space
To generate a Universum sample between a majority sample and a minority sample , the following equation can be used:
where δ represents a random number in the uniform distribution U [0, 1].
3.2. Generation of Universum Samples in the Feature Space
To generate a Universum sample in the feature space between a majority sample and a minority sample the following equation can be utilized:
where, is the feature transformation function which is generally unknown and is a random number between [0, 1]. The proposed work uses = 0.5. Similarly to SVM, LS-SVM, and PSVM, the transformation function need not be known to users; instead, its kernel function can be deployed. If a feature mapping is unknown to users, one can apply Mercer’s conditions on ELM to define a kernel matrix for KELM [17] as follows:
In the proposed work, we have to calculate the kernel function , where represents the original target training sample and is the Universum sample. According to [23] without computing and , we can obtain the corresponding kernel using the following equation:
3.3. Proposed Reduced-Kernel Weighted Extreme Learning Machine Using Universum Samples in Feature Space (RKWELM-UFS)
Training of an ELM [42] based classifier requires the computation of the output layer weight matrix β. The proposed RKWELM-UFS uses the same equation as RKWELM [21] to obtain the output layer weight matrix β which is reproduced below:
where, W is the diagonal weight matrix, which gives different weights to the majority class, the minority class, and the Universum instances using Equation (10), T is the target vector in which the class label for Universum samples is set to 0 (given the class label of majority and minority class are +1 and −1 respectively), and is the kernel matrix of the proposed RKWELM-UFS.
In the proposed work, the Universum instances are added to the training process along with the original training instances. The reason behind computing β in the same manner as RKWELM is that the proposed RKWELM-UFS computes the kernel matrix by deploying the original training instances excluding the Universum instances as centroids. The value of is obtained by augmentation of the two matrices . The following subsections describe the computation of , and .
3.3.1. Computation of
The proposed work computes the kernel matrix for the N number of original training instances termed as in the same manner as it was computed in the KELM [42], which is represented as:
3.3.2. Computation of
Equation (20) can be used to create a Universum sample between two original training samples and in feature space. As we have discussed the transformation function is unknown to the user, so the computation of is not possible here. For convenience, we will refer to the Universum sample as . In the proposed work without computing , we can directly compute the corresponding kernel . ) is calculated using Equation (22). In the proposed algorithm, only the original training samples are used as centroids, so the matrix for p number of Universum samples and N number of original training samples can be represented as:
3.3.3. Computation of
The addition of Universum samples in the training process requires that the original kernel matrix i.e., be augmented to include the matrix . The final hidden layer output kernel matrix of the proposed RKWELM-UFS is obtained by augmentation of the two matrices and which is denoted as .
The output of RKWELM-UFS can be obtained using Equation (18) used in RKWELM, which is reproduced below:
Here represents the test instance and represent the training instance for i = 1, 2, …, N.
| Algorithm 1 Pseudocode of the proposed RKWELM-UFS |
| INPUT: Training Dataset |
| Number of Universum samples to be generated: p |
| OUTPUT: |
| 1: Calculate the kernel matrix as shown in Equation (24) for the N number of original training instances using Equation (21). |
| 2: Calculate the kernel matrix as shown in Equation (25) for the N number of training instances and p number of Universum instances as follows. |
| for j = 1 to p |
| Randomly select one majority instance |
| Randomly select one minority instance |
| for i = 1 to N |
| calculate using Equation (22) |
| End |
| End |
| 3: Augment the matrix with the matrix to obtain the reduced kernel matrix using Universum samples shown in Equation (26). |
| 4: To obtain the output weight matrix β use the Equation (23). |
| 5: To determine the class label of an instance x use the Equation (27). |
3.4. Computational Complexity
For training of the ELM-based classification algorithm, it is necessary to obtain the output layer weight matrix i.e., . For the proposed RKWELM-UFS is obtained using Equation (23) which is reproduced below:
Here, is a matrix of size , where N is the number of training instances and p is the number of Universum samples. The weight matrix, i.e., W, is of size and the target matrix i.e., T is of size where c is the number of target class labels; here, the number of target class labels is 2 because we are using the binary classification problems. To compute first we need to compute the and . In the following steps the computational complexity of computing is identified step by step:
- The computational complexity of calculating i.e., the kernel matrix shown in Equation (24) is , where n is the number of features of training data in input space.
- The computational complexity of calculating matrix shown in Equation (25) is .
- The computational complexity of the output weights can be calculated as
- 3.1
- Matrix multiplications:Computational complexity:
- 3.2
- Computational complexity of computing the inverse of N × N matrix computed in Step 3.1 is
- 3.3
- Computational complexity of matrix multiplications is
- 3.4
- Computational complexity of matrix multiplication of 2 matrices obtained in Step 3.1 and Step 3.3 is
The final computational complexity of calculating β is . The computational complexity can be simplified to because the value of c is 2, the value of n is smaller than N, and the maximum value of p can be N.
4. Experimental Setup and Result Analysis
This section provides the experiments performed to evaluate the proposed work, which includes the specification of the datasets used for experimentation, the parameter settings of the proposed algorithm, the evaluation metrics used for performance comparison, and the results obtained through experiments and performance comparison with the state-of-the-art classifiers.
4.1. Dataset Specifications
The proposed work uses 44 binary class-imbalanced datasets for performing the experiments. These datasets are downloaded from the KEEL dataset repository [44,45] in 5-fold cross-validation format. Table 2 provides the specification of these datasets. In Table 2, # Attributes denote the number of features, # Instances denotes the number of instances and, IR denotes the class imbalance ratio in the presented datasets. The class imbalance ratio (IR) for the binary class dataset can be defined as follows:

Table 2.
Specification of 44 benchmark datasets from KEEL dataset repository.
The datasets used for the experiments are normalized using min-max normalization in the range [1, −1] using the following equation:
Here, the original feature value of nth feature is denoted by x, minimum value of nth feature is denoted by minn and the maximum value of nth feature is denoted by maxn.
4.2. Evaluation Matrix
The confusion matrix, also called the error matrix, can be employed to evaluate the performance of a classification model. It allows the visualization of the performance of an algorithm. In a confusion matrix TP denotes True Positive, TN denotes True Negative, FP denotes False Positive, and FN denotes False Negative.
Accuracy is not a suitable measure to evaluate the performance of a classifier when dealing with a class-imbalanced problem. The other performance matrices used for the performance evaluation in such problems are G-mean and AUC (area under the ROC curve). The AUC defines the measure of the entire area under the ROC curve in two dimensions. The ROC known as receiver operating characteristic curve is a graph that shows the performance of the model by plotting and on the graph.
Here,
4.3. Parameter Settings
The proposed RKWELM-UFS creates Universum samples between randomly selected pairs of majority and minority samples. Because of the randomness, this work presents the mean (denoted as tstR or TestResult) and standard deviation (denoted as std) of the test G-mean and test AUC obtained for 10 trials. The proposed RKWELM-UFS has two parameters, namely the regularization parameter C and the Kernel width parameter σ (denoted as KP). The optimal values of these parameters are obtained using grid search, by varying them on the range respectively.
4.4. Experimental Results and Performance Comparison
The proposed RKWELM-UFS is compared with three sets of algorithms used to handle class imbalance learning. The first set contains the existing approaches which use Universum samples in the classification model generation to handle class-imbalanced problems such as MUEKL [16] and USVM [9]. The second set of approaches consists of the single classifiers such as KELM [46], WKELM [18], CCR-KELM [19], and WKSMOTE [23] which are used to handle class-imbalanced problems. The third set contains the popular ensemble classifiers such as RUSBoost [36], BWELM [41], UBRKELMMV [21], UBRKELM-SV [21], UBKELM-MV [22], and UBKELM-SV [22].
The statistical t-test and Wilcoxon signed-rank test are used to evaluate the performance of the proposed RKWELM-UFS and other methods in consideration. In the t-test result, the value of H (null hypothesis) is 1 if the test rejects the null hypothesis at the 5% significance level, and 0 otherwise.
In the Wilcoxon signed-rank test result, the value of H (null hypothesis) is 1 if the test rejects the null hypothesis that there is no difference between the grade medians at the 5% significance level. In the statistical tests, the p-value indicates the level of significant difference between the compared algorithms; the lower the p-value, the higher the significant difference between the compared algorithms. This work uses AUC and G-mean as the measures of the performance evaluation. The AUC results of classifiers MUEKL and USVM shown in Table 3 are obtained from the work MUEKL [16].

Table 3.
Performance comparison of the proposed RKWELM-UFS with other existing Universum-based classifiers in terms of average AUC (std, KP, and C denote the standard deviation, Kernel width parameter, and regularization parameter, respectively.
4.4.1. Performance Analysis in Terms of AUC
Table 3, Table 4 and Table 5 provide the performance of the proposed RKWELM-UFS and other classification models in terms of AUC. The reported test AUC of the proposed RKWELM-UFS given in Table 3, Table 4 and Table 5 is the averaged test AUC obtained in 10 trials, using 5-fold cross-validation in each trial. Table 3 provides the performance of the proposed RKWELM-UFS and the existing Universum-based classifiers MUEKL and USVM on 35 datasets in terms of average AUC, where the RKWELM outperforms the other classifiers on 32 datasets. Table 4 provides the performance of the proposed RKWELM-UFS and the existing single classifiers like KELM, WKELM, CCR-KELM, and WKSMOTE on 21 datasets in terms of average AUC, where the RKWELM outperforms the other classifiers on 14 datasets. Table 5 provides the performance of the proposed RKWELM-UFS and the existing ensemble of classifiers such as RUSBoost, BWELM, UBRKELM-MV, UBRKELMSV, UBKELM-MV, UBKELM-SV on 21 datasets in terms of average AUC, where the RKWELM outperforms the other classifiers on 10 datasets.

Table 4.
Performance comparison of the proposed RKWELM-UFS with existing single classifiers in terms of average AUC (std., KP, and C denotes the standard deviation, Kernel width parameter, and regularization parameter, respectively.

Table 5.
Performance Comparison of the proposed RKWELM-UFS with existing ensemble of classifiers in terms of average AUC (std, KP, and C denote the standard deviation, Kernel width parameter, and regularization parameter, respectively).
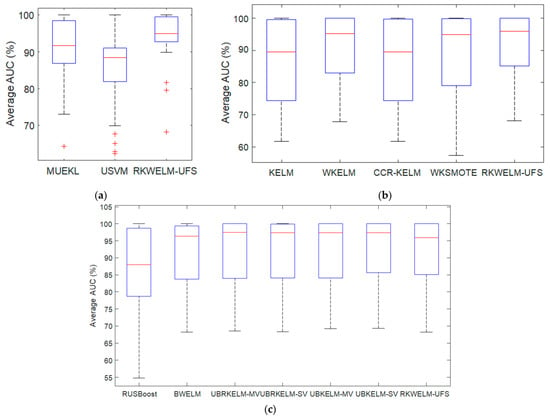
Figure 1a–c shows the boxplot diagram for the AUC results of the classifiers on various datasets shown in Table 3, Table 4 and Table 5 respectively. The boxplot creates a visual representation of the data to visualize the performance. It can be seen in Figure 1a,b that the proposed method RKWELM-UFS has the highest median value and smallest inter-quantile range, which shows that the RKWELM-UFS is performing better than MUEKL, USVM, KELM, WKELM, CCR-KELM, and WKSMOTE. It can be seen in Figure 1c that RKWELM-UFS is performing better than RUSBoost. Table 6 provides the t-test results and Table 7 provides the Wilcoxon Signed-rank test results on the AUC of various algorithms provided in Table 3, Table 4 and Table 5 for comparison. The results provided in Table 6 and Table 7 suggest that the proposed RKWELM-UFS performs significantly better than MUEKL, USVM, KELM, WKELM, CCR-KELM, RUSBoost, and BWELM, and its performance is approximately similar to that of WKSMOTE, UBRKELM-MV, UBRKELM-SV, UBKELMMV, and UBKELM-SV in terms of AUC.

4.4.2. Performance Analysis in Terms of G-mean
Table 8 and Table 9 provide the performance of the proposed RKWELM-UFS and other classification models in terms of the G-mean. The reported test G-mean of the proposed RKWELM-UFS given in Table 8 and Table 9 is the averaged test g-mean obtained in 10 trials, using 5-fold cross-validation in each trial. Table 8 provides the performance of the proposed RKWELM-UFS and the existing single classifiers such as KELM, WKELM, CCR-KELM, and WKSMOTE on 21 datasets in terms of average G-mean, where the RKWELM outperforms the other classifiers on 16 datasets. Table 9 provides the performance of the proposed RKWELM-UFS and the existing ensemble of classifiers such as RUSBoost, BWELM, UBRKELM-MV, UBRKELMSV, UBKELM-MV, and UBKELM-SV on 21 datasets in terms of average Gmean, where the RKWELM outperforms the other classifiers on seven datasets.

Table 8.
Performance Comparison of the proposed RKWELM-UFS with existing single classifiers in terms of average G-mean (std., KP, and C denote the standard deviation, Kernel width parameter, and regularization parameter, respectively).

Table 9.
Performance Comparison of the proposed RKWELM-UFS with existing ensemble of classifiers in terms of average G-mean (std., KP, and C denote the standard deviation, Kernel width parameter, and regularization parameter, respectively).
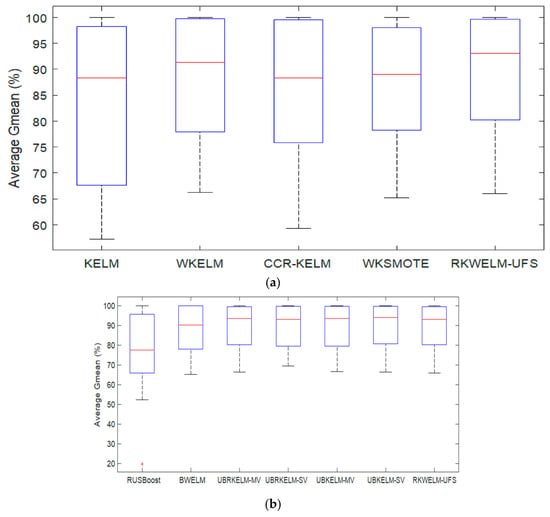
Figure 2a,b shows the boxplot diagram for the G-mean results of the classifiers on various datasets shown in Table 8 and Table 9, respectively. It can be seen in Figure 2a that the proposed RKWELM-UFS has the highest median value and smallest inter-quantile range, which shows that the RKWELM-UFS is performing better than KELM, WKELM, CCR-KELM, and WKSMOTE in terms of the G-mean. It can be seen in Figure 2b that RKWELM-UFS is performing better than RUSBoost and BWELM in terms of the G-mean. Table 10 provides the t-test results and Table 11 provides the Wilcoxon signed-rank test results on the G-mean of various algorithms provided in Table 8 and Table 9 for comparison. The results provided in Table 10 and Table 11 suggest that the proposed RKWELM-UFS performs significantly better than KELM, CCR-KELM, WKSMOTE, and RUSBoost, and performs approximately similarly to WKELM, BWELM, UBRKELM-MV, UBRKELM-SV, UBKELM-MV, UBKELM-SV in terms of the G-mean.

5. Conclusions and Future Work
The use of additional data for training along with the original training data has been employed in many approaches. The Universum data are used to add prior knowledge about the distribution of data in the classification model. Various ELM-based classification models have been suggested to handle the class imbalance problem, but none of these models use prior knowledge. The proposed RKWELM-UFS is the first attempt that employs Universum data to enhance the performance of the RKWELM classifier. This work generates the Universum samples in the feature space using the kernel trick. The reason behind the creation of the Universum instances in the feature space is that the mapping of input data to the feature space is not conformal. The proposed work is evaluated on 44 benchmark datasets with an imbalance ratio between 0:45 to 43:80 and a number of instances between 129 to 2308. The proposed method is compared with 10 state-of-the-art methods used for class-imbalanced dataset classification. G-mean and AUC are used as metrics to evaluate the performance of the proposed method. The paper also incorporates statistical tests to verify the significant performance difference between the proposed and compared methods.
In Universum data-based learning, it has been observed that the efficiency of such classifiers depends on the quality and volume of Universum data created. The methodology of choosing or creating the appropriate Universum samples should be the subject of further research. In the proposed work, the Universum samples are created between randomly selected pairs of majority and minority class samples. In the future, some strategic concepts can be used to select the majority and minority samples instead of random selection. In the future, Universum data can be incorporated in other ELM-based classification models to enhance their learning capability on class imbalance problems. The future work also includes the development of a multi-class variant of the proposed RKWELM-UFS.
Author Contributions
Conceptualization, R.C.; methodology, R.C. and S.S.; Software, R.C.; validation, R.C. and S.S.; formal analysis, R.C.; investigation, R.C.; resources, data curation, R.C.; writing—original draft preparation, R.C.; writing—review and editing, S.S. supervision, S.S. All authors have read and agreed to the published version of the manuscript.
Funding
The work received no funding from any organization, institute, or person.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schaefer, G.; Nakashima, T. Strategies for addressing class imbalance in ensemble classification of thermography breast cancer features. In Proceedings of the 2015 IEEE Congress on Evolutionary Computation (CEC), Sendai, Japan, 25–28 May 2015; pp. 2362–2367. [Google Scholar]
- Sadewo, W.; Rustam, Z.; Hamidah, H.; Chusmarsyah, A.R. Pancreatic Cancer Early Detection Using Twin Support Vector Machine Based on Kernel. Symmetry 2020, 12, 667. [Google Scholar] [CrossRef] [Green Version]
- Hao, W.; Liu, F. Imbalanced Data Fault Diagnosis Based on an Evolutionary Online Sequential Extreme Learning Machine. Symmetry 2020, 12, 1204. [Google Scholar] [CrossRef]
- Mulyanto, M.; Faisal, M.; Prakosa, S.W.; Leu, J.-S. Effectiveness of Focal Loss for Minority Classification in Network Intrusion Detection Systems. Symmetry 2020, 13, 4. [Google Scholar] [CrossRef]
- Tahvili, S.; Hatvani, L.; Ramentol, E.; Pimentel, R.; Afzal, W.; Herrera, F. A novel methodology to classify test cases using natural language processing and imbalanced learning. Eng. Appl. Artif. Intell. 2020, 95, 103878. [Google Scholar] [CrossRef]
- Furundzic, D.; Stankovic, S.; Jovicic, S.; Punisic, S.; Subotic, M. Distance based resampling of imbalanced classes: With an application example of speech quality assessment. Eng. Appl. Artif. Intell. 2017, 64, 440–461. [Google Scholar] [CrossRef]
- Mariani, V.C.; Och, S.H.; dos Santos Coelho, L.; Domingues, E. Pressure prediction of a spark ignition single cylinder engine using optimized extreme learning machine models. Appl. Energy 2019, 249, 204–221. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Weston, J.; Collobert, R.; Sinz, F.; Bottou, L.; Vapnik, V. Inference with the universum. In Proceedings of the 23rd International Conference on Machine Learning, New York, NY, USA, 25 June 2006; pp. 1009–1016. [Google Scholar]
- Qi, Z.; Tian, Y.; Shi, Y.J. Twin support vector machine with Universum data. Neural Netw. 2012, 36, 112–119. [Google Scholar] [CrossRef]
- Dhar, S.; Cherkassky, V. Cost-sensitive Universum-svm. In Proceedings of the 2012 11th International Conference on Machine Learning and Applications, Boca Raton, FL, USA, 12–15 December 2012; pp. 220–225. [Google Scholar]
- Richhariya, B.; Tanveer, M.J. EEG signal classification using Universum support vector machine. Expert Syst. Appl. 2018, 106, 169–182. [Google Scholar] [CrossRef]
- Qi, Z.; Tian, Y.; Shi, Y. A nonparallel support vector machine for a classification problem with Universum learning. J. Comput. Appl. Math. 2014, 263, 288–298. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, Y.; Fujita, H.J. An improved non-parallel Universum support vector machine and its safe sample screening rule. Knowl. Based Syst. 2019, 170, 79–88. [Google Scholar] [CrossRef]
- Tencer, L.; Reznáková, M.; Cheriet, M.J. Ufuzzy: Fuzzy models with Universum. Appl. Soft Comput. 2017, 59, 1–18. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, S.; Yao, L.; Li, D.; Du, W.; Zhang, J. Multiple universum empirical kernel learning. Eng. Appl. Artif. Intell. 2020, 89, 103461. [Google Scholar] [CrossRef]
- Huang, G.-B.; Zhu, Q.-Y.; Siew, C.-K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Zong, W.; Huang, G.-B.; Chen, Y.J. Weighted extreme learning machine for imbalance learning. Neurocomputing 2013, 101, 229–242. [Google Scholar] [CrossRef]
- Xiao, W.; Zhang, J.; Li, Y.; Zhang, S.; Yang, W. Class-specific cost regulation extreme learning machine for imbalanced classification. Neurocomputing 2017, 261, 70–82. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Class-specific kernelized extreme learning machine for binary class imbalance learning. Appl. Soft Comput. 2018, 73, 1026–1038. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Underbagging based reduced kernelized weighted extreme learning machine for class imbalance learning. Eng. Appl. Artif. Intell. 2018, 74, 252–270. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Class imbalance learning using UnderBagging based kernelized extreme learning machine. Neurocomputing 2019, 329, 172–187. [Google Scholar] [CrossRef]
- Mathew, J.; Pang, C.K.; Luo, M.; Leong, W.H. Classification of imbalanced data by oversampling in kernel space of support vector machines. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 4065–4076. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, C. Selecting informative Universum sample for semi-supervised learning. In Proceedings of the Twenty-First International Joint Conference on Artificial Intelligence, Pasadena, CA, USA, 11–17 July 2009. [Google Scholar]
- Zhao, J.; Xu, Y.J. A safe sample screening rule for Universum support vector machines. Knowl. Based Syst. 2017, 138, 46–57. [Google Scholar] [CrossRef]
- Cherkassky, V.; Dai, W. Empirical study of the Universum SVM learning for high-dimensional data. In Proceedings of the International Conference on Artificial Neural Networks, Limassol, Cyprus, 14–17 September 2009; pp. 932–941. [Google Scholar]
- Hamidzadeh, J.; Kashefi, N.; Moradi, M. Combined weighted multi-objective optimizer for instance reduction in two-class imbalanced data problem. Eng. Appl. Artif. Intell. 2020, 90, 103500. [Google Scholar] [CrossRef]
- Lin, W.-C.; Tsai, C.-F.; Hu, Y.-H.; Jhang, J.-S. Clustering-based undersampling in class-imbalanced data. Inf. Sci. 2017, 409, 17–26. [Google Scholar] [CrossRef]
- Ofek, N.; Rokach, L.; Stern, R.; Shabtai, A. Fast-CBUS: A fast clustering-based undersampling method for addressing the class imbalance problem. Neurocomputing 2017, 243, 88–102. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Zhu, T.; Lin, Y.; Liu, Y. Synthetic minority oversampling technique for multiclass imbalance problems. Pattern Recognit. 2017, 72, 327–340. [Google Scholar] [CrossRef]
- Agrawal, A.; Viktor, H.L.; Paquet, E. SCUT: Multi-class imbalanced data classification using SMOTE and cluster-based undersampling. In Proceedings of the 2015 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K), Lisbon, Portugal, 12–14 November 2015; pp. 226–234. [Google Scholar]
- Wang, Z.; Chen, L.; Fan, Q.; Li, D.; Gao, D. Multiple Random Empirical Kernel Learning with Margin Reinforcement for imbalance problems. Eng. Appl. Artif. Intell. 2020, 90, 103535. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Class-specific extreme learning machine for handling binary class imbalance problem. Neural Netw. 2018, 105, 206–217. [Google Scholar] [CrossRef]
- Guo, W.; Wang, Z.; Hong, S.; Li, D.; Yang, H.; Du, W. Multi-kernel Support Vector Data Description with boundary information. Eng. Appl. Artif. Intell. 2021, 102, 104254. [Google Scholar] [CrossRef]
- Seiffert, C.; Khoshgoftaar, T.M.; Van Hulse, J.; Napolitano, A. RUSBoost: A hybrid approach to alleviating class imbalance. IEEE Trans. Syst. Man Cybern.-Part A Syst. Hum. 2009, 40, 185–197. [Google Scholar] [CrossRef]
- Haixiang, G.; Yijing, L.; Yanan, L.; Xiao, L.; Jinling, L. BPSO-Adaboost-KNN ensemble learning algorithm for multi-class imbalanced data classification. Eng. Appl. Artif. Intell. 2016, 49, 176–193. [Google Scholar] [CrossRef]
- Shen, C.; Wang, P.; Shen, F.; Wang, H. UBoost: Boosting with theUniversum. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 34, 825–832. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.; Abe, N.J. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 1612. [Google Scholar]
- Zhang, Y.; Liu, B.; Cai, J.; Zhang, S. Ensemble weighted extreme learning machine for imbalanced data classification based on differential evolution. Neural Comput. Appl. 2017, 28, 259–267. [Google Scholar] [CrossRef]
- Li, K.; Kong, X.; Lu, Z.; Wenyin, L.; Yin, J. Boosting weighted ELM for imbalanced learning. Neurocomputing 2014, 128, 15–21. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhou, H.M.; Ding, X.J.; Zhang, R. Extreme Learning Machine for Regression and Multiclass Classification. IEEE Trans. Syst. Man Cybern. Part B 2012, 42, 513–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.Y.; Ong, Y.S.; Zheng, Q.H. A Fast Reduced Kernel Extreme Learning Machine. Neural Netw. 2016, 76, 29–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcala-Fdez, J.; Fernandez, A.; Luengo, J.; Derrac, J.; Garcia, S.; Sanchez, L.; Herrera, F. KEEL Data-Mining Software Tool: Data Set Repository, Integration of Algorithms and Experimental Analysis Framework. J. Mult.-Valued Log. Soft Comput. 2011, 17, 255–287. [Google Scholar]
- Alcala-Fdez, J.; Sanchez, L.; Garcia, S.; del Jesus, M.J.; Ventura, S.; Garrell, J.M.; Otero, J.; Romero, C.; Bacardit, J.; Rivas, V.M.; et al. KEEL: A software tool to assess evolutionary algorithms for data mining problems. Soft Comput. 2009, 13, 307–318. [Google Scholar] [CrossRef]
- Zeng, Y.J.; Xu, X.; Shen, D.Y.; Fang, Y.Q.; Xiao, Z.P. Traffic Sign Recognition Using Kernel Extreme Learning Machines with Deep Perceptual Features. IEEE Trans. Intell. Transp. Syst. 2017, 18, 1647–1653. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).