Abstract
MBRs (Membrane bioreactors) have been increasingly employed for municipal and industrial wastewater treatment in the last decades for their small footprint and excellent effluent quality. However, microorganisms are often detected in the permeates of microfiltration (MF) membranes even with small pore sizes. Coliform bacteria are known for indicating the potential presence of pathogenic bacteria that cause infectious disease such as bacteremia, respiratory tract infections, and urinary tract infections. Thus, the retention of coliform bacteria by membrane processes is important when the membrane process is utilized in water reclamation. In this study, a microbial community of coliform bacteria in the permeates of MF membranes with different pore sizes (0.2, 0.4, and 0.8 µm) was identified. The results showed that the dominant coliform bacteria changed from Enterobacter spp. and Citrobacter spp. in the activated sludge to Enterobacter spp. and Klebsiella spp. in the permeate of MF membranes, while some pieces of membranes showed complete retention. The bacterial regrowth on the surface of the piping system on the permeate side could be a significant factor contributing to the frequent and exclusive detection of Enterobacter spp. and Klebsiella spp. in the case of membranes with small pore size (0.2 and 0.4 µm) after a long continuous filtration time. To indicate the public health-related risk of treated wastewater by MF, Escherichia coli may not be a suitable indicator species because E. coli is relatively retentive in MF compared to other coliforms.
1. Introduction
The membrane bioreactor (MBR) system has been a promising technology utilized for water reclamation and purification since the late 1960s, and it has been increasingly applied in municipal and industrial wastewater treatment due to a higher degradation capability for organic compounds and smaller space requirements in comparison to conventional activated sludge (CAS) treatment [1,2]. A variety of pollutants, e.g., pathogens, nitrogen-containing [3,4] and phosphorus-containing compounds [5,6], bioaccumulating heavy metals [7], emerging pollutants such as microplastics [8], and pharmaceutical and personal care products (PPCPs) [9,10], have proven to be removed efficiently by MBR. It is accepted that membranes with pore sizes smaller than 0.22 µm can be used for clearance of biological particle contamination. Thus, the MBR system with microfiltration (MF) membranes is often utilized as the pre-treatment for the latter reverse osmosis (RO) process when an excellent effluent quality is needed in municipal and industrial wastewater treatment [11,12]. A high retention of microorganisms in MBR is preferable for the stable operation of the latter stage of RO because the presence of bacteria in the RO stage is likely to cause biological fouling of RO membranes. However, recent studies have shown that microorganisms were detected in the permeate of MF membranes even when the membranes with pore sizes significantly smaller than the bacterial sizes were used [13,14]. Several reasons for bacterial permeation during membrane filtration can be considered: (1) The loss of membrane integrity and aging of the membrane [15,16]; (2) bacterial deformation [17]; (3) ultra-small bacterial cell size during adaptation responses to starvation/survival stresses [18]; (4) bacterial regrowth on the permeate side after penetration; (5) shape of the bacteria cell [19]; and (6) the Gram type [14,20] are reported to be important parameters that play a key role in the particle retention.
In the field of microbiology, 0.22 μm filterable (permeable) bacteria, termed ultramicrobacteria [21], are characterized as having a small bacterial morphology size (cell volume < 0.1 µm3), having a streamlined genome size, and lacking several biosynthetic pathways and ribosomal proteins [22]. Ultramicrobacteria are shown to be omnipresent in the environment [23,24]. A study on a freshwater lake showed that as many as 141 permeable bacteria affiliated to the phyla Proteobacteria, Bacteroidetes, Firmicutes, or Actinobacteria were collected in the permeate of membranes of pore size 0.22 μm [25]. The presence of these ultramicrobacteria could induce incomplete retention of microorganisms by MF membranes.
The filterability of bacteria by MF membranes has been investigated using a few bacterial strains: Brevundimonas diminuta (a small, Gram-negative bacterium formerly named Pseudomonas diminuta), Staphylococcus epidermidis (a Gram-positive bacterium with spherical shape), and Acholeplasma laidlawii (mycoplasma, a small bacterium lacking a cell wall) have been used for the investigation on the effect of bacterial cell structures on the permeation through MF membranes [26]. Among these species, Acholeplasma laidlawii was earlier demonstrated capable of penetrating 0.2 µm membranes and even some 0.1 µm rated membranes [27,28]. Ralstonia spp., Achromobacter spp., Methylobacterium spp., and Methylorubrum spp. were detected in the permeate of a laboratory MBR with membranes of pore size 0.2 µm in our previous study [29]. In addition to bacterial sizes, flow interruption and pressure release could cause leakage of bacteria through MF membranes [26].
In order to use MF membranes in water and wastewater treatment, the retention of coliform bacteria is important for discussing health-related issues. Although most coliform bacteria are not harmful themselves, their presence is used as an indicator for the potential presence of other pathogenic organisms [30]. Fecal coliform bacteria (FCB), affiliated with coliform bacteria, are well known for their involvement in food spoilage and causing diseases in both humans and animals [31], such as typhoid fever [32], gastrointestinal diseases [33] m and hepatitis [34]. Escherichia coli (E. coli) O157:H7 can cause infection when present at a level fewer than 10 to 100 CFU [35]. The World Health Organization (WHO) recommends that potable water should have below 20 CFU/mL heterotrophic bacterial counts with no coliform bacteria, fecal coliforms, E. coli, Enterococci, and P. aeruginosa [36]. The retention of coliform bacteria by MF in water and wastewater treatment is important due to their indication role for pathogens, although only limited literature has addressed the bacterial species of coliforms in the permeate of MF.
Therefore, this study adopted membrane filtration and cultivation methods to (1) identify the permeable coliform bacteria capable of penetrating membranes with pore sizes ranging from 0.2 to 0.8 µm and (2) determine the bacterial community changes of coliform bacteria contained in the permeate of membranes of various pore sizes after membrane filtration. This study showed that E. coli may not be a suitable species to assess the retention of coliform bacteria by MF membranes because E. coli is relatively retentive in MF compared to other coliforms. In addition, a possibility was suggested that the regrowth on the surface of the piping system is the reason for the frequent and exclusive detection of Enterobacter spp. and Klebsiella spp. from the samples taken from the permeate after long continuous filtration time.
2. Materials and Methods
2.1. Preparation and Filtration of Activated Sludge
Mixed liquor of activated sludge was taken from a wastewater treatment plant in Tokyo University of Technology (Tokyo, Japan). It was aerated for 24 h at room temperature before the start of the filtration experiment. The MBR system consisted of a 9L volume reactor and an acrylic board with four pieces polycarbonate MF membranes of 47 mm diameter (ADVANTEC Toyo Roshi Kaisha, Ltd., Tokyo, Japan) with respective pore sizes of 0.2, 0.4, and 0.8 µm, as shown in Figure 1. The peripherals of the membranes were fixed with epoxy resin to an acrylic base board. The membranes with pore sizes of 0.2 and 0.4 µm were used because they are used for the elimination of bacteria, while the membranes with pore sizes of 0.8 µm were used to obtain more permeable bacteria for characterization. These polycarbonate membranes were used because track-etched membranes have relatively uniform cylindrical pores of definite size, although a certain heterogeneity is involved [37].
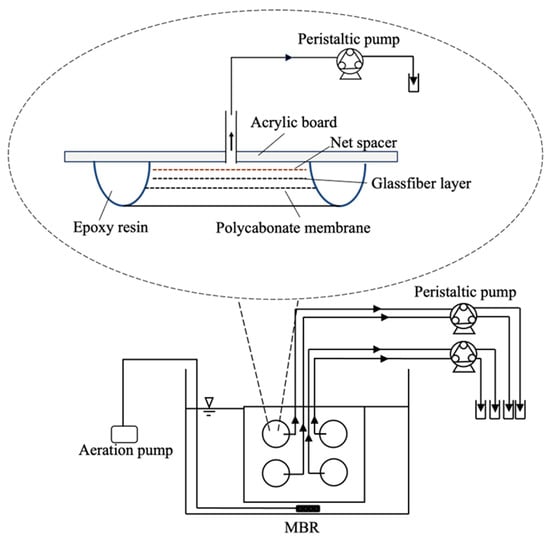
Figure 1.
Schematic diagram of the experimental MBR setup and membrane binding to the base board.
In the laboratory MBR, aeration was continuously applied on the surface of membranes to minimize membrane fouling. The permeates were collected by peristaltic pumps. When the fouling of the membrane proceeded, the volume flux decreased even with the constant speed operation of the pumps. Before membrane filtration conducted, the MBR system was disinfected with 250 ppm sodium hypochlorite solution, with suction filtration applied for a period of 18 h. Then, sodium hypochlorite solution was replaced with 0.02 g/L sodium thiosulfate for 2 h to eliminate residual chlorine in the MBR system. The presence of coliform bacteria after the disinfection of the MBR setup was checked at this point. After that, MBR was operated with the mixed liquor of activated sludge. For preventing microbial contamination from the air, the very edge of each tube connected to the permeate side of the membranes was emerged in the chlorine solution before the start of the experiment.
Table 1 shows the list of nominal pore sizes of the MF membranes used in each experimental run (R1–R5) and the number of Enterobacteriaceae in the laboratory MBR setup filled with the mixed liquor of activated sludge. In R3, R4, and R5, two pieces of membranes of identical pore size (0.4 µm) were tested in the filtration process to obtain more detailed information for the performance of the membranes of this pore size. In the case of R1, 1L activated sludge was diluted 10 times with 9 L sodium thiosulfate solution. In the case of R2–R5, 4.5 L activated sludge was diluted 2 times with 4.5 L sodium thiosulfate solution. The coliform numbers after the dilution are shown in Table 1. The feed-activated sludge in the case of R1 contained only a small quantity of E. coli because there were fewer students in the campus during the sampling time. The proportions of E. coli in total Enterobacteriaceae were in the range 19% to 45% in R2, R3, R4, and R5, while that in R1 was 6.5%.

Table 1.
The number of pieces of MF membranes with various pore sizes examined in each run and occurrences of E. coli and other Enterobacteriaceae for the feed-activated sludge.
2.2. Permeate Collecting and Cultivation of Enterobacteriaceae
Before the collection of permeates, medium bottles were autoclaved for 15 min at 121 °C with 200 µL sodium thiosulfate solution contained at the concentration of 0.02 g/mL for the purpose of complete removal of residual chlorine. The edge of the permeate-collection tube was cut at 1 cm length from the edge of the tube by heated scissors before collection of each permeate. The cap and the bottleneck of the medium bottle were heated again after collecting permeate. A volume of 100 µL permeate was poured onto Chromagar ECC medium (CHROMagar Co., Ltd., Paris, France) for the simultaneous enumeration of E. coli (blue-colored colonies) and other Enterobacteriaceae (mauve-colored colonies) by incubating for 24 h at 37 °C. The coliform retention was defined as 1 − (np/nf), where np is the coliform numbers in the permeate (CFU/mL), and nf is the coliform numbers in the feed-activated sludge tank (CFU/mL). If no colonies were found in the permeate sample, “1/total sampling volume” was used for np to avoid over-estimation of the retention. In this case, the retention was shown with “>”, for example, “>99.94%”.
2.3. Identification of Isolated Bacteria
2.3.1. DNA Extraction
Isolates were obtained by picking up the colonies on Chromagar ECC agar plates and streaking on new agar plates with the same composition. DNA was extracted from the isolated colonies on the agar plates using the Cica geneus DNA extraction reagent (Kanto Chemical Co., INC., Tokyo, Japan), followed by incubation for 6 min at 72 °C and 3 min at 94 °C successively. The supernatant of this reaction solution was utilized as template DNA. The extracted DNA was reserved at −20 °C before the PCR process.
2.3.2. PCR Amplification, Purification, and Sequencing
Premixed PCR enzyme TaKaRa Ex Taq HS (Takara Bio Inc., Kusatsu, Japan) and universal primers 27F(5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGYTACCTTGTTACGACTT-3′) [38] were utilized in the PCR process to amplify the 16S rRNA gene for the identification of bacterial species. A total reaction volume of 25 µL for each sample consisted of 12.5 µL the premixed PCR enzyme, 1 µL of 27F and 1492R primer, 9.5 µL dH2O, and 1 μL template DNA. The PCR condition consisted of initial denaturation at 94 °C for 1 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min. Pure water was templated as negative control to check for contaminations in each PCR reaction. After amplification, gel electrophoresis with Gene Ladder Fast 1 (Nippon Gene Co., Ltd., Tokyo, Japan) was performed on a 1% QA-Agarose (MP Biomedicals, Inc., Santa Ana, CA, USA) gel to verify the size quality of the PCR products. The verified PCR products were purified using the MonoFas DNA purification kit (ANIMOS Inc., Saitama, Japan) according to the manufacturer instruction. Sanger sequencing with primers of 518F (5′-CCAGCAGCCGCGGTAATACG-3′) and 800R (5′-TACCAGGGTATCTAATCC-3′) was conducted by Macrogen Japan Corp. (Tokyo, Japan). In the cases of isolates obtained in R1, R2, and R3, the full-length 16S rRNA gene sequences merged from the reads by both 518F and 800R primers were used for the identification by the BLASTN program (https://blast.ncbi.nlm.nih.gov/, accessed on 27 February 2024) provided by National Center for Biotechnology Information (NCBI), National Institute of Health, U.S., while the gene sequences obtained by only the 800R primer were used in the case of isolates obtained in R4 and R5. The obtained sequences were registered, and the issued accession numbers are shown in Tables S1 and S2.
2.4. Size Measure and Gram Staining of Target Bacteria
The isolated colonies were dissolved in 500 µL phosphate-buffered saline solution and stirred evenly. Then, 30 µL of the suspension was added on the slide glass and desiccated by heat. The desiccated samples were immersed in methanol for 1 min, followed by staining with Gram’s staining solution No.1, No.2, and No.3, as described in [39]. The morphology of the bacteria was observed by an optical microscope (DM 750, Leica Microsystems GmbH, Wetzlar, Germany). The bacterial sizes were measured using Image J, which is an image processing program developed at the National Institutes of Health and the Laboratory for Optical and Computational Instrumentation (LOCI, University of Wisconsin). A number of 30 bacterial cells were measured, and the mean value was considered as the bacterial size.
3. Results
3.1. Effect of Pore Size on the Bacterial Presence in the Permeate
Table 2 shows the time-average bacterial count number in the permeate of each membrane piece during the filtration of mixed liquor of the activated sludge in each experimental run. A total of 18 pieces of membranes were examined in this experiment, in which 6 membranes showed complete retention of the coliform bacteria contained in the activated sludge. Bacterial leakages were detected in the permeate of all pieces of 0.8 µm membranes in a range from 2.5 to 45 CFUs/mL (retention from 99.93% to 99.99%). With regard to the membranes with a pore size of 0.4 µm, three pieces out of eight pieces showed complete bacterial retention (from >99.77% to >99.997%), and the rest of the membranes showed different levels of bacterial penetration (from 1.4 to 52 CFUs/mL, retention from 99.79% to 99.998%). In the case of membranes with 0.2 µm pore size, three pieces out of four pieces of membranes showed complete bacterial retention (from >99.77% to >99.997%), and the remaining one piece showed sporadic bacterial penetration (3.8 CFUs/mL, corresponding with removal rate of 99.98%). Unsurprisingly, membranes of pore size 0.2 µm yielded a good performance on retention of coliform bacteria, as the 0.2 µm membrane is often used in sterilization and clarification processes in pharmaceutical and biotechnology industries [40]. The result of the membrane with the pore size of 0.2 µm during R4 was cancelled due to damage of the membrane.

Table 2.
Time-average coliform count numbers in the permeate of membranes with pore sizes of 0.2, 0.4, and 0.8 μm. (The membrane with 0.2 µm pore size in the R4 was discarded because of membrane breach.) If no colonies were found in the sample, “<1/total sampling volume” is shown in the table. The results for membranes with pore size of 0.4 μm were shown separately for each piece (1) (2).
3.2. Filtration Volume Flux and Time-Dependent Bacterial Leakage
The filtration volume flux (m/d) and enumeration of leakage bacteria (CFU/mL) are shown in Figure 2. The volume flux decreased with time, especially in R3, R4, and R5, due to clogging of the membranes, although the pump was operated with a constant speed. The presence of bacteria in the permeate in this study originated from the penetration of bacteria from the feed side through the membranes because no bacteria were detected during chlorination of the apparatus before the start of the filtration with activated sludge. Bacterial leakages were constantly detected when membranes of 0.8 µm pore size were examined, indicating that the pore size of membranes is an important factor affecting the retention of microorganisms. In R1 and R2, bacterial colonies were only detected in the permeate of the 0.8 µm membrane, which could be attributed to a low coliform concentration in the activated sludge and low applied trans-membrane pressure in the operation under constant filtration volume flux. On the other hand, in the case of filtration by membranes of smaller pore size, a longer filtration time was necessary for bacterial leakage; frequent bacterial leakage and intense decrease in filtration volume flux were observed simultaneously during R3, R4, and R5, which indicated that a higher applied trans-membrane pressure across the membrane facilitated the bacterial penetration.
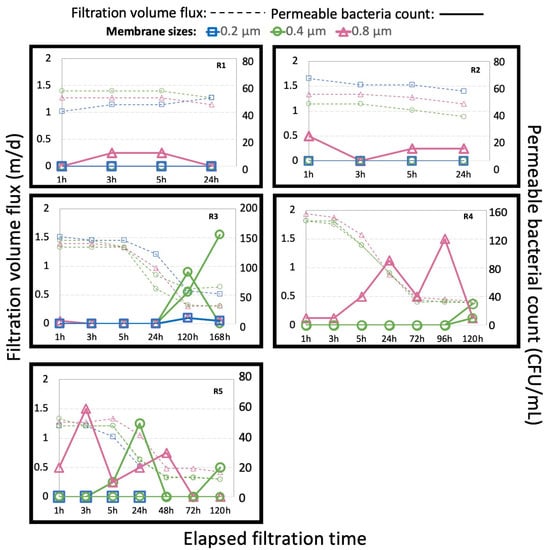
Figure 2.
Filtration volume flux and bacterial count of leakage bacteria by time elapsed.
3.3. Bacterial Community Change of Enterobacteriaceae during MBR Process
E. coli could be distinguished from other Enterobacteriaceae by the color of the colonies on the agar plates used in this study. The results of 16S rRNA sequencing showed that all examined isolates of blue colonies were identified as Escherichia/Shigella spp. (n = 9; accession numbers PP396542, PP396546, PP396547, PP396548, PP396549, PP396552, PP396559, PP396570, and PP396576), while none of examined isolates of mauve colonies were identified as Escherichia/Shigella spp. These results indicate that the identification and numeration of E. coli by Chromagar ECC was completely accurate for the examined samples.
As mentioned in the materials and methods section, the ratio of E. coli to total Enterobacteriaceae contained in the mixed liquor of activated sludge in this study were in the range of 19% to 45% (except for R1, when the count numbers of E. coli were few due to fewer students in the campus at that period). In total, 131 colonies were obtained from the tested permeates (including membranes of pore sizes 0.2, 0.4, and 0.8 µm) through five runs (R1–R5) in this study. Excluding the colonies observed in R1, a total of 129 colonies were observed. The number of blue colonies (presumptive E. coli) was 4 out of the observed 129 colonies (3.1%). The population of E. coli in the total Enterobacteriaceae was remarkably decreased with filtration. In addition, all of the blue colonies were obtained from the permeates of membranes of pore size 0.8 µm. These results indicate that E. coli was more retentive than other Enterobacteriaceae in MF, probably due to the size exclusion mechanism for the larger size of the species (2.0–6.0 µm in length and 1.1–1.5 µm wide [41]). The retention of E. coli by the membranes of pore sizes 0.4 µm or 0.2 µm would have been >99.99% considering the permeate E. coli numbers smaller than 1/100 μL in R3, R4, and R5. (Accurate estimation of E. coli retention was difficult in R1 and R2 because of low count numbers of feed-activated sludge.)
Figure 3 shows the microbial community change of Enterobacteriaceae excluding E. coli after MF. The results on the identification of 46 isolates randomly picked up from 131 permeable isolates obtained from FW (filtrated water) penetrated through membranes of pore sizes 0.2, 0.4, and 0.8 µm are shown in the figure together with the results on 40 isolates obtained from the activated sludge samples with mauve-colored colonies on ECC agar plates. It can be inferred that genera Enterobacter and Citrobacter dominated in the activated sludge before filtration, and genera Enterobacter and Klebsiella dominated in the permeate of membrane filtration, although the bacterial diversity at the genus level was comparable before and after the filtration process. Meanwhile, the presence of genera Serratia, Klebsiella, Citrobacter, and Enterobacter was detected in both the activated sludge and permeate, which indicates their prevalence in activated sludge and their capability of penetration. In addition, relatively superior bacterial penetration was shown for certain genera, as the proportion of bacteria affiliated to genera Enterobacter (30.4–39.6%), Klebsiella (13.0–27.1%), and Serratia (4.3–10.4%) increased along with the membrane filtration process. The proportion of bacteria affiliated to genus Citrobacter decreased after filtration (28.2–12.5%). Kluyvera spp. and Yokenella sp. were detected in the permeate of the membrane but not detected in the activated sludge, as shown in Tables S1 and S2, possibly due to the limitation of the number of isolates examined. Although selective agar medium was used for the enumeration of Enterobacteriaceae in this study, four isolates were identified as members outside Enterobacteriaceae (Aeromonas spp. for two isolates from the feed-activated sludge and Pseudomonas spp. for two isolates from the permeates).
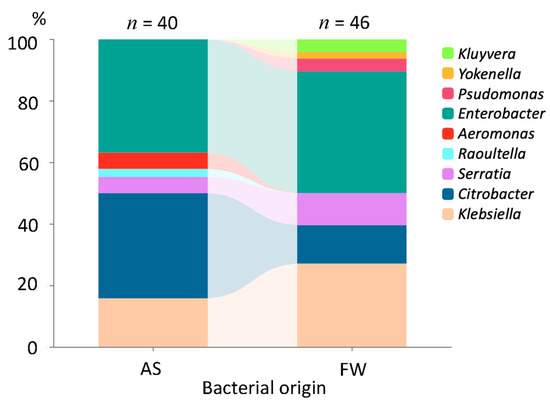
Figure 3.
The bacterial community change of Enterobacteriaceae cultivated on ECC medium. (AS, activated sludge; FW, filtrated water. Among a total of 46 isolates obtained in the FW, 1, 21, and 24 isolates originated from membranes of pore sizes 0.2, 0.4, and 0.8 µm, respectively).
3.4. Detected Species in the Permeate of Specific Membrane
The detailed results of bacterial identification in the permeate and the permeable bacterial origin in each experimental run are shown in Figure 4 and Table S1. The results on different pieces of the membranes of pore size 0.4 µm are shown separately in the table. Coliforms were detected in the permeate of membranes of the large pore size of 0.8 µm in each experimental run with a diversity of bacterial species. On the other hand, the permeate of respective membranes of pore size 0.4 µm consisted of a single bacterial species in R3 and R4, although different species were detected in R5. For example, Enterobacter spp. dominated in the permeate of a piece of membrane of pore size 0.4 µm in the case of R3, while Klebsiella spp. dominated the permeate of another piece of the membrane with the same pore size. Klebsiella spp. was detected exclusively in the permeates through the membranes with pore size of 0.4 µm after a long continuous filtration time (>120 h), as shown in Figure 4 and Table S1.
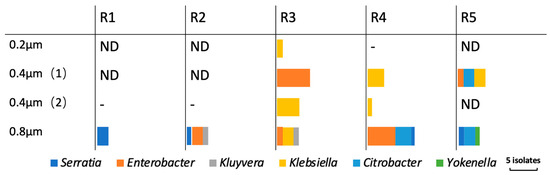
Figure 4.
Detected species in the permeate of respective membranes in each experimental run. The results for membranes with pore size of 0.4 μm were shown separately for each piece (1) (2).
3.5. Bacterial Sizes Found in the Permeate
Figure 5 shows the morphological sizes of permeable coliforms in the permeate of membranes of pores sizes 0.2, 0.4, and 0.8 µm. A total of 46 isolates were classified into six genera affiliated to the family Enterobacteriaceae. The microscopic observation (detailed sizes and Gram stainability results shown in Table S1) shows that bacterial species of Enterobacteriaceae generally have rod shapes with Gram-negative cell walls, which may have also helped them penetrate through the membrane [20]. The length and width of permeable bacteria in this experiment ranges from 0.5 to 1.7 µm and 0.3 to 1.1 µm, respectively, with the most frequent length being from 0.9 to 1.1 µm and width from 0.7 to 0.9 µm. The measured bacterial size for the isolates within Enterobacteriaceae was independent from bacterial species. Our observation does not exclude the possibility that bacterial species with smaller cell sizes preferentially penetrated the membranes because the bacterial sizes were determined after the formation of colonies.

Figure 5.
Bacterial length (a) and width (b) of permeable coliforms in the permeate of membrane filtration.
4. Discussion
4.1. Factors Affecting Bacterial Penetration through Microfiltration Membranes
The presence of coliform bacteria at a high count number in an MBR effluent have been considered to be an indication of a membrane breach or post-membrane biological regrowth [15]. However, as shown in this study, MF membranes cannot be a complete barrier for bacterial penetration. In order to identify the key coliform bacterial species with high ability for penetration through membranes in wastewater treatment, polycarbonate membranes with definite cylindrical pores having diameters of 0.2, 0.4, and 0.8 µm were used in our experiment on the membrane bioreactor. Membranes of larger pore size (0.8 µm) showed a lower retention (leakage was observed for all tested pieces), as can be expected. In the cases of membranes of smaller pore size (0.2 µm and 0.4 µm), bacterial leakage was observed for some pieces of these membranes over time, especially at higher trans-membrane pressures caused by fouling, although most of the tested pieces showed more than 99.99% retentions. The level of leakage was consistent with a previous report in which E. coli could penetrate nucleopore membranes of the pore size 0.4 µm, with a retention level of approximately 99.9% [20]. The decrease in retention with the increase in applied pressure difference was also reported for Acholeplasma laidlawii across track-etched membranes rated at 100 nm and 220 nm [42] and for H. gracilis and B. diminuta through membranes with a pore size of 0.45 µm [19]. In the case of Gram-negative bacteria (including Enterobacteriaceae), the deformation of cell walls partly explained the effect on retention [14].
4.2. Bacterial Community Change by Filtration
The permeate of the membranes of smaller pore size often consisted of a single species (mainly Enterobacter or Klebsiella) in this study. Frequent and exclusive detection of Klebsiella spp. on the permeate side of the 0.2 and 0.4 µm membranes after a long continuous operation time suggests the contribution of regrowth on the surface of the piping system, while a variety of species penetrated the membranes of larger pore size (0.8 µm). The ability of Klebsiella (and possibly as well as Enterobacter) to regrow on the surface of the piping system on the permeate side of membranes could cause continuous detection of these bacterial species in the treated water because biofilm formation can impart protective effects, increasing disinfectant resistance [43,44].
As to the change in the bacterial community due to MF, a report on surface water filtration showed that the permeable bacteria were mainly affiliated to the Cytophagales and the genus Hylemonella, outside the members of Enterobacteriaceae [45]. Bacteria species affiliated to Alpha- and Beta-proteobacteria were detected on the RO membrane biofilm of an UF–RO system, although UF was able to remove microorganisms almost completely [46]. As far as we know, there is no literature on the preferential penetration of species affiliated with Enterobacteriaceae (Gamma-proteobacteria) in MF. Further investigation is needed for the preferential penetration of specific coliform species. At this moment, the difference in size and flexibility of different species of Enterobacteriaceae is not well known.
4.3. Health-Related Risks of Water Treated by Microfiltration
Membrane filtration is often coupled with disinfection processes like UV irradiation, chlorination, or ozonation to provide a multi-barrier approach for pathogen removal and inactivation because an MF membrane cannot be a complete barrier for bacteria, as shown in this study. The effectiveness of disinfection processes like UV irradiation, chlorination, and ozonation can vary significantly depending on bacterial species and strains. A study found that E. coli was more sensitive to UV irradiation than another fecal coliform strain and Bacillus subtilis [47]. Chlorine disinfection is effective against most coliforms, although some disinfectants are more effective for E. coli than for Klebsiella sp. [48]. Ozone has been demonstrated to rapidly inactivate E. coli and other coliforms, but certain strains may possess oxidative stress-response mechanisms conferring some resistance [49].
The MF process in general can reduce potential public health risks associated with coliforms as well as other disinfection processes. However, the presence of Klebsiella spp. and Enterobacter spp. in the permeate of the MF process, as shown in this study, must be noted, though these species are generally not as virulent as pathogenic E. coli strains [31].
E. coli, in spite of its wide use as a fecal indicator organism, exhibited relatively high retention compared to other coliforms in MF in this study. This questions its suitability for indicating the microbiological quality of membrane-treated waters. The high retention for E. coli and frequent detection of Klebsiella and Enterobacter are consistent with a study on molecular-based detection of pathogenic bacteria in MBR, showing that the effluent contained Klebsiella pneumoniae genes, while it contained minimal E. coli genes [13]. The monitoring of not only E. coli but Klebsiella spp. and Enterobacter spp. is recommended to assess the health-related risk of MF-treated water, although more accumulation of information is needed.
5. Conclusions
In this study, we profiled the permeable coliforms that are capable of penetrating various pore sizes of membranes in a membrane bioreactor. The results indicated that coliforms in the activated sludge mixed liquor were retained at the percentage level of 99.99% by MF with various pore sizes (i.e., 0.2, 0.4, and 0.8 µm), and the bacterial count number in the feed water could affect that in the permeate, although some pieces of membranes with pore sizes of 0.2 and 0.4 µm showed complete retention for coliforms. It is noteworthy that high trans-membrane pressure caused by membrane fouling contributed to the increase in the count numbers of coliforms in the permeate. The result of bacterial identification by 16S rRNA gene sequencing showed that the genera Enterobacter and Citrobacter dominated in the activated sludge before filtration, and the genera Enterobacter and Klebsiella dominated in the permeate of membrane filtration. Bacterial regrowth on the piping system could be the reason for the detection of these genera, considering the simple constituents of bacterial species in the permeate. To indicate the public health risk of MF-treated wastewater, the monitoring of not only E. coli but Klebsiella spp. is recommended because E. coli is relatively retentive in MF.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/w16091269/s1, Table S1: Permeable coliform bacterial species identified in the permeate of membrane and the characterization; Table S2: Coliform bacterial species identified in the activated sludge.
Author Contributions
S.Z., data curation and writing—original draft preparation; T.U. and S.G., writing—review and editing; T.U., supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Japan Society for the Promotion of Science, category of scientific research (C), grant number 20K04758.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Acknowledgments
The authors thank K. Nakagawa for his help in the experimental works.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Judd, S.J. The status of industrial and municipal effluent treatment with membrane bioreactor technology. J. Chem. Eng. 2016, 305, 37–45. [Google Scholar] [CrossRef]
- Xiao, K.; Xu, Y.; Liang, S.; Lei, T.; Sun, J.; Wen, X.; Zhang, H.; Chen, C.; Huang, X. Engineering application of membrane bioreactor for wastewater treatment in China: Current state and future prospect. Front. Environ. Sci. Eng. 2014, 8, 805–819. [Google Scholar] [CrossRef]
- Ren, Y.; Ngo, H.H.; Guo, W.; Wang, D.; Peng, L.; Ni, B.J.; Wei, W.; Liu, Y. New perspectives on microbial communities and biological nitrogen removal processes in wastewater treatment systems. Bioresour. Technol. 2020, 297, 122491. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Rajta, A.; Setial, H.; Bhatia, R. Simultaneous nitrification–denitrification by phosphate accumulating microorganisms. World J. Microbiol. Biotechnol. 2020, 36, 151. [Google Scholar] [CrossRef]
- Dorofeev, A.G.; Nikolaev, Y.A.; Mardanov, A.V.; Pimenov, N.V. Role of phosphate-accumulating bacteria in biological phosphorus removal from wastewater. Appl. Biochem. Microbiol. 2020, 56, 1–14. [Google Scholar] [CrossRef]
- Qiu, G.; Zuniga-Montanez, R.; Law, Y.; Thi, S.S.; Nguyen, T.Q.N.; Eganathan, K.; Liu, X.; Nielsen, P.H.; Williams, R.B.H.; Wuertz, S. Polyphosphate-accumulating organisms in full-scale tropical wastewater treatment plants use diverse carbon sources. Water Res. 2019, 149, 496–510. [Google Scholar] [CrossRef]
- Diep, P.; Mahadevan, R.; Yakunin, A.F. Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front. Bioeng. Biotechnol. 2018, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Lares, M.; Ncibi, M.C.; Sillanpää, M.; Sillanpää, M. Occurrence, identification and removal of microplastic particles and fibers in conventional activated sludge process and advanced MBR technology. Water Res. 2018, 133, 236–246. [Google Scholar] [CrossRef]
- Sipma, J.; Osuna, B.; Collado, N.; Monclús, H.; Ferrero, G.; Comas, J.; Rodriguez-Roda, I. Comparison of removal of pharmaceuticals in MBR and activated sludge systems. Desalination 2010, 250, 653–659. [Google Scholar] [CrossRef]
- Anand, U.; Reddy, B.; Singh, V.K.; Singh, A.K.; Kesari, K.K.; Tripathi, P.; Kumar, P.; Tripathi, V.; Simal-Gandara, J. Potential environmental and human health risks caused by antibiotic-resistant bacteria (ARB), antibiotic resistance genes (ARGs) and emerging contaminants (ECs) from municipal solid waste (MSW) landfill. Antibiotics 2021, 10, 374. [Google Scholar] [CrossRef]
- Dolar, D.; Gros, M.; Rodriguez-Mozaz, S.; Moreno, J.; Comas, J.; Rodriguez-Roda, I.; Barceló, D. Removal of emerging contaminants from municipal wastewater with an integrated membrane system, MBR–RO. J. Hazard. Mater. 2012, 239, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Hai, F.I.; Yamamoto, K. Membrane biological reactors. In Treatise on Water Science; Wilderer, P., Ed.; Elsevier: Oxford, UK, 2011; pp. 571–613. [Google Scholar]
- Harb, M.; Hong, P.Y. Molecular-based detection of potentially pathogenic bacteria in membrane bioreactor (MBR) systems treating municipal wastewater: A case study. Environ. Sci. Pollut. Res. Int. 2017, 24, 5370–5380. [Google Scholar] [CrossRef] [PubMed]
- Helling, A.; Kubicka, A.; Schaap, I.A.T.; Polakovic, M.; Hansmann, B.; Thiess, H.; Strube, J.; Thom, V. Passage of soft pathogens through microfiltration membranes scales with transmembrane pressure. J. Membr. Sci. 2017, 522, 292–302. [Google Scholar] [CrossRef]
- Hirani, Z.M.; Bukhari, Z.; Oppenheimer, J.; Jjemba, P.; LeChevallier, M.W.; Jacangelo, J.G. Characterization of effluent water qualities from satellite membrane bioreactor facilities. Water Res. 2013, 47, 5065–5075. [Google Scholar] [CrossRef] [PubMed]
- Stockner, J.G.; Klut, M.E.; Cochlan, W.P. Leaky filters: A warning to aquatic ecologists. Can. J. Fish. Aquat. Sci. 1990, 47, 16–23. [Google Scholar] [CrossRef]
- Arthur, G.; Clémence, C.; Christine, R.; Patrice, B.; Etienne, D.; Christel, C. Bacteria transfer by deformation through microfiltration membrane. J. Membr. Sci. 2017, 523, 446–455. [Google Scholar]
- Velimirov, B. Nanobacteria, ultramicrobacteria and starvation forms: A search for the smallest metabolizing bacterium. Microbes Environ. 2001, 16, 67–77. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Hammes, F.; Düggelin, M.; Egli, T. Influence of size, shape, and flexibility on bacterial passage through micropore membrane filters. Environ. Sci. Technol. 2008, 42, 6749–6754. [Google Scholar] [CrossRef] [PubMed]
- Lebleu, N.; Roques, C.; Aimar, P.; Causserand, C. Role of the cell-wall structure in the retention of bacteria by microfiltration membranes. J. Membr. Sci. 2009, 326, 178–185. [Google Scholar] [CrossRef]
- Hood, M.A.; MacDonell, M.T. Distribution of ultramicrobacteria in a gulf coast estuary and induction of ultramicrobacteria. Microb. Ecol. 1987, 14, 113–127. [Google Scholar] [CrossRef]
- Ghuneim, L.A.J.; Jones, D.L.; Golyshin, P.N.; Golyshina, O.V. Nano-sized and filterable bacteria and archaea: Biodiversity and function. Front. Microbiol. 2018, 9, 1971. [Google Scholar] [CrossRef]
- Suzina, N.E.; Duda, V.I.; Esikova, T.Z.; Shorokhova, A.P.; Gafarov, A.B.; Oleinikov, R.R.; Akimov, V.N.; Abashina, T.N.; Polivtseva, V.N.; Boronin, A.M. Novel ultramicrobacteria, strains NF4 and NF5, of the genus Chryseobacterium: Facultative epibionts of Bacillus subtilis. Microbiology 2011, 80, 535–548. [Google Scholar] [CrossRef]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.Y.N.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015, 6, 6372. [Google Scholar] [CrossRef]
- Maejima, Y.; Kushimoto, K.; Muraguchi, Y.; Fukuda, K.; Miura, T.; Yamazoe, A.; Kimbara, K.; Shintani, M. Proteobacteria and Bacteroidetes are major phyla of filterable bacteria passing through 0.22 μm pore size membrane filter, in Lake Sanaru, Hamamatsu, Japan. Biosci. Biotechnol. Biochem. 2018, 82–87, 1260–1263. [Google Scholar] [CrossRef]
- Helling, A.; Grote, C.; Büning, D. Ulbricht Mathias, Wessling Matthias, Polakovic Milan, Thom Volkmar. Influence of flow alterations on bacteria retention during microfiltration. J. Membr. Sci. 2019, 575, 147–159. [Google Scholar] [CrossRef]
- Cronholm, K.; Bates, S.; Nguyen, N.; Leahy, A.; Blanchard, M.; Lentine, K.R. Validation of a microbiological method using Acholeplasma laidlawii for assessing performance of microporous membranes for mycoplasma clearance. PDA J. Pharm. Sci. Technol. 2009, 63, 438–461. [Google Scholar]
- Folmsbee, M.; Howard, G.; McAlister, M. Nutritional effects of culture media on mycoplasma cell size and removal by filtration. Biologicals 2010, 38, 214–217. [Google Scholar] [CrossRef]
- Zhou, S.; Ninoseki, M.; Kusaba, A.; Nakagawa, K.; Urase, T. Bacterial species identified in the filtrate of microfiltration membranes in the separation of activated sludge. JWET 2021, 19, 294–301. [Google Scholar] [CrossRef]
- Li, D.L.; Liu, S.Y. Water Quality Monitoring in Aquaculture. In Water Quality Monitoring and Management; Academic Press: Cambridge, MA, USA, 2019; pp. 303–328. [Google Scholar]
- Patel, A.K.; Singhania, R.R.; Pandey, A.; Joshi, V.K.; Nigam, P.S.; Soccol, C.R. Introduction Enterobacteriaceae, Coliforms and E. coli. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Elsevier Inc.: Exeter Devon, UK, 2014; Volume 1, pp. 659–666. [Google Scholar]
- Mermin, J.H.; Villar, R.; Carpenter, J.; Roberts, L.; Samaridden, A.; Gasanova, L.; Lomakina, S.; Bopp, C.; Hutwagner, L.; Mead, P.; et al. A massive epidemic of multidrug-resistant typhoid fever in tajikistan associated with consumption of municipal water. J. Infect. Dis. 1999, 179, 1416–1422. [Google Scholar] [CrossRef]
- Weissman, J.B.; Craun, G.F.; Lawrence, D.N.; Pollard, R.A.; Saslavv, M.S.; Gangarosa, E.J. An epidemic of gastroenteritis traced to a contaminated public water supply. Am. J. Epidemiol. 1976, 103, 391–398. [Google Scholar] [CrossRef]
- Laborde, D.J.; Weigle, K.A.; Weber, D.J.; Kotch, J.B. Effect of fecal contamination on diarrheal illness rates in day-care centers. Am. J. Epidemiol. 1993, 138, 243–255. [Google Scholar] [CrossRef]
- Greig, J.D.; Todd, E.C.D.; Bartleson, C.; Michaels, B. Infective Doses and Pathogen Carriage. In Proceedings of the USDA 2010 Food Safety Education Conference, Atlanta, GA, USA, 25 March 2010. [Google Scholar]
- Khatoon, A.; Pirzada, Z.A. Bacteriological quality of bottled water brands in Karachi. Pakistan. Biologia (Pakistan) 2010, 56, 137–143. [Google Scholar]
- Apel, P.Y.; Blonskaya, I.V.; Dmitriev, S.N.; Orelovitch, O.L.; Sartowska, B. Structure of polycarbonate track-etch membranes: Origin of the “paradoxical” pore shape. J. Membr. Sci. 2006, 282, 393–400. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt., E., Goodfellow, M., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Gram Staining. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562156/ (accessed on 14 August 2023).
- Lee, A.; McVey, J.; Faustino, P.; Lute, S.; Sweeney, N.; Pawar, V.; Khan, M.; Brorson, K.; Hussong, D. Use of Hydrogenophaga pseudoflava penetration to quantitatively assess the impact of filtration parameters for 0.2-micrometer-pore-size filters. Appl. Environ. Microbiol. 2010, 76, 695–700. [Google Scholar] [CrossRef]
- Percival, S.L.; Williams, D.W. Escherichia coli. In Microbiology of Waterborne Diseases, 2nd ed.; Percival, S.L., Yates, M.V., Williams, D.W., Chalmers, R.M., Gray, N.F., Eds.; Elsevier Ltd.: Amsterdam, The Netherland; Academic Press: Cambridge, MA, USA, 2014; pp. 89–117. [Google Scholar]
- Kim, K.; Baltus, R.E.; Chellam, S. Rejection and fouling of track-etched microfiltration membranes by Acholeplasma laidlawii: Clues to mycoplasma behavior during “sterile” dead-end filtration. J. Membr. Sci. 2023, 685, 121925. [Google Scholar] [CrossRef]
- Flemming, H.C.; Percival, S.L.; Walker, J.T. Contamination potential of biofilms in water distribution systems. Water Supply 2002, 2, 271–280. [Google Scholar] [CrossRef]
- Abebe, G.M. The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020, 2020, 1705814. [Google Scholar] [CrossRef]
- Liu, J.; Li, B.; Wang, Y.Y.; Zhang, G.J.; Jiang, X.T.; Li, X.Y. Passage and community changes of filterable bacteria during microfiltration of a surface water supply. Environ. Int. 2019, 131, 104998. [Google Scholar] [CrossRef]
- Bereschenko, L.A.; Stams, A.J.M.; Heilig, G.H.J.; Euverink, G.J.W.; Nederlof, M.M.; Van Loosdrecht, M.C.M. Investigation of microbial communities on reverse osmosis membranes used for process water production. Water Sci Technol. 2007, 55, 181–190. [Google Scholar] [CrossRef]
- Guo, M.; Huang, J.; Hu, H.; Liu, W. Growth and repair potential of three species of bacteria in reclaimed wastewater after UV disinfection. Biomed. Environ. Sci. 2011, 24, 400–407. [Google Scholar]
- Silveira, A.B.D.; Bechtlufft, M.D.P.; Van Der Sand, S.T.; Corçã, G. Evaluation of the activity of disinfectants against coliform bacteria group strains isolated from a sewage treatment plant (ETE-Ipanema). Acta Sci. Vet. 2006, 34, 71–76. [Google Scholar] [CrossRef]
- Cho, M.; Kim, J.; Kim, J.Y.; Yoon, J.; Kim, J.H. Mechanisms of Escherichia coli inactivation by several disinfectants. Water Res. 2010, 44, 3410–3418. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).