Abstract
Khao Dawk Mali 105 (KDML105) rice is one of the most important crops of Thailand. It is a challenging task to identify the genes responding to salinity in KDML105 rice. The analysis of the gene co-expression network has been widely performed to prioritize significant genes, in order to select the key genes in a specific condition. In this work, we analyzed the two-state co-expression networks of KDML105 rice under salt-stress and normal grown conditions. The clustering coefficient was applied to both networks and exhibited significantly different structures between the salt-stress state network and the original (normal-grown) network. With higher clustering coefficients, the genes that responded to the salt stress formed a dense cluster. To prioritize and select the genes responding to the salinity, we investigated genes with small partners under normal conditions that were highly expressed and were co-working with many more partners under salt-stress conditions. The results showed that the genes responding to the abiotic stimulus and relating to the generation of the precursor metabolites and energy were the great candidates, as salt tolerant marker genes. In conclusion, in the case of the complexity of the environmental conditions, gaining more information in order to deal with the co-expression network provides better candidates for further analysis.
1. Introduction
Rice is one of the most important agricultural products of Thailand with the highest domestic consumption and exportation. Khao Dawk Mali 105 (KDML105) rice, known as jasmine rice, is considered a high-quality rice from Thailand. It is grown mainly in the northeastern region of Thailand, where the soil contains a high salinity. This poor soil quality affects plant growth and productivity. With the ability to grow in high salinity areas, the salinity tolerance score of KDML105 is lower than some other rice varieties, such as IR29, KKU-LLR-039 and Niewdam Gs. No. 88084 [1]. This information suggested that KDML105 rice should contain some important salt tolerant genes.
In order to identify the key gene(s) playing an important role in salt tolerance, the high-throughput technology of RNA sequencing (RNA-seq) has been applied. New transcripts, single nucleotide polymorphisms (SNP), and gene and splice variants could be identified using this technique [2,3]. In addition, this technology is widely used for the study of differential gene expressions during plant responses to various stressors [2,3,4,5,6,7,8]. For example, the study of Zhang and his coworkers [3] reported the transcriptome analysis of rice roots in response to potassium deficiency. Furthermore, beyond the gene expression level, the analysis of a gene co-expression network has become a broadly used technique for searching for important markers or genes that are in cooperation with other genes. However, this network analysis is quite rare and needs a more elaborate method to represent as much as information regarding the strength of the connections. Subsequently, various network-based analyses have been applied to the gene co-expression network [9,10,11,12,13,14,15,16,17]. Some of the simple and widely used local network measurements are the degree centrality (or hub centrality) [13,14,17,18,19] and clustering coefficient [17,20,21,22,23,24,25]. The study of Chen and his colleagues employed a co-expression network analysis to identify the hub genes associated with metastasis risk and prognosis in cancer cells [9]. Zhang and Horvath applied clustering coefficient on a gene co-expression network of cancer in humans, and found that the clustering coefficient could be used for distinguishing the difference between normal genes and cancer genes [17]. The method using the clustering coefficient gave better results, as compared to other methods using a simple measure of degree of connectivity [17]. The development of the clustering coefficients has been implemented for the RNA-seq dataset [15,16]. However, the constructed co-expression networks are typically performed as a binary graph by ignoring the strength of the connections between the genes. The relevance of genes can be derived from the correlations of the gene expression levels of all of the gene pairs in the network. However, calculating the correlations of all of the gene pairs in the network causes a high computational complexity in terms of the network analysis. Fortunately, the computations with greater data can presently be performed with high performance computing technologies. Thus, the analysis of a weighted network is possible, and is of interest for its application in the real biological network. Li and his coworkers used a weighted gene co-expression network analysis to identify the novel potential genes involved in the early metamorphosis process in sea cucumbers [11]. Jianqiang Li and his colleagues developed a weighted gene co-expression network analysis framework to analyze forty oral squamous cell carcinoma (OSCC) specimens as well as their matched non-tumorous epithelial counterparts [10].
For the network analysis, several topological features are widely employed to identify new target genes. Degree is one of the most commonly used features to identify a local connection. It represents the number of neighboring nodes in the network. The hub genes in the network are the genes with high degrees, which are mostly known to be important genes for the structure of the network and biological functions [13,14,26]. This means that the higher the number of neighbors for a considered gene, the more implied importance the gene has than others in term of maintaining the network structure as well as the organisms’ lives [13,14,26]. However, information regarding the connections among neighbors is still lacking. The clustering coefficient is another local density-connection measure that focuses on connections among neighbors, and it has been developed for various kinds of networks from a simple binary network to a weighted network, as well as a recent version for a signed weighted network, which mostly fulfils with any real-world networks [13,17,23,27,28]. Thus, the clustering coefficients have been used in many fields [20,21,22,23,24,28,29,30,31,32,33]. The clustering coefficient for a weighted network was applied to the financial and metabolic networks by using the geometric mean of the weights [28]. The application of the clustering coefficient was also done for a gene co-expression network of simulated data, a cancer microarray data set, and a yeast microarray data set [17]. The results show that, in a scale-free network, the weighted clustering coefficient was not inversely related to the connectivity, while the binary clustering coefficient was inversely related to connectivity [17].
In this work, we studied the whole transcriptome sequencing data of KDML105 under salt-stress and control situations. The complete signed gene co-expression network was constructed for both conditions, and the clustering coefficients were applied to the networks. The aim is to compare the different structures of the normal-state network and salinity-state network using a clustering coefficient and degree on the weighted co-expression network. Finally, potential target genes were selected and proposed for further study and experimental validation.
2. Materials and Methods
2.1. Plant Material Preparation
KDML105 rice at seedling and booting stages were used as the plant materials. In rice, the seedling stage is a part of the vegetative phase of rice growth, developing more leaves and seminal roots after the first root and shoot of the rice emerge following germination. The booting stage is the first step in the reproductive phase of rice. In this stage, a bulging of the leaf stem conceals the developing panicle. Rice during both seedling and booting stages is sensitive to the salt stress. The experiments were conducted using three biological replicates, each consisting of one plant. The KDML seeds were soaked in distilled water for five days in plastic cups until germination. The germinated seeds were transplanted into the plastic trays for the seedling samples, and the booting samples were transplanted into the plastic plots. All plants were grown in a Bangsai nutrient solution that contained 50 g/L of MgSO4, 80 g/L of KNO3, 12.5 g/L of NH4H2PO4, 8.5 g/L of KH2PO4, 0.4 g/L of Manganese chelate of ethylenediaminetetraacetic acid (Mn-EDTA), 0.8 g/L of micronutrient, 100 g/L of Ca(NO3)2, and 3 g/L of Ferric iron chelate of ethylenediaminetetraacetic acid (Fe-EDTA). For the seedling stage of the experiment, the plants were grown in soil, in the pot 3 × 3 × 7 cm3 in size for 18 days after germination. Then, they were separated into two groups. One group was grown in normal condition by flooding with water, while the other was flooded with 75 mM NaCl. After 21 days, the plants were collected and recorded as day 0 plants under normal condition, and the samples that were under the stress condition were also collected, and two days later they were treated with 75 mM of NaCl for RNA analysis, and were recorded as day 2. For the flower stage of the experiment, the rice plants were grown in soil in a 25 cm diameter pot, with one plant per pot. In the booting stage, for the salt stress treatment, 75 mM of NaCl was flooded over the soil, while the plants flooded with water were used as the controls. After three days, flag leaves were collected for RNA extraction, from the day 0 plants and the day 2 plants, after being exposed to the treatment. Table 1 displays the number of samples and conditions used in this study.

Table 1.
Number of samples and conditions.
2.2. RNA Extraction and Sequencing
The total RNA was extracted from the plant tissues using the PureLink™ Plant RNA Reagent (Invitrogen, Carlsbad, CA, USA). The RNA-seq libraries were prepared according to the study of Udomchalothorn et al., [34]. The contaminated genomic DNA was treated with DNaseI (NEB), and the complementary DNA (cDNA) libraries were prepared using the KAPA stranded RNA-seq library preparation kit for Illumina® (Kapa Biosystem, Davis, CA, USA). Fragments of 300 bp in size were chosen and connected with the adaptors. Then, all of the fragments were enriched using PCR for 12 cycles. All of the cDNA libraries of each treatment were sequenced using the Illumina HiSeqTM 3000 sequencing system (Illumina, Davis, CA, USA). Three replicates of the RNA-seq libraries were used for the analysis. The complete genomic sequence of rice (Oryza sativa L.) was used as a reference [35]. The RNA-seq data consisted of 50 bp single-end reads (SRA BioProject ID: PRJNA507040). On average, there were approximately 20,665,662 reads per sample for the seedling stage and about 28,691,447 reads per sample for the booting stage. The quality control was performed according to the program of Pipeline of Parentally-Biased Expression (POPE) [36].
For the transcriptome data analysis, the adaptors of all of the short-sequence reads from the sequencing were removed, and then the sequence reads were categorized using the pipeline developed by Missirian et al. (2011) [37]. The sequence reads were aligned and mapped to the rice genome database (RGAP 6.0) (http://rice.plantbiology.msu.edu/, [35]) using Bowtie 2 [38] and TopHat [39]. The differentially expressed genes were determined using the DESeq package (version 1.24.0) [40].
2.3. Construction of a Gene Go-Expression Network
In order to construct a gene co-expression network, the RNA-seq read counts were analyzed and normalized using R package DESeq [40]. Non-informative genes, which were the genes whose RNA-seq read counts were zero for all of its replicates, were removed from the analysis. The gene co-expression networks were constructed for normal and salt-stress conditions, namely, normal-state and salinity-state networks, respectively. The gene expression levels of both the seedling and booting stages were combined for each condition. For the whole-state networks, all of the expression levels for the normal and salinity conditions were mixed together. Then nodes represent the genes and the edges represent the level of co-expression for any of the pairs of genes, which were the correlation values of the Pearson correlation coefficient of the expression levels. This resulted in a complete graph with edge weights of the correlation scores that were either positive or negative. When only high co-expression levels for constructing a binary network were concerned, the connections of any two genes existed when their absolute correlation coefficients were greater than or equal to 0.9. Finally, only the connected nodes were used to build up the networks.
2.4. Local Node and Network Properties
2.4.1. Degree and Degree Assortativity
Degree is a commonly used measure for calculating the connectivity of a gene in a network. A high value degree can interpret how important an interesting gene is in the connection of a network [13]. Assuming that a gene node, i, is in a binary network, its’ degree can be calculated as the number of edges connected to it. For a weighted network, all of the connections in the network have their own values, rather than 0 or 1 as with a binary network. Therefore, a weighted degree was introduced as the sum of all of the weights of the edges connected to node i.
The Degree Assortativity is the Pearson correlation coefficient of the degree between pairs of connected nodes in a network [41]. It quantifies the tendency of the nodes being connected to other nodes that have similar degree or not. Positive values of this correlation mean any two connected nodes possessing a similar degree, while a negative correlation is represented by a network containing most of connected nodes with different degrees. All of the possible values of this correlation lie between −1 and 1, indicating the level of assortative patterns. When the correlation is 1, the network is said to be perfectly assortative, but when the correlation is −1, the network is completely disassortative. At the correlation of zero, the network is said to be non-assortative.
2.4.2. Diameter Length
Diameter is defined as the maximal shortest path of all of the shortest paths between any two nodes of a network, which is the largest number of nodes to be traversed in order to travel from one node to another. For any disconnected graph, the diameter is defined as infinite [42].
2.4.3. Clustering Coefficient
The clustering coefficient is a measure of the proportion of true connections and the number of all possible connections among the neighbors of a gene node. Assume that a gene node, is an observed node while node and node are its neighbors. If there is connection between and , the connection between node , and occurs. According to Watts and Strogatz [43], the definition of the original clustering coefficient for a binary network is as follows.
where is a binary value indicating the connection between node and node . is degree of node . has value in range . It is if the neighbors of do not connect to each other and it is if all neighbor are pairwise connecting to each other. Based on a real-world network, mostly their nodes are connected with some level of strength connections or weights. Onnela et al. [27,28] was introduced the clustering coefficient for a weighted graph by taking a geometric mean of total weights of the neighbors as follows.
where is the weight of the edge connecting node of and . These weights can be positive or negative. The value of is in the range where means there are no neighbors of connected to each other, and means that all of the neighbors are connected pairwise to each other with the highest weight. This formula uses the real weights in the network to compute both the numerator and denominator, while the original formula uses a cut-off to approximate the weights into a binary class, either connected or not connected.
3. Results
3.1. Overview of the Workflow
In order to identify the genes responding to salt tolerance, we considered two different states of a co-expression network. An overview of the workflow is shown in Figure 1. Firstly, all of the transcripts of genes under the salinity and normal conditions for the seedling and booting stages were sequenced using RNA-seq technology. The raw data were manipulated in the data preprocessing so as to retrieve the expression levels (read counts) for each transcript and then for the differential expression analysis. Later, the functional and gene ontology (GO) annotations were performed for all of the differentially expressed genes; either during the booting or seedling stages, and a GO enrichment analysis was performed. Then, the co-expression networks for salinity and normal conditions were constructed, and their network structures and properties were compared. The analysis results of the local network connections, such as the degree and clustering coefficient, were used to identify the genes related to salt tolerance.
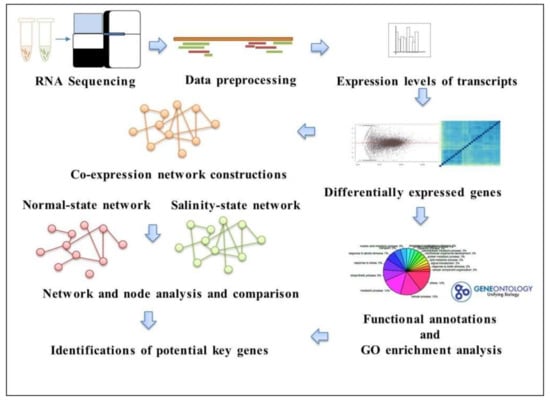
Figure 1.
The analysis workflow to identify key genes with different state networks.
3.2. Identify Differentially Expressed Genes Sensitive to Salinity in Rice
The rice stain KDML105, in the seedling and booting stages, was observed under salt-stress and control conditions, with three replicates each. The high-throughput transcriptome analysis of control and salt stress-treated rice samples was conducted using an Illumina HiSeqTM 3000. We used the complete genomic sequence of rice (O. sativa L.) as a reference [35]. The comparison of the transcriptome differences of the reads between the control and the treated group was performed using DESeq package (version 1.24.0) [40] in R-programming language. The p-value was adjusted using Benjamini–Hochberg method for multiple-test correction. Finally, we obtained a total of 788 differentially expressed genes during the seedling stage, and 759 differentially expressed genes during the booting stage (with the adjusted p-value of less than 0.01), as described in Table 2. In total, there were 1472 differentially expressed genes (713 genes during the seedling stage only, 684 genes during the booting stage only, and 75 genes in both stages). In the seedling stage, salt stress caused the significant changes of 788 genes (Supplementary Table S1), while in the booting stage, 759 genes were found to have the differentially expression level due to salt stress (Supplementary Table S2). Among the differentially expressed genes, more than 50% of them were down-regulated by salt stress. Although the number of differentially expressed genes at the booting stage was lower than the number of differentially expressed genes at the seedling stage, stronger effects of the salt stress were found in the booting stage changing the level of all of the affected genes by more than two-fold (Figure 2).

Table 2.
Number of differentially expressed genes under salt stress for the seedling and booting stages.
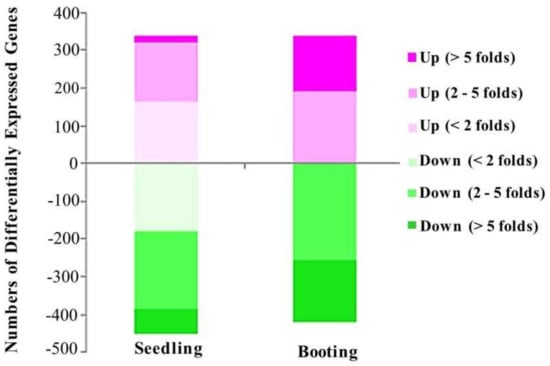
Figure 2.
Fold change plot. Number of differentially expressed genes caused by salt stress treatment at the seedling stage and booting stage.
3.3. Functional Annotations of Differentially Expressed Genes
To classify the function of the differentially expressed genes affected by salinity, the rice information and GO annotations for each gene locus were retrieved from http://rice.plantbiology.msu.edu/ [35]. In total, we annotated the GO terms for 718 differentially expressed genes during the seedling stage, and 666 genes during the booting stage (Supplementary Tables S3 and S4). The proportions of GO terms for both stages were similar (Supplementary Figure S1). Some differences were of interest and were discussed. The large proportions belonged to the genes that encoded the proteins with catalytic activity and binding in the molecular functions. The genes that encoded the proteins with hydrolase activity accounted for the next largest proportion during the seedling stage, but the proteins with a structural molecule activity were found more during the booting stage (Figure 3). Notice that during the booting stage, the differentially expressed genes encoded for proteins in the structural molecule activity were found more than these genes during the seedling stage. In both stages, the encoded proteins of the differentially expressed genes distributed through many cellular organelles. Furthermore, among the biological processes, many of these genes participated in cellular processes, metabolic processes, biosynthesis and the process of response to stress as expected. In order to better gain the enhanced functions of the differentially expressed genes, a GO enrichment analysis was performed using the Fisher’s exact test under a R programming environment. The GO enrichment showed that the differentially expressed genes of the seedling stage and the booting stage accounted for different functions as shown in Table 3. Obviously, the genes responding to the abiotic stimulus and stress were enriched during the seedling stage, and the genes related to the catalytic activity and the genes whose encoded proteins were localized in the extracellular region and cell wall were found most significantly during the seedling stage. During the booting stage, the genes whose encoded proteins were localized in the ribosome, nucleolus, cytosol, and vacuole, were found most when compared to the other gene ontology terms. Proteins with a structural molecule activity were enriched in this period as well. Furthermore, the genes responding to the translation process and the abiotic stimulus were found to be enriched during this period. Notice that genes responding to stress were found during the booting stage, but it is not significant after adjusting the p-value with multiple test correction when comparing with the other terms (as shown in Table 3 with the adjusted p-value by Benjamini–Hochberg correction for multiple testing). Since during the seedling stage, leaves are developing and grow, the process of metabolism is important; especially, carbohydrate metabolic process. The booting stage is the first step to develop rice into a reproductive phase in which rice need to develop seeds and flowers. Genes whose encoded proteins located in nucleolus, ribosome, vacuole and cytosol were found most and related to the translation of these genes.
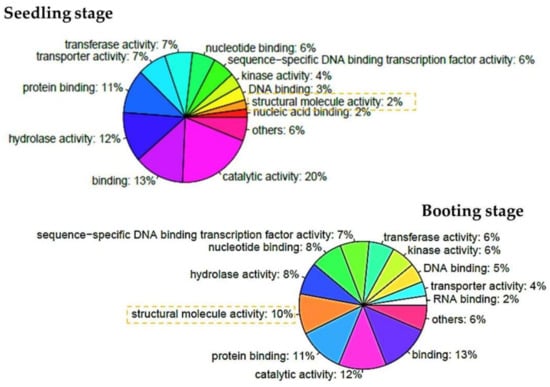
Figure 3.
Gene ontology classification of differentially expressed genes among the molecular functions. The top-left of the pie chart represents the gene proportion of each function during the seedling stage. The bottom-right of the pie chart represents the gene proportion of each function during the booting stage.

Table 3.
Gene ontology (GO) enrichment analysis for differentially expressed genes.
3.4. Analysis of Co-Expression Networks in the Comparison of Salinity and Normal States of Rice
To better understand how genes work together in a rice cell under different conditions, co-expression networks were built separately for the salinity and normal states. The expression levels of both the seedling and booting stages were used to calculate the correlations between the gene pairs of all of the differentially expressed genes. Interestingly, as shown in Figure 4, these genes under a salinity condition were more cooperated to each other than the genes under a normal situation, both in terms of negative and positive correlations. Notice that the negative correlations were found more under the salinity condition. This signified that these genes’ encoded proteins might have an “on” switch during the seedling stage, but “off” switch during the booting stage, or the other way round. Then, the binary co-expression networks for the salinity state and normal state were constructed by inferring network edges based on the high absolute correlation (>0.9). An analysis of the node properties for the different network structures of the salinity-state network and the normal-state network is shown in Table 4 and Figure 5 and Figure 6. In total, there were 1472 differentially expressed genes (713 genes during the seedling stage only, 684 genes during the booting stage only, and 75 genes were found in both stages). The salinity-state network contains 1446 genes connected to each other while the normal-state network contains 1443 genes connected to each other. Notice that in Table 4, the salinity-state network had more connections and was denser than the normal-state network as shown in their connections per node, average degree, number of hub and end nodes, and their diameter length. The Degree Assortativity showed that under both stages, these genes had similar degrees but were found much more in the normal situation than in the salinity. Global cluster densities of both networks were similar, with a bit more density in the salinity-state network. Many genes with a low degree in the normal-state network behaved differently and had more partners (or high degree) in the salinity state, as seen in Figure 5.
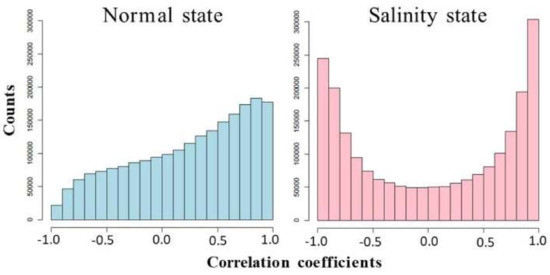
Figure 4.
Distributions of correlation coefficients between the gene expression levels under normal and salinity states.

Table 4.
Node and network properties.
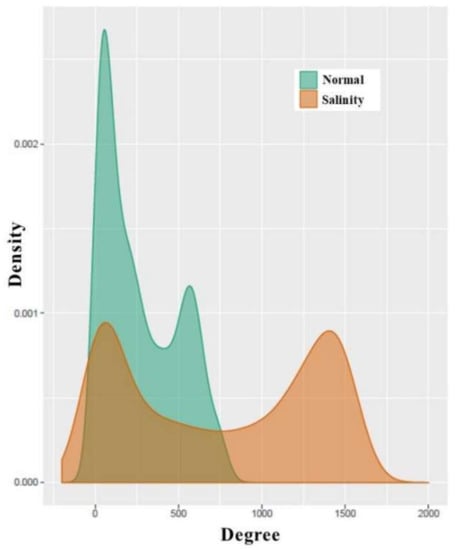
Figure 5.
Degree distributions of normal-state and salinity-state networks.
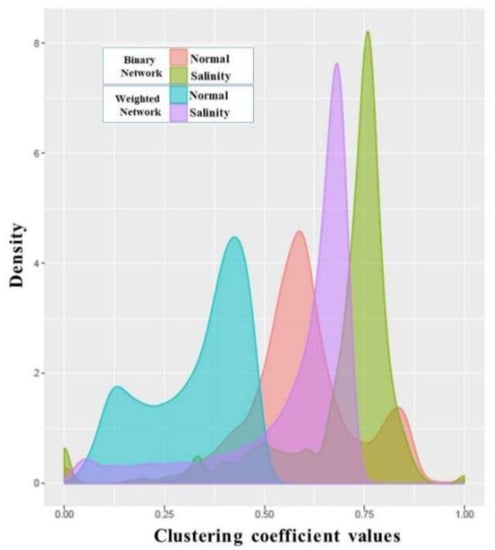
Figure 6.
Clustering coefficient distributions of normal-state and salinity-state networks. Pink and green plots are the clustering coefficient distributions of the binary networks for normal and salinity states, respectively. The blue and purple plots are the clustering coefficient distributions of the weighted signed networks for the normal and salinity states.
In general, the degree distribution of many real-world networks, including the co-expression network, follows the power-law distribution. Interestingly, in Figure 5, the degree distributions of both the normal and salt-stress networks follows a bimodal distribution, especially, in the salt-stress network. However, the normal network trended to follow the power-law distribution, as the majority of nodes had low degrees, even if the number of nodes with high degrees was more than the number of nodes expected by the power law. The salt-stress network clearly showed the bimodal distribution. This showed that a greater number of genes tended to have more co-partners during the salt stress.
To analyze how well each gene connected to its partners, rather than the utilization of the degree, examining the clustering coefficient is an alternative way. The distributions of the binary clustering coefficients (pink and green plots in Figure 6 for normal and salinity states, respectively) showed that most of the genes came to form a dense local cluster in the salinity condition rather than in the normal situation. The same tendency was also found when applying the weighted clustering coefficients (blue and purple plots in Figure 6 for the normal and salinity states, respectively). This means that under the normal state, most of the genes connected to each other were quite dense and had nearly the same local structure (similar clustering coefficient values), while under the salinity state, some genes behaved differently, and most turned out to have more connections among their partners. As expected, under the salt-stress condition, most of the genes behaved differently and needed to express more in order to release the stress, as well as to return to normal as fast as possible. The weighted clustering coefficients were smaller than the binary clustering coefficients as the network edges had weights ranging from −1 to 1 (the correlation values as shown in Figure 4). The negative weights had a direct effect on the clustering values.
3.5. Candidate Genes Responding to the Salinity Are Less Cooperative in the Normal
As shown in Table 4 and Figure 5, under salt-stress conditions more genes were co-expressed as many genes having more degrees. It is quite interesting that a certain gene with small partners in a normal situation gains more partners when it faces the stress. Therefore, our first list of candidate genes comprised genes showing small partners in a normal situation, but showing much higher partners in the salinity conditions. Initially, 484 genes were identified as small-degree genes (with degree < 50) under the normal situation. Then, the list was filtered to select the high hub genes (degree > 700) in the salinity-state network. Finally, the 135 hub genes and their corresponding degrees, clustering coefficients, and functional annotations are presented in Supplementary Table S5. This candidate list was enriched with the functions of the response to abiotic stimulus (GO:0009628) and the generation of precursor metabolites and energy (GO:0006091).
Of these genes, seventeen genes were found to be significantly differentially expressed genes under transcription levels that might directly affect the whole stress-release process (all of the list with their associated GO terms can be found in Table 5). Fifteen genes were found to have differential expressions in the booting stage, while only two genes were found in the seedling stage.LOC_Os05g45810, LOC_Os02g06330, LOC_Os09g29130, LOC_Os07g47140, LOC_Os05g37690, LOC_Os04g32460, LOC_Os05g02500, LOC_Os11g44810, and LOC_Os08g07970 were found to be down regulated in the booting stage. LOC_Os02g38040, LOC_Os04g32590, LOC_Os10g31850, LOC_Os12g06340, LOC_Os01g39770, and LOC_Os08g35190 were found to be up-regulated in booting stage. LOC_Os11g47920 and LOC_Os06g14750 were found to be upregulated in the seedling stage.

Table 5.
List of candidate key genes responding to the salinity.
With the literature search, the candidates in Table 5 were grouped according to the evidence, if there was high evidence showing the function of salt stress in the plant, it is indicated as *** (>10 pieces of literature); ** for moderate evidence (from 5 to 10 pieces of literature); and * for some evidence (from 1 to 5 pieces of literature). Lastly, “-” means that is for no present evidence.
*** LOC_Os01g39770 (calcineurin B; CBL9), LOC_Os02g06330 (AP2 domain containing protein), LOC_Os02g38040 (leucine-rich repeat family protein), LOC_Os05g45810 (calcineurin B; CBL4), LOC_Os08g07970 (OsbZIP64), and LOC_Os10g31850 (RING finger and CHY zinc finger domain-containing protein 1) were found to be related to performing the function of responding to salt, in various plants such as peanut, wheat, and Arobidopsis thaliana. For example, recently, CBL4 and CBL9 were found to be related to drought stress in wheat [44]. Interestingly, LOC_Os02g06330 (AP2 domain containing protein) was found in the transcriptional regulation under salinity stress in African rice [63].
** LOC_Os04g32460 (OsFBL16), LOC_Os05g37690 (OsFBL23) and LOC_Os07g47140 (CCT/B-box zinc finger protein) were found to be related to salt and drought stress in apple, soybean and Arabidopsis. OsFBL16 and OsFBL23 encoded for F-Box proteins, which play an important role in controlling the degradation of unneeded proteins during the cell cycle [134].
* LOC_Os05g02500 (OsMKP1, GSN1, dual specificity protein phosphatase) was studied in rice to regulate the activities of stress-related mitogen-activated protein kinases [91]. LOC_Os06g14750 (phosphatidylinositol-4-phosphate 5-Kinase family protein) and LOC_Os11g47920 (SCARECROW) were analyzed as salt-inducible genes in barley roots [93]. LOC_Os09g29130 (ZF-HD protein dimerization region containing protein) is a zinc-finger transcription factor that was studied recently as a novel regulator of the stress responsive gene. LOC_Os12g06340 (OsBLH1, BEL1-like homeodomain transcription factor) was studied in rice for the floret meristem [133].
- Currently, there is no study found for transcription factor LOC_Os04g32590 and auxin-repressed proteins LOC_Os08g35190 and LOC_Os11g44810.
The constructed co-expression network is shown in Figure 7. Due to the large and dense network, approximately 10% of all nodes plus 17 candidates were selected to plot the networks for each condition. Each node in the figure represents a differentially expressed gene, and each edge represents a connection between the two differentially expressed genes under different conditions. Notice that all seventeen candidates (red nodes in Figure 7) dispersed in the normal-state network while there were more connections in the salinity-state network.
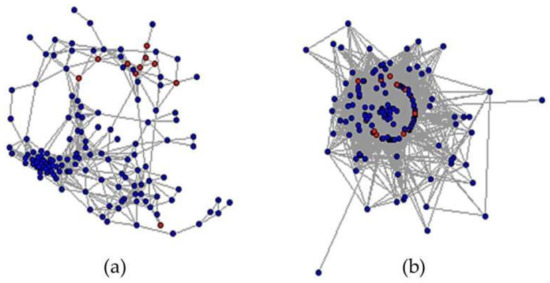
Figure 7.
Co-expression networks under different states. Red nodes represent all seventeen candidates while blue nodes represent all of the other nodes. (a) Co-expression network of genes under normal situation. (b) Co-expression network of genes under salt stress.
4. Discussion and Conclusions
With the importance of Thai rice and the serious problem of saline soil in the northeastern part of Thailand, we proposed the two-state co-expression networks under normally grown and salt-stress conditions, in order to identify the key KDML105 gene markers responding to the salt stress. These two co-expression networks were constructed based on the gene expression levels investigated by RNA-seq technology during the seedling and booting stages, and then these data were compared in terms of the node and network properties. The results showed that the structure of the salinity-state network was denser than that of the normal-state network. Especially, when we applied the weighted clustering coefficient to the signed co-expression networks, more genes operated more partners in responding to the stress. This is clearly because under high salinity conditions there are more connections compared to the normal state. These changes in the gene regulation reveal that KDML105 rice under salt stress could be damaged, as salt or NaCl could stimulate a large change in cooperation among genes. This would help us to find a better list of genes whose encoded proteins were activated to interact and function more under salinity.
To select a potential list of genes for responding to the salt, the genes with few partners under normal state, but with many more partners under the salinity state were selected as the candidate key genes. Seventeen genes out of these genes were found in the transcription levels, and were validated with the literature search. Notice that out of these genes, OsFBL23 and OsFBL16 (LOC_Os05g37690 and LOC_Os04g32460, respectively), encoded for F-box proteins, were found down regulated during the booting stage. According to the analysis of their degrees and weighted clustering coefficients on the salinity-state network, they demonstrated a regulating role to control many other partners whose expression levels were correlated. F-Box proteins were found to be important for controlling the degradation of cellular proteins when they formed the Skp1p-cullin-F-box complex [134]. This protein complex plays an important role for unneeded protein degradation during the cell cycle. It has been found that several publications have reported that these proteins respond to salt stress [71,84,85,86,87,88,89]. Thus, it is crucial to investigate this degradation process as a pathway system in which OsFBL23 and OsFBL16 are involved.
Interestingly, the genes encoded for calcineurin B-like proteins, LOC_Os01g39770 (CBL9) and LOC_Os05g45810 (CBL4) were regulated differently. CBL9 was upregulated while CBL4 was downregulated during the booting stage. Both calcineurin genes were found in fourteen publications, which recently supported that they responded salt stress [44,45,46,47,48,49,50,51,52,53,54,55,56,57]. CBL4 has been found to be acting as a calcium sensor involved in the regulatory pathway for the control of the intracellular Na+ and K+ homeostasis and salt tolerance [135]. For further experimental investigation and modeling, it would be interesting to focus on the effect of salinity in KDML105 rice when CBL9 on chromosome 1 was switched on while CBL4 on chromosome 5 was switched off during the salt stress.
In terms of the computational analysis, we found that the use of a network analysis is still a great tool to filter and select key genes under a specific condition. The construction of a co-expression network is the first and most important task of concern. In this work, the co-expression networks were built from the calculation of the Pearson correlation on the RNA-seq data. Small replicates and features directly affected the correlation values which also affected the network structure. It has been known that rice is sensitive to salt stress during the seedling and booting stages. Therefore, this work used the transcriptomic data of both the seedling and booting stages of KDML105 rice to construct the networks under normal-grown and salinity states. To distinguish the different structures of both networks, we found that the utility of the weighted networks revealed more curated structures and the change before and after the KDML105 rice was affected by the stress. In addition, the selection of the best key genes was performed with the use of degree because it obviously established the number of co-partners, while the clustering coefficient in this work showed a more or less similar local density. In principle, the framework of the analysis in this work can be applied to many other applications that are not only for KDML105 rice, but also for other plants and organisms as well as any other conditions. In addition, the development of the network analysis tool has become an important task to crop a large amount of genomic and transcriptomic data.
All in all, gaining more information in order to deal with the gene co-expression network at different stages can provide a better understanding of the complex regulatory systems responding to the environment, and also better candidates for further analysis and investigation.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/12/594/s1, Figure S1: Gene ontology classification of differentially expressed genes, Table S1: Differentially expressed genes under seedling stage, Table S2: Differentially expressed genes under booting stage, Table S3: Gene ontology (GO) mapping for differentially expressed genes under seedling stage, Table S4: Gene ontology (GO) mapping for differentially expressed genes under booting stage, Table S5: Candidate key genes related with transcription level.
Author Contributions
Conceptualization, S.C. and K.P.; methodology and formal analysis, A.S., C.C. and K.P., resources and data curation, P.C. and N.K.; writing—original draft preparation, A.S., C.C., and K.P.; writing—review and editing, T.B., S.C. and K.P.
Funding
This research work was supported by the Ratchadaphiseksompoj Research Fund (CU-59-012-FW), Chulalongkorn University. A.S. was supported by King Mongkut’s University of Technology North Bangkok (Contract Number: KMUTNB-ART-60-061). C.C. was supported by Development and Promotion of Science and Technology Talents Project (DPST), Thailand.
Conflicts of Interest
The authors declare no conflict of interest and the sponsors had no role in the design, execution, interpretation, or writing of the study.
References
- Chunthaburee, S.; Dongsansuk, A.; Sanitchon, J.; Pattanagul, W.; Theerakulpisut, P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J. Biol. Sci. 2016, 23, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Cloonan, N.; Grimmond, S.M. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, H.; Wang, H.; Cui, J.; Wang, J.; Hu, J.; Guo, L.; Qian, Q.; Xue, D. Transcriptome analysis of rice seedling roots in response to potassium deficiency. Sci. Rep. 2017, 7, 5523. [Google Scholar] [CrossRef] [PubMed]
- Oshlack, A.; Robinson, M.D.; Young, M.D. From RNA-seq reads to differential expression results. Genome Biol. 2010, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Postnikova, O.A.; Shao, J.; Nemchinov, L.G. Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol. 2013, 54, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yoon, J.; Choi, H.; Fan, Y.; Chen, R.; An, G. Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol. 2015, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; He, X.; Wu, D.; Zhu, B.; Cai, S.; Nadira, U.A.; Jabeen, Z.; Zhang, G. Comparative transcriptome profiling of two Tibetan wild barley genotypes in responses to low potassium. PLoS ONE 2014, 9, e100567. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.Q.; Luo, X.M.; Wang, Y.L.; Xu, Q.J.; Bai, L.J.; Yuan, H.J.; Tashi, N. Transcriptome sequencing in a Tibetan barley landrace with high resistance to powdery mildew. Sci. World J. 2014, 2014, 594579. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wang, F.; Feng, J.; Zhou, R.; Chang, Y.; Liu, J.; Zhao, Q. Co-expression network analysis identified six hub genes in association with metastasis risk and prognosis in hepatocellular carcinoma. Oncotarget 2017, 8, 48948–48958. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, D.; Qiu, W.; Shi, Y.; Yang, J.J.; Chen, S.; Wang, Q.; Pan, H. Application of weighted gene co-expression network analysis for data from paired design. Sci Rep. 2018, 8, 622. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kikuchi, M.; Li, X.; Gao, Q.; Xiong, Z.; Ren, Y.; Zhao, R.; Mao, B.; Kondo, M.; Irie, N.; et al. Weighted gene co-expression network analysis reveals potential genes involved in early metamorphosis process in sea cucumber Apostichopus japonicus. Biochem. Biophys. Res. Commun. 2018, 495, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wu, J. A new algorithm for essential proteins identification based on the integration of protein complex co-expression information and edge clustering coefficient. Int. J. Data Min. Bioinform. 2015, 12, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Plaimas, K.; Eils, R.; Konig, R. Identifying essential genes in bacterial metabolic networks with machine learning methods. BMC Syst. Biol. 2010, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- Plaimas, K.; Mallm, J.P.; Oswald, M.; Svara, F.; Sourjik, V.; Eils, R.; Konig, R. Machine learning based analyses on metabolic networks supports high-throughput knockout screens. BMC Syst. Biol. 2008, 2, 67. [Google Scholar] [CrossRef] [PubMed]
- Proost, S.; Krawczyk, A.; Mutwil, M. LSTrAP: Efficiently combining RNA sequencing data into co-expression networks. BMC Bioinform. 2017, 18, 444. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Dean, A.K.; Zhang, W. A general co-expression network-based approach to gene expression analysis: Comparison and applications. BMC Syst. Biol. 2010, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Ekman, D.; Light, S.; Bjorklund, A.K.; Elofsson, A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006, 7, R45. [Google Scholar] [CrossRef] [PubMed]
- The International Cancer Genome Consortium; Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabe, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; et al. International network of cancer genome projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Vitevitch, M.S. The influence of the phonological neighborhood clustering coefficient on spoken word recognition. J. Exp. Psychol. Hum. Percept. Perform. 2009, 35, 1934–1949. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, R.; Vitevitch, M.S. The influence of clustering coefficient on word-learning: How groups of similar sounding words facilitate acquisition. Front. Psychol. 2014, 5, 1307. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Ren, C.; Li, C. Revisiting the variation of clustering coefficient of biological networks suggests new modular structure. BMC Syst. Biol. 2012, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Kalna, G.; Higham, D.J. A clustering coefficient for weighted networks, with application to gene expression data. AI Commun. Netw. Anal. Nat. Sci. Eng. 2007, 20, 263–271. [Google Scholar]
- Lakizadeh, A.; Jalili, S.; Marashi, S.A. CAMWI: Detecting protein complexes using weighted clustering coefficient and weighted density. Comput. Biol. Chem. 2015, 58, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Lee, C.H.; Fuh, C.S.; Juan, H.F.; Huang, H.C. Link clustering reveals structural characteristics and biological contexts in signed molecular networks. PLoS ONE 2013, 8, e67089. [Google Scholar] [CrossRef] [PubMed]
- Fatumo, S.; Plaimas, K.; Mallm, J.P.; Schramm, G.; Adebiyi, E.; Oswald, M.; Eils, R.; Konig, R. Estimating novel potential drug targets of Plasmodium falciparum by analysing the metabolic network of knock-out strains in silico. Infect. Genet. Evol. 2009, 9, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Saramaki, J.; Kivela, M.; Onnela, J.P.; Kaski, K.; Kertesz, J. Generalizations of the clustering coefficient to weighted complex networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2007, 75, 027105. [Google Scholar] [CrossRef] [PubMed]
- Onnela, J.; Saramäki, J.; Kertész, J.; Kaski, K. Intensity and coherence of motifs in weighted complex networks. Phys. Rev. E 2005, 71, 065103(R). [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, G.; Wang, Y.; Blanzieri, E.; Su, C. Link clustering with extended link similarity and EQ evaluation division. PLoS ONE 2013, 8, e66005. [Google Scholar] [CrossRef] [PubMed]
- Kuperman, M.N.; Risau-Gusman, S. Relationship between clustering coefficient and the success of cooperation in networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2012, 86, 016104. [Google Scholar] [CrossRef] [PubMed]
- Mcassey, M.P.; Bijma, F. A clustering coefficient for complete weighted networks. Netw. Sci. 2015, 3, 183–195. [Google Scholar] [CrossRef]
- Rodriguez-Mendez, V.; Ser-Giacomi, E.; Hernandez-Garcia, E. Clustering coefficient and periodic orbits in flow networks. Chaos 2017, 27, 035803. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Qiao, Y.; Wu, Y. lustering analysis of karyotype resemblance-near coefficient for 6 Bupleurum species. Zhongguo Zhong Yao Za Zhi 2012, 37, 1157–1160. [Google Scholar] [PubMed]
- Udomchalothorn, T.; Plaimas, K.; Sripinyowanich, S.; Boonchai, C.; Kojonna, T.; Chutimanukul, P.; Comai, L.; Buaboocha, T.; Chadchawan, S. OsNucleolin1-L expression in Arabidopsis enhances photosynthesis via transcriptome modification under salt stress conditions. Plant Cell Physiol. 2017, 58, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Missirian, V.; Henry, I.; Comai, L.; Filkov, V. POPE: Pipeline of Parentally-Biased Expression. In Bioinformatics Research and Applications; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2012; pp. 177–188. [Google Scholar]
- Missirian, V.; Comai, L.; Filkov, V. Statistical mutation calling from sequenced overlapping DNA pools in TILLING experiments. BMC Bioinform. 2011, 12, 287. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E. Assortative mixing in networks. Phys. Rev. Lett. 2002, 89, 208701. [Google Scholar] [CrossRef] [PubMed]
- West, D.B. Introduction to Graph Theory, 2nd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2001; 588p. [Google Scholar]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Du, Y.T.; Fu, J.D.; Yu, T.F.; Wang, C.T.; Chen, M.; Chen, J.; Ma, Y.Z.; Xu, Z.S. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 2018, 18, 93. [Google Scholar] [CrossRef] [PubMed]
- Egea, I.; Pineda, B.; Ortiz-Atienza, A.; Plasencia, F.A.; Drevensek, S.; Garcia-Sogo, B.; Yuste-Lisbona, F.J.; Barrero-Gil, J.; Atares, A.; Flores, F.B.; et al. The SlCBL10 calcineurin B-like protein ensures plant growth under salt stress by regulating Na+ and Ca2+ homeostasis. Plant Physiol. 2018, 176, 1676–1693. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Kayum, M.A.; Thamilarasan, S.K.; Nath, U.K.; Park, J.; Chung, M.; Nou, I. Molecular characterisation and expression profiling of calcineurin B-like (CBL) genes in chinese cabbage under abiotic stresses. Funct. Plant. Biol. 2017, 44, 739–750. [Google Scholar] [CrossRef]
- Hima Kumari, P.; Anil Kumar, S.; Ramesh, K.; Sudhakar Reddy, P.; Nagaraju, M.; Bhanu Prakash, A.; Shah, T.; Henderson, A.; Srivastava, R.K.; Rajasheker, G.; et al. Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in sorghum bicolor. Genes 2018, 9, 236. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, M.M.; Ren, L.; Liu, Y.; Chen, H.Y. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genomics 2016, 291, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Wei, Q.; Wang, R.; Zhang, Y.; Zhang, F.; He, Y.; Zhou, S.; Feng, J.; Yang, G.; He, G. BdCIPK31, a calcineurin B-like protein-interacting protein kinase, regulates plant response to drought and salt stress. Front. Plant Sci. 2017, 8, 1184. [Google Scholar] [CrossRef] [PubMed]
- Miranda, R.S.; Alvarez-Pizarro, J.C.; Costa, J.H.; Paula, S.O.; Prisco, J.T.; Gomes-Filho, E. Putative role of glutamine in the activation of CBL/CIPK signalling pathways during salt stress in sorghum. Plant Signal. Behav. 2017, 12, e1361075. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Wan, S.; Xia, Y.; Ren, N.; Zhou, Y.; Jiang, X. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 2018, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Jiang, P.; Qin, G.; Gai, S.; Zhu, D.; Qiao, L.; Wang, J. Transcriptome profiling and digital gene expression analysis of genes associated with salinity resistance in peanut. Electron. J. Biotechnol. 2018, 32, 19–25. [Google Scholar] [CrossRef]
- Wang, J.J.; Lu, X.K.; Yin, Z.J.; Mu, M.; Zhao, X.J.; Wang, D.L.; Wang, S.; Fan, W.L.; Guo, L.X.; Ye, W.W.; et al. Genome-wide identification and expression analysis of CIPK genes in diploid cottons. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, D.; Xu, Z.; Huang, Y.; He, X.; Wang, J.; Gu, M.; Li, J.; Shao, H. Comparative expression analysis of Calcineurin B-like family gene CBL10A between salt-tolerant and salt-sensitive cultivars in B. oleracea. Sci. Total Environ. 2016, 571, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, Q.; Chen, Q.; Xiang, N.; Yang, Y.; Yang, Y. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip (Brassica rapa var. rapa). Front. Plant Sci. 2017, 8, 1191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, L.; Jiao, Z.; Chen, Y.; Liu, H.; Chen, X.; Fu, J.; Wang, G.; Zheng, J. Characterization of the calcineurin B-Like (CBL) gene family in maize and functional analysis of ZmCBL9 under abscisic acid and abiotic stress treatments. Plant Sci. 2016, 253, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Linghu, J.; Wang, D.; Liu, X.; Yu, A.; Li, F.; Zhao, T. Foxtail millet CBL4 (SiCBL4) interacts with SiCIPK24, modulates plant salt stress tolerance. Plant Mol. Biol. Report. 2017, 35, 634–646. [Google Scholar] [CrossRef]
- Chen, H.; Liu, L.; Wang, L.; Wang, S.; Cheng, X. VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 2016, 129, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Cheng, Y.; Wu, J.; Cheng, Q.; Li, W.; Fan, S.; Jiang, L.; Xu, Z.; Kong, F.; Zhang, D.; et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 2015, 66, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Abdullah, S.N.A.; Aziz, M.A.; Namasivayam, P. A novel CBF that regulates abiotic stress response and the ripening process in oil palm (Elaeis guineensis) fruits. Tree Genet. Genomes 2015, 11. [Google Scholar] [CrossRef]
- Gao, W.; He, M.; Liu, J.; Ma, X.; Zhang, Y.; Dai, S.; Zhou, Y. Overexpression of Chrysanthemum lavandulifolium ClCBF1 in Chrysanthemum morifolium ‘White Snow’ improves the level of salinity and drought tolerance. Plant Physiol. Biochem. 2018, 124, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Gumi, A.M.; Guha, P.K.; Mazumder, A.; Jayaswal, P.; Mondal, T.K. Characterization of OglDREB2A gene from African rice (Oryza glaberrima), comparative analysis and its transcriptional regulation under salinity stress. 3 Biotech 2018, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.L.; Xue, N.D.; Zhi, L.; Yong, L.L.; Pei, Y.Y.; Fei, T.; Liang, Z.; Shi, G.L.; Lin, F.D.; Ji, R.S.; Yan, M.W.; et al. Simultaneous overexpression of the HhERF2 and PeDREB2a genes enhanced tolerances to salt and drought in transgenic cotton. Protein Pept. Lett. 2016, 23, 450–458. [Google Scholar]
- Li, X.J.; Li, M.; Zhou, Y.; Hu, S.; Hu, R.; Chen, Y.; Li, X.B. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS ONE 2015, 10, e0118056. [Google Scholar] [CrossRef] [PubMed]
- Mondini, L.; Nachit, M.M.; Pagnotta, M.A. Allelic variants in durum wheat (Triticum turgidum L. var. durum) DREB genes conferring tolerance to abiotic stresses. Mol. Genet. Genomics 2015, 290, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.L.; Wang, Y.S.; Cheng, H.; Wang, L.Y. Characterization and expression analysis of a gene encoding CBF/DREB1 transcription factor from mangrove Aegiceras corniculatum. Ecotoxicology 2015, 24, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Liu, X.; Deng, H.; Shen, S. Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. Plant Sci. 2011, 181, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, X.; Tang, T.; Zhou, R.; Chen, S.; Li, W.; Shi, S. Comparative genomics of two ecologically differential populations of Hibiscus tiliaceus under salt stress. Funct. Plant Biol. 2011, 38, 199–208. [Google Scholar] [CrossRef]
- Yang, S.; Luo, C.; Song, Y.; Wang, J. Two groups of Thellungiella salsuginea RAVs exhibit distinct responses and sensitivity to salt and ABA in transgenic Arabidopsis. PLoS ONE 2016, 11, e0153517. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, M.; Yuan, Y.; Hu, J.; Yang, Y.; Pang, J.; Wang, L. Ectopic expression of cucumber (Cucumis sativus L.) CsTIR/AFB genes enhance salt tolerance in transgenic arabidopsis. Plant Cell Tissue Organ Cult. 2017, 131, 107–118. [Google Scholar] [CrossRef]
- Li, W.; Zhao, F.; Fang, W.; Xie, D.; Hou, J.; Yang, X.; Zhao, Y.; Tang, Z.; Nie, L.; Lv, S. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front. Plant Sci. 2015, 6, 732. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Moon, J.C.; Park, Y.C.; Kim, J.H.; Kim, D.S.; Jang, C.S. Molecular dissection of the response of a rice leucine-rich repeat receptor-like kinase (LRR-RLK) gene to abiotic stresses. J. Plant Physiol. 2014, 171, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Reza, K.A.; Sharmin, S.; Moosa, M.M.; Mahmood, N.; Ghosh, A.; Begum, R.A.; Khan, H. Identification and molecular characterization of a receptor-like protein kinase gene from Corchorus capsularis. Turkish J. Biol. 2013, 37, 11–17. [Google Scholar] [CrossRef]
- Shi, C.C.; Feng, C.C.; Yang, M.M.; Li, J.L.; Li, X.X.; Zhao, B.C.; Huang, Z.J.; Ge, R.C. Overexpression of the receptor-like protein kinase genes AtRPK1 and OsRPK1 reduces the salt tolerance of Arabidopsis thaliana. Plant Sci. 2014, 217–218, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Sharma, S.; Kumar, R.; Mendu, V.; Singh, K.; Upadhyay, S.K. Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front. Plant Sci. 2016, 7, 1374. [Google Scholar] [CrossRef]
- Van der Does, D.; Boutrot, F.; Engelsdorf, T.; Rhodes, J.; McKenna, J.F.; Vernhettes, S.; Koevoets, I.; Tintor, N.; Veerabagu, M.; Miedes, E.; et al. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 2017, 13, e1006832. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.; Yao, X.; Liu, S.; Zhang, P.; Chen, K. The Antarctic moss leucine-rich repeat receptor-like kinase (PnLRR-RLK2) functions in salinity and drought stress adaptation. Polar Biol. 2018, 41, 353–364. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Li, C.; Wang, T.; Zhang, P.; Chen, K. PnLRR-RLK27, a novel leucine-rich repeats receptor-like protein kinase from the Antarctic moss Pohlia nutans, positively regulates salinity and oxidation-stress tolerance. PLoS ONE 2017, 12, e0172869. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, P.; Liu, S.; Cong, B.; Chen, K. A leucine-rich repeat receptor-like kinase from the Antarctic moss Pohlia nutans confers salinity and ABA stress tolerance. Plant Mol. Biol. Report. 2016, 34, 1136–1145. [Google Scholar] [CrossRef]
- Wu, F.; Sheng, P.; Tan, J.; Chen, X.; Lu, G.; Ma, W.; Heng, Y.; Lin, Q.; Zhu, S.; Wang, J.; et al. Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice. J. Exp. Bot. 2015, 66, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Yuan, S.; Li, Z.; Zhou, M.; Wu, P.; Hu, Q.; Mendu, V.; Wang, L.; Luo, H. STRESS INDUCED FACTOR 2, a leucine-rich repeat kinase regulates basal plant pathogen defense. Plant Physiol. 2018, 176, 3062–3080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pei, C.; Ma, J. Screening of candidate salt tolerance-related genes in alfalfa based on transcriptome-proteome correlation research techniques. Zhiwu Shengli Xuebao/Plant Physiol. J. 2016, 52, 317–324. [Google Scholar] [CrossRef]
- Huo, D.; Zheng, W.; Li, P.; Xu, Z.; Zhou, Y.; Chen, M.; Zhang, X. Identification, classification, and drought response of F-box gene family in foxtail millet. Acta Agron. Sin. (China) 2014, 40, 1585–1594. [Google Scholar] [CrossRef]
- Jia, Q.; Xiao, Z.X.; Wong, F.L.; Sun, S.; Liang, K.J.; Lam, H.M. Genome-wide analyses of the soybean F-box gene family in response to salt stress. Int. J. Mol. Sci. 2017, 18, 818. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Y.; Cui, M.H.; Noh, M.S.; Jung, K.W.; Shin, J.S. The FBA motif-containing protein AFBA1 acts as a novel positive regulator of ABA response in Arabidopsis. Plant Cell Physiol. 2017, 58, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Koops, P.; Pelser, S.; Ignatz, M.; Klose, C.; Marrocco-Selden, K.; Kretsch, T. EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5547–5560. [Google Scholar] [CrossRef] [PubMed]
- Song, J.B.; Wang, Y.X.; Li, H.B.; Li, B.W.; Zhou, Z.S.; Gao, S.; Yang, Z.M. The F-box family genes as key elements in response to salt, heavy mental, and drought stresses in Medicago truncatula. Funct. Integr. Genomics 2015, 15, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xing, S.; Cui, H.; Chen, X.; Wang, X. Genome-wide identification and characterization of the apple (Malus domestica) HECT ubiquitin-protein ligase family and expression analysis of their responsiveness to abiotic stresses. Mol. Genet. Genomics 2016, 291, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Chen, K.; Dong, N.Q.; Shi, C.L.; Ye, W.W.; Gao, J.P.; Shan, J.X.; Lin, H.X. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Katou, S.; Kuroda, K.; Seo, S.; Yanagawa, Y.; Tsuge, T.; Yamazaki, M.; Miyao, A.; Hirochika, H.; Ohashi, Y. A calmodulin-binding mitogen-activated protein kinase phosphatase is induced by wounding and regulates the activities of stress-related mitogen-activated protein kinases in rice. Plant Cell Physiol. 2007, 48, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Mikami, K.; Katagiri, T.; Iuchi, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Shi, W.; Nakamura, T.; Takabe, T. Analysis of salt-inducible genes in barley roots by differential display. J. Plant Res. 2002, 115, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the apple (Malus domestica Borkh.) genome. Mol. Genet. Genomics 2018, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Song, A.; Zhu, X.; Chen, F.; Jiang, J.; Chen, Y.; Sun, Y.; Shan, H.; Gu, C.; Li, P.; et al. The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 2012, 235, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Kielbowicz-Matuk, A.; Rey, P.; Rorat, T. Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann. Bot. 2014, 113, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Horiuchi, H.; Fujita, K.; Takuhara, Y.; Suzuki, S. Characterization of grape C-repeat-binding factor 2 and B-box-type zinc finger protein in transgenic Arabidopsis plants under stress conditions. Mol. Biol. Rep. 2012, 39, 7933–7939. [Google Scholar] [CrossRef] [PubMed]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Araki, T.; Meshi, T.; Iwabuchi, M. Expression of a subset of the Arabidopsis Cys2/His2-type zinc-finger protein gene family under water stress. Gene 2000, 248, 23–32. [Google Scholar] [CrossRef]
- Tian, Z.D.; Zhang, Y.; Liu, J.; Xie, C.H. Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biol. (Stuttg.) 2010, 12, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Bevan, M.; Brutnell, T.; Buell, C.R.; Cone, K.; Hake, S.; Jackson, D.; Kellogg, E.; Lawrence, C.; McCouch, S.; et al. A recommendation for naming transcription factor proteins in the grasses. Plant Physiol. 2009, 149, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, L.; Qian, W.; Hao, X.; Yang, Y.; Wang, X. Positive regulation of CsbZIP4 transcription factor on salt stress response in transgenic Arabidopsis. Acta Agron. Sin. (China) 2017, 43, 1012–1020. [Google Scholar] [CrossRef]
- Castro, P.H.; Lilay, G.H.; Munoz-Merida, A.; Schjoerring, J.K.; Azevedo, H.; Assuncao, A.G.L. Phylogenetic analysis of F-bZIP transcription factors indicates conservation of the zinc deficiency response across land plants. Sci. Rep. 2017, 7, 3806. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Singh, S.; Singh, M.; Srivastava, P.K.; Mishra, R.K.; Singh, V.P.; Prasad, S.M. Transcriptional regulation of salinity stress in plants: A short review. Plant Gene 2017, 11, 160–169. [Google Scholar] [CrossRef]
- Lapham, R.; Lee, L.Y.; Tsugama, D.; Lee, S.; Mengiste, T.; Gelvin, S.B. VIP1 and its homologs are not required for Agrobacterium-mediated transformation, but play a role in Botrytis and salt stress responses. Front. Plant Sci. 2018, 9, 749. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. Correction to: OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2018, 97, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hu, X.; Li, C.; Xu, X.; Su, C.; Li, J.; Song, H.; Zhang, X.; Pan, Y. SlbZIP38, a tomato bZIP family gene downregulated by abscisic acid, is a negative regulator of drought and salt stress tolerance. Genes 2017, 8, 402. [Google Scholar] [CrossRef] [PubMed]
- Sunitha, M.; Srinath, T.; Reddy, V.D.; Rao, K.V. Expression of cold and drought regulatory protein (CcCDR) of pigeonpea imparts enhanced tolerance to major abiotic stresses in transgenic rice plants. Planta 2017, 245, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Guo, Y.; Zhao, J.; Cheng, H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, J.; Li, X.; Wang, S.; Wu, J. Salt and drought stress and ABA responses related to bZIP genes from V. radiata and V. angularis. Gene 2018, 651, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Pan, X.; Zhang, J.; Ma, Y.; He, S.; Zheng, Y.; Ma, Y. Effect of salt-stress on gene expression in citrus roots revealed by RNA-seq. Funct. Integr. Genomics 2018, 18, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Gao, L.; Hao, C.; Zhao, G.; Jia, J.; Kong, X. A novel wheat C-bZIP gene, TabZIP14-B, participates in salt and freezing tolerance in transgenic plants. Front. Plant Sci. 2017, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lian, C.; Duan, H.; Lu, X.; Xia, X.; Yin, W. Analysis of PebZIP26 and PebZIP33 transcription factors from populus euphratica. Beijing Linye Daxue Xuebao/J. Beijing For. Univ. 2017, 39, 18–30. [Google Scholar] [CrossRef]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, D.D.; Barros, P.M.; Cordeiro, A.M.; Serra, T.S.; Lourenco, T.; Chander, S.; Oliveira, M.M.; Saibo, N.J. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 2012, 63, 3643–3656. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gao, Y.; Zhao, T.; Li, J.; Yao, M.; Xu, X. Genome-wide identification and expression pattern analysis of zinc-finger homeodomain transcription factors in tomato under abiotic stress. J. Am. Soc. Hortic. Sci. 2018, 143, 14–22. [Google Scholar] [CrossRef]
- An, J.; Liu, X.; Song, L.; You, C.; Wang, X.; Hao, Y. Apple RING finger E3 ubiquitin ligase MdMIEL1 negatively regulates salt and oxidative stresses tolerance. J. Plant Biol. 2017, 60, 137–145. [Google Scholar] [CrossRef]
- Chapagain, S.; Park, Y.C.; Kim, J.H.; Jang, C.S. Oryza sativa salt-induced RING E3 ligase 2 (OsSIRP2) acts as a positive regulator of transketolase in plant response to salinity and osmotic stress. Planta 2018, 247, 925–939. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Chapagain, S.; Han, A.R.; Park, Y.C.; Park, H.M.; Kim, Y.H.; Jang, C.S. Molecular characterization of rice arsenic-induced RING finger E3 ligase 2 (OsAIR2) and its heterogeneous overexpression in Arabidopsis thaliana. Physiol. Plant. 2017, 161, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Kim, J.J.; Lim, S.D.; Park, Y.C.; Moon, J.C.; Jang, C.S. Molecular dissection of Oryza sativa salt-induced RING Finger Protein 1 (OsSIRP1): Possible involvement in the sensitivity response to salinity stress. Physiol. Plant. 2016, 158, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Park, H.M.; Han, A.R.; Jang, C.S. Molecular characterization of Oryza sativa arsenic-induced RING E3 ligase 1 (OsAIR1): Expression patterns, localization, functional interaction, and heterogeneous overexpression. J. Plant Physiol. 2016, 191, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.D.; Jung, C.G.; Park, Y.C.; Lee, S.C.; Lee, C.; Lim, C.W.; Kim, D.S.; Jang, C.S. Molecular dissection of a rice microtubule-associated RING finger protein and its potential role in salt tolerance in Arabidopsis. Plant Mol. Biol. 2015, 89, 365–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Zhu, S.; Zhang, H.; Li, Y.; Zhang, T.; Sun, J. Overexpression of GhSARP1 encoding a E3 ligase from cotton reduce the tolerance to salt in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 478, 1491–1496. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Chapagain, S.; Jang, C.S. A negative regulator in response to salinity in rice: Oryza sativa Salt-, ABA- and drought-induced RING Finger Protein 1 (OsSADR1). Plant Cell Physiol. 2018, 59, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Moon, J.-C.; Chapagain, S.; Oh, D.G.; Kim, J.J.; Jang, C.S. Role of salt-induced RING finger protein 3 (OsSIRP3), a negative regulator of salinity stress response by modulating the level of its target proteins. Environ. Exp. Bot. 2018, 155, 21–30. [Google Scholar] [CrossRef]
- Qi, S.; Lin, Q.; Zhu, H.; Gao, F.; Zhang, W.; Hua, X. The RING Finger E3 Ligase SpRing is a positive regulator of salt stress signaling in salt-tolerant wild tomato species. Plant Cell Physiol. 2016, 57, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Xing, Y.; Munir, S.; Yu, C.; Song, L.; Li, H.; Wang, T.; Ye, Z. An ATL78-Like RING-H2 finger protein confers abiotic stress tolerance through interacting with RAV2 and CSN5B in tomato. Front. Plant Sci. 2016, 7, 1305. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Lou, L.; Liu, L.; Yu, F.; Zhao, Q.; Zhang, H.; Wu, Y.; Tang, S.; Xia, R.; Zhu, B.; et al. The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J. 2015, 82, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, Q.; Liu, Z.; Yang, H.; Wang, J.; Li, X.; Yang, Y. Arabidopsis C3HC4-RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress-responsive abscisic acid signaling. J. Integr. Plant Biol. 2016, 58, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Zang, D.; Li, H.; Xu, H.; Zhang, W.; Zhang, Y.; Shi, X.; Wang, Y. An Arabidopsis zinc finger protein increases abiotic stress tolerance by regulating sodium and potassium homeostasis, reactive oxygen species scavenging and osmotic potential. Front. Plant Sci. 2016, 7, 1272. [Google Scholar] [CrossRef] [PubMed]
- Khanday, I.; Ram Yadav, S.; Vijayraghavan, U. Rice LHS1/UsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol. 2013, 161, 1970–1983. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).