Abstract
Both X-linked hypohidrotic ectodermal dysplasia (XLHED) and non-syndromic tooth agenesis (NSTA) result in symptoms of congenital tooth loss. This study investigated genetic causes in two families with XLHED and four families with NSTA. We screened for mutations of WNT10A, EDA, EDAR, EDARADD, PAX9, MSX1, AXIN2, LRP6, and WNT10B through Sanger sequencing. Whole exome sequencing was performed for the proband of NSTA Family 4. Novel mutation c.1051G>T (p.Val351Phe) and the known mutation c.467G>A (p.Arg156His) of Ectodysplasin A (EDA) were identified in families with XLHED. Novel EDA receptor (EDAR) mutation c.73C>T (p.Arg25*), known EDA mutation c.491A>C (p.Glu164Ala), and known Wnt family member 10A (WNT10A) mutations c.511C>T (p.Arg171Cys) and c.742C>T (p.Arg248*) were identified in families with NSTA. The novel EDA and EDAR mutations were predicted as being pathogenic through bioinformatics analyses and structural modeling. Two variants of WNT10A, c.374G>A (p.Arg125Lys) and c.125A>G (p.Asn42Ser), were found in patients with NSTA. The two WNT10A variants were predicted to affect the splicing of message RNA, but minigene experiments showed normal splicing of mutated minigenes. This study uncovered the genetic foundations with respect to six families with XLHED or NSTA. We identified six mutations, of which two were novel mutations of EDA and EDAR. This is the first report of a nonsense EDAR mutation leading to NSTA.
1. Introduction
Tooth agenesis (TA) is a disorder characterized by congenital tooth loss. According to a meta-analysis, the mean prevalence of TA is 6.53 ± 3.33% [1]. The high prevalence of TA, as well as the esthetic and functional problems it causes, make it a significant dental issue [2]. There are two sub-types of TA: non-syndromic TA (NSTA) and syndromic TA (STA). In NSTA, the patients do not have anomalies other than congenital tooth loss [3]. In STA, tooth agenesis is a symptom of a syndrome [4].
NSTA is a disorder with great genetic heterogeneity. Many genes have been reported as being associated with NSTA. Wnt family member 10A (WNT10A) is a key mediator of WNT signaling. Suppression of WNT signaling arrests tooth development [5]. Yamashiro et al. reported that Wnt10a may have a link with the differentiation of odontoblasts and cusp morphogenesis [6]. Zhu et al. found that WNT/β-catenin activity is required for tooth development at the early cap stage [7]. In recent years, WNT10A was found to be mutated in 28–62% of patients with NSTA [8,9,10]. Tardieu et al. found that heterozygous WNT10A mutations resulted in relatively fewer missing teeth, while compound heterozygous WNT10A mutations caused more missing teeth [11]. In addition to WNT signaling, EDA/EDAR/EDARADD signaling has been shown to play a significant role in NSTA [12,13]. Tucker et al. showed that Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis [14]. MSX1 and PAX9 play an important role in tooth development. In Pax9−/− or Msx1−/− mice, tooth development is arrested in the bud stage [15,16]. This is because the two genes can synergistically activate the BMP4 gene, which is vital for the evolution from the bud to cap stage in tooth budding [17]. Mutations of MSX1 and PAX9 account for a proportion of NSTA cases [18,19]. Mutations of some other genes, such as AXIN2, WNT10B, LRP6, EDAR, and EDARADD, have also been identified in a few patients [19,20].
STA is comprised of many diseases, including hypohidrotic ectodermal dysplasia (HED) and odonto-onycho-dermal dysplasia (OODD). Hypohidrotic ectodermal dysplasia is characterized by sparse hair, oligodontia, and reduced sweating [21]. The inheritance of HED can be X-linked, autosomal dominant, or autosomal recessive [22]. X-linked HED (XLHED) is caused by mutations of Ectodysplasin A (EDA) [23,24]. Mutations of both EDA receptor (EDAR) and EDAR-associated death domain (EDARADD) genes are reported to cause autosomal-dominant (ADHED) and autosomal-recessive HED (ARHED) [25,26]. Cluzeau et al. and our group reported that WNT10A also mutated in some patients with ADHED or ARHED [24,27]. Odonto-onycho-dermal dysplasia (OODD), another kind of STA characterized by massive loss of teeth, hyperhidrosis, palmoplantar hyperkeratosis, and nail dystrophy, is caused by mutations of WNT10A [28].
From our previous introduction, we know that, although NSTA and STA are two different subtypes of tooth agenesis, they can be caused by mutations of the same genes. EDA, EDAR, EDARADD, and WNT10A are candidate genes of both NSTA and STA. In our previous study, we found that patients with XLHED suffer only from oligodontia and sparse hair, representing a middle stage between XLHED and NSTA [29]. Such patients presenting with a middle status between XLHED and NSTA have been reported by other researchers as well [30,31]. We proposed that XLHED and EDA-related NSTA are possibly the same disease, caused by EDA mutations but with different degrees of severity [29]. Bohring et al. and Vink et al. reported that WNT10A mutations can cause a broad spectrum of phenotypic variability, from OODD to NSTA [32,33]. The phenotypic variability exists at both inter- and intra-familial levels [33]. We hypothesize that some cases of STA and NSTA, caused by mutations of the same gene, represent the same disease but with phenotypic variability.
In this study, to further investigate the genetic basis and relationship of STA and NSTA, we studied genetic defects in two families with XLHED and four families with NSTA through Sanger sequencing, whole exome sequencing, and bioinformatics analyses. Two novel and four known pathogenic mutations were identified.
2. Materials and Methods
2.1. Subjects
The study aimed to uncover the genetic basis and relationship of XLHED and NSTA. The study was approved by the Ethical Review Committee at the Guanghua School and Hospital of Stomatology at Sun Yat-Sen University (ERC-[2013]-9, date of approval: 1 December 2013, and ERC-[2014]-25, date of approval: 1 November 2014). Recommendations from the Declaration of Helsinki were followed. We obtained informed consent from all of the participants. The inclusion criterion for XLHED was the presence of at least two out of the three following symptoms: sparse hair, oligodontia, and reduced sweating [27]. The patient was excluded if the diagnosis of another kind of ectodermal dysplasia was made [22]. The inclusion criterion of NSTA was the presence of congenital loss of teeth. The exclusion criterion of NSTA was the involvement of other symptoms, such as hypohidrosis, hypotrichosis, and cleft lip and palate. In this project, all of the participants were assessed clinically except the uncle of the proband in XLHED1. Two patients with XLHED, four patients with NSTA, and their family members were recruited, and 4 mL of peripheral blood were collected for each participant at the Department of Pediatric Dentistry, Guanghua Hospital of Stomatology. The uncle of the proband of XLHED Family 1 lives in another city. Therefore, his blood sample was collected in a local hospital and mailed to us.
2.2. Mutation Detection of WNT10A, EDA, EDAR, EDARADD, PAX9, MSX1, AXIN2, LRP6, and WNT10B
We extracted genomic DNA from the blood. Primers covering coding and flanking sequences of WNT10A, EDA, EDAR, and EDARADD were reported in our previous studies [24,29]. We designed primers to amplify PAX9, MSX1, AXIN2, LRP6, and WNT10B. The primer sequences are listed in Tables S1–S5. We then performed Polymerase Chain Reaction (PCR) in order to amplify the exons of nine genes, and the products were sequenced on an ABI 3730XL genetic analyzer (Applied Biosystems, Foster City, CA, USA). Mutation nomenclature was used, with +1 corresponding to the A of the ATG translation initiation codon of the reference sequence NM_025216.2 (WNT10A), NM_001399.4 (EDA). The population genetics data from 1000 Genomes (http://browser.1000genomes.org/index.html) and Database of Single Nucleotide Polymorphisms (dbSNP, https://www.ncbi.nlm.nih.gov/snp) were used as control to rule out population polymorphism.
2.3. Whole Exome Sequencing and Data Analysis
Whole exome sequencing was performed for the proband of Family 4, who did not have any mutations detected with respect to WNT10A, EDA, PAX9, and MSX1. Whole exome sequences were enriched with xGen® Exome Research Panel v1.0 (Integrated DNA Technologies, Coralville, IA, USA). The enriched samples were sequenced with an Illumina HiSeq 2000 to generate 150 bp paired-end reads. Reads were aligned to the human reference genome hg19 using the Burrows–Wheeler Aligner [34]. Single Nucleotide Polymorphisms (SNPs) and Insertions and Deletions (InDels) were identified by the Genome Analysis Toolkit (GATK) and annotated by ANNOVAR [35,36]. Variants of EDAR, EDARADD, AXIN2, WNT10B, LRP6, and LTBP3 genes were extracted and filtered with the criteria of “minor allele frequency (MAF) < 1%” and “exonic”. The candidate mutation of EDAR was verified with PCR, followed by Sanger sequencing. PCR was performed, and the PCR products were sequenced as described in Section 2.2. The reference sequence for EDAR was NM_022336.3.
2.4. Bioinformatics Analyses
Domain information of EDA, WNT10A, and EDAR was retrieved from the Human Protein Reference Database (http://hprd.org/query). PolyPhen 2 [37], Sorting Intolerant from Tolerant (SIFT) [38], Mutation Taster [39], and Human Splicing Finder [40] were used to predict the effect of novel missense mutation. CLUSTAL X (1.83) [41] was used to compare the human wild-type WNT10A protein (CCDS2426.1) with those of the chimpanzee (ENSPTRT00000023964.3), dog (ENSCAFT00000023775.4), rat (NP_001101697), mouse (CCDS15057), chicken (NP_001006590), and zebrafish (NP_571055). A cross-species alignment of EDA protein was performed as described in a previous study [29].
2.5. Structural Modeling
The structures of wild-type and mutated EDA and WNT10A proteins were modeled with Swiss Pdb Viewer v4.1 [42]. The structure of EDA (PDB ID 1RJ7; X-ray, resolution 2.3Å) and Wnt-8 (PDB ID 4F0A; X-ray, resolution 3.25Å) was used as templates for homology modeling. Visualization of the three-dimensional (3D) structures were performed with PyMol v1.5.0.3 (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC., Cambridge, MA, USA).
2.6. Minigene Study
A minigene was designed to analyze whether WNT10A variants affected the splicing of message RNA. Exon 1, Intron 1, Exon 2, Partial Intron 2 (containing the first 203 bp and the last 210 bp) and Exon 3 of WNT10A were amplified by PCR from the genomic DNA of the probands of NSTA Families 1 and 3, who carried the heterozygous WNT10A variants c.374G>A (p.Arg125Lys) and c.125A>G (p.Asn42Ser). The primers were as follows: E1E2-F: 5′-ACTTAAGCTTGCCCGTCAGGGCCTGCGCG-3′; E1E2-R: 5′-AGGCTTCCCACCAGTCTGGAGACACATTTG-3′; E3-F: 5′-TCTCCAGACTGGTGGGAAGCCTCCCTCCCA-3′; E3-R: 5′-GCCCTCTAGATGAAGCAGACCCAGGGGTGG-3′. The underlined sequences are restriction endonuclease sites for HindIII and XbaI. The sequences in bold are homologous sequences for overlap PCR. The PCR products of E1E2 and E3 were assembled together by overlap PCR with E1E2-F and E3-R.
The PCR products of E1E2-F and E3-R were digested and ligated to the pcDNA3.1(+) vector (Invitrogen Corporation, Carlsbad, CA, USA) at the HindIII and XbaI cloning sites. The plasmid constructs containing wild-type, c.374G>A, or c.125A>G variants were examined by PCR and Sanger sequencing. The primers were as follows: pcDNA-F: 5′-CGCAAATGGGCGGTAGGCGTG-3′; WNT10A-F: 5′-CAGGAGTAAGCACAAGCTG-3′; WNT10A-R: 5′-GGGAGGGAGGCTTCCCACCAGTCTGGAGACACATTTG-3′; pcDNA-R: 5′-CCAGGGTCAAGGAAGGCACG-3′.
HEK-293T and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum at 37 °C, with 5% CO2. Two micrograms of pcDNA3.1(+) negative control vector, wild-type minigene construct, c.374G>A minigene construct, and c.125A>G minigene construct were transfected into HEK-293T and HeLa cells with LipofectamineTM 2000 (Invitrogen, CA, USA).
At 48 h after transfection, total RNA was extracted with Trizol reagents (Invitrogen, CA, USA). Reverse transcription was performed with PrimeScript™RT reagent Kit (TaKaRa, Beijing, China). First strand complementary DNA (cDNA) was amplified by PCR and followed by Sanger sequencing. The primers were as follows: forward: 5′-CGTTTAAACTTAAGCTTGCCCGTCAGGGCCTGCGCG-3′; reverse: 5′-TAGAAGGCACAGTCGAGG-3′.
3. Results
3.1. Clinical Report
The proband (III:1, Table 1) of XLHED Family 1 was a two-year-old boy with no history of eruption of primary teeth. We attempted to assess the missing teeth of the patient by panoramic radiograph, but we failed because the patient was too young to cooperate. His hair and eyebrows were sparse. He was hypohidrotic and presented with intolerance to heat. The uncle (II:3) of the proband had 22 missing permanent teeth, as he himself reported. He was an adult without a history of tooth extraction. His teeth were congenitally missing. He also suffered from hypohidrosis and hypotrichosis. A 6-year-old boy (II:1, Table 1) was the only patient of XLHED Family 2. Panoramic radiograph showed that he had 18 missing primary teeth and 23 missing permanent teeth (excluding the third molars) (Figure 1A, Table 2). The shape of the incisors was conic. He was hypohidrotic, and his hair was sparse.

Table 1.
Summary of clinical data and mutations in EDA, EDAR, and WNT10A genes.
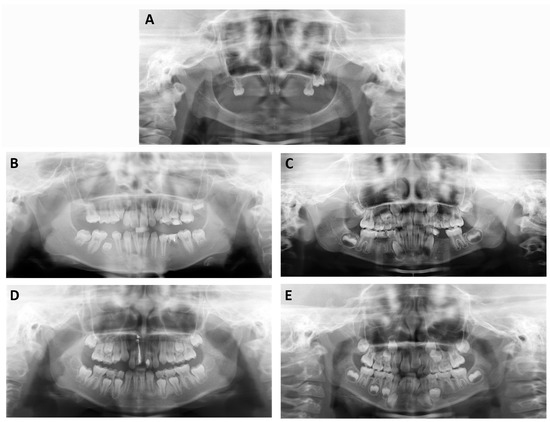
Figure 1.
Panoramic radiograph of the probands of X-linked hypohidrotic ectodermal dysplasia (XLHED) Family 2 (A); non-syndromic tooth agenesis (NSTA) Family 1 (B); NSTA Family 2 (C); NSTA Family 3 (D); and NSTA Family 4 (E). They were missing 23, 4, 9, 2, and 6 permanent teeth, respectively. Because the proband of XLHED1 was too young to cooperate, we were not able to assess the missing teeth of the patient by panoramic radiograph. For the uncompressed Figure 1 file, please download the Supplementary Materials.

Table 2.
Summary of missing permanent teeth of the five probands whose panoramic radiographs are available.
The probands of the four NSTA families presented with congenital loss of teeth, without any other accompanying symptoms. The shape of the teeth was normal. The numbers of missing permanent teeth are listed in Table 1. The diagnosis of tooth agenesis was confirmed by panoramic radiographs (Figure 1B–E, Table 2). We did not evaluate the number of missing primary teeth of the patients because some primary teeth may be replaced by permanent teeth. The pedigrees are reported in Figure 2. There was only one patient per family in NSTA Families 1–3. In NSTA Family 4, the mother of the proband had six congenitally missing teeth.
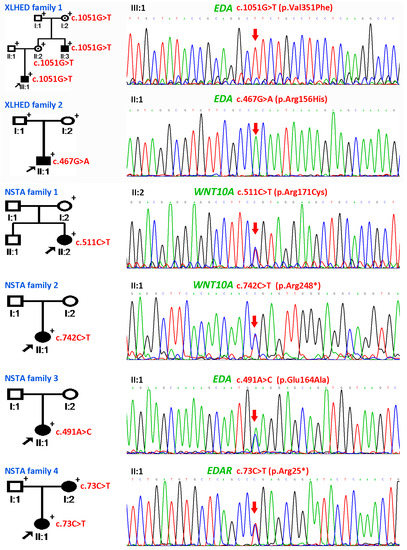
Figure 2.
Pedigrees and mutation information of X-linked hypohidrotic ectodermal dysplasia (XLHED) Families 1–2 and non-syndromic tooth agenesis (NSTA) Families 1–4. In XLHED Family 1, a novel Ectodysplasin A (EDA) mutation c.1051G>T (p.Val351Phe) was cosegregated in the family. In XLHED Family 2, the known de novo EDA mutation c.467G>A (p.Arg156His) was identified in the genome of the proband. In NSTA Family 1, the proband carried the heterozygous Wnt family member 10A (WNT10A) mutation c.511C>T (p.Arg171Cys). In NSTA Family 2, the heterozygous WNT10A mutation c.742C>T (p.Arg248*) was identified in the genome of the proband. In NSTA Family 3, the proband was found to have the heterozygous EDA mutation c.491A>C (p.Glu164Ala). In NSTA Family 4, the proband and her mother shared the EDA receptor (EDAR) mutation c.73C>T (p.Arg25*), while her father was wild-type at this location. Black arrows pointed to the probands. The individuals, whose blood samples were available, are marked by “+”. Red arrows indicate the mutations.
3.2. Genetic Findings of WNT10A, EDA, PAX9, and MSX1
Sequencing of the EDA gene in XLHED Family 1 revealed novel mutation c.1051G>T (p.Val351Phe) shared by the proband and his uncle (Figure 2). His mother and grandmother were asymptomatic heterozygous carriers of the mutation. His father and grandfather were wild-type at c.1051. The mutation was located at the Tumour Necrosis Factor (TNF) homology domain of the EDA protein, and replaced a valine with a phenylalanine at amino acid 351. The mutation was not reported by dbSNP, 1000 Genomes, the Human Gene Mutation Database (public version), or PubMed. A cross-species alignment of the EDA protein showed that p.Val351 was conserved (Figure 3A). SIFT, PolyPhen2, and the Mutation Taster predicted the mutation as being “damaging”, “probably damaging”, and “disease causing”, respectively. Structural modeling of the p.Val351Phe-mutated TNF homology domain of the EDA protein showed that the mutation was located on the surface of the three monomers (Figure 4A). Compared with the wild-type p.Val351 (Figure 4B), the mutated p.351Phe was too large and extruded with p.Val250 of the adjacent monomer (Figure 4C).

Figure 3.
A cross-species alignment of amino acid sequence of Ectodysplasin A (EDA) showed that p.Val351 was conserved.
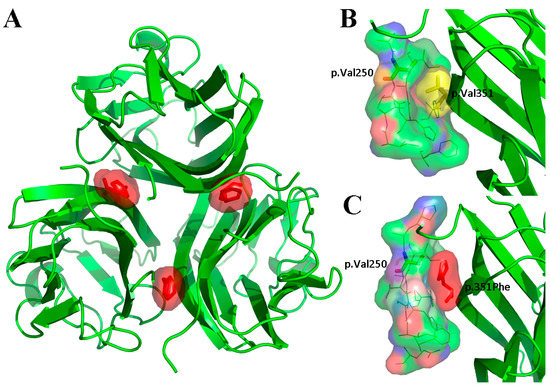
Figure 4.
Structural modeling results of the Tumour Necrosis Factor (TNF) homology domain of p.Val351Phe-mutated (A, C) and wild-type (B) Ectodysplasin A (EDA) proteins. (A) The p.Val351Phe mutation was located on the monomer–monomer interaction surface of the three monomers. (B) In the wild-type EDA protein, there was a small gap between p.Val351 and p.Val250 of the adjacent monomer. (C) In the p.Val351Phe-mutated EDA protein, the mutated p.351Phe was too large and extruded with p.Val250 of the adjacent monomer.
Screening of EDA in XLHED Family 2 showed a known de novo mutation c.467G>A (p.Arg156His) in the genome of the patient (Figure 2). The parents of the patient were normal and did not carry the mutation. The mutation c.467G>A is one of the most prevalent mutations found in patients with XLHED [43].
We screened for mutation of WNT10A and EDA genes in the probands of four NSTA families. In the genome of the proband of NSTA Family 1, the known mutation c.511C>T (p.Arg171Cys) and the variant c.374G>A (p.Arg125Lys) were identified in the WNT10A gene (Figure 2). The mother of the proband carried a c.374G>A variant but was free of the c.511C>T mutation. In Family 2, the known heterozygous WNT10A mutation c.742C>T (p.Arg248*) was identified in the genome of the proband (Figure 2). Blood samples of the parents were not available. In Family 3, we found the known heterozygous EDA mutation c.491A>C (p.Glu164Ala) and the heterozygous WNT10A variant c.125A>G (p.Asn42Ser, rs149865858) in the genome of the proband (Figure 2). The parents of the proband were not willing to participate in this study, so we were not able to analyze the genotype of the parents.
Because we did not identify any mutations of WNT10A and EDA genes in Family 4, we performed Sanger sequencing for the PAX9 and MSX1 genes. The proband of Family 4 was negative for PAX9 and MSX1 mutation.
3.3. Results of Whole Exome Sequencing
Subsequently, the proband of Family 4 was assigned for whole exome sequencing. In total, 27,013,053 reads were obtained. The average coverage of the target sequences was 64.18X. All in all, 94.9% of the target sequences were covered with at least 20X. There were 64,338 SNPs and 9797 indels identified. The variants of EDAR, EDARADD, AXIN2, WNT10B, LRP6, and LTBP3 genes were called, and we obtained 43 SNPs and 3 indels. After filtering with “minor allele frequency (MAF) < 1%” and “exonic”, we obtained the novel EDAR mutation c.73C>T (p.Arg25*). This mutation was confirmed by Sanger sequencing. The mother of the proband carried the mutation, while the father was wild-type at the locus (Figure 2). The mutation induced a termination codon at the amino acid 25 of the encoded protein. The mutated protein was unlikely to have any function as the majority of the protein was truncated (Figure 5). This mutation was not reported in 1000 Genomes, the Human Gene Mutation Database (HGMD; public version), or PubMed.
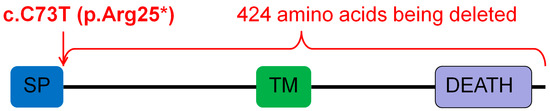
Figure 5.
Ectodysplasin A receptor (EDAR) mutation c.73C>T (p.Arg25*) introduced a stop codon at amino acid 25 of the encoded protein, truncating 424 amino acids of the protein. SP: signal peptide; TM: transmembrane domain; DEATH: death domain.
3.4. Bioinformatics Analyses and Minigene Results of WNT10A Variants c.374G>A and c.125A>G
We identified WNT10A variants c.374G>A in NSTA Family 1, and c.125A>G in NSTA Family 3, as described in Section 3.2. Some analyses and experiments were done to find out whether they were pathogenic or not.
The WNT10A variant c.374G>A (p.Arg125Lys) was not reported in 1000 Genomes, the Human Gene Mutation Database (HGMD; public version), or PubMed. A cross-species alignment of the amino acid sequence of WNT10A showed that p.Arg125 was conserved (Figure 6A). The variant led to a substitution of an arginine to a lysine at the position. It was predicted as being “benign” by PolyPhen 2, and “tolerated” by SIFT. Mutation Taster predicted the mutation as being “disease causing” because it changed the amino acid sequence, and it may disrupt the splicing of the second exon with the third exon. Human Splicing Finder (HSF) also predicted that the mutation could affect the splicing of mRNA by creating a new exonic splicing silencer and breaking down an exonic splicing enhancer. Structural modeling results showed that there was a hydrogen bond between p.Arg125 and p.Glu340 in the wild-type WNT10A protein (Figure 7A). This hydrogen bond vanished in the p.Arg125Lys-mutated WNT10A protein, and a new hydrogen bond between p.125Lys and p.Leu352 arose (Figure 7B).
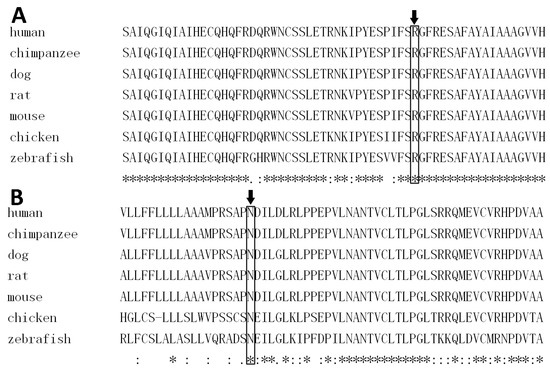
Figure 6.
A cross-species alignment of amino acid sequence of Wnt family member 10A (WNT10A) indicated that p.Arg125 (A) and p.Asn42 (B) were conserved.
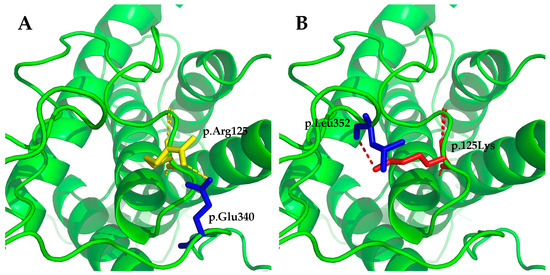
Figure 7.
Structural modeling results of the wild-type Wnt family member 10A (WNT10A) protein and the p.Arg125Lys-mutated WNT10A protein. (A) There was a hydrogen bond between p.Arg125 and p.Glu340 in the wild-type WNT10A protein. (B) The original hydrogen bond vanished in the p.Arg125Lys-mutated WNT10A protein, and a new hydrogen bond between p.125Lys and p.Leu352 arose. The changes may lead to instability of the structure and affect the affinity to the receptor of WNT10A.
The c.125A>G mutation occurred at the evolutionary conserved p.Asn42 (Figure 6B) and changed the amino acid to serine. It was predicted to be “tolerated” by SIFT. However, PolyPhen 2 predicted it as being “probably damaging”, and Mutation Taster predicted it as being “disease causing”. The splicing of Exons 1 and 2 may be affected by the mutation, predicted by Mutation Taster and HSF. This mutation was not reported by the HGMD public version or PubMed, but the 1000 Genomes database recorded three heterozygous allele carriers of the WNT10A mutation c.125A>G. Structural modeling for the p.Asn42Ser-mutated WNT10A protein was not feasible due to the fact that the homologous template 4F0A does not cover p.Asn42.
A minigene splicing assay was used to analyze the mRNA-splicing effect of WNT10A variants c.374G>A and c.125A>G. The minigenes, containing wild-type, c.374G>A, or c.125A>G of WNT10A Exons 1–3, were cloned into the pcDNA3.1 plasmid. HeLa and HEK-293T cells were used to investigate the splicing effect of the variants. As shown in Figure 8A, no abnormal splicing product was found. Sequencing results of the splicing products showed normal splicing of the two minigenes, which contained variant c.374G>A or c.125A>G (Figure 8B–D).
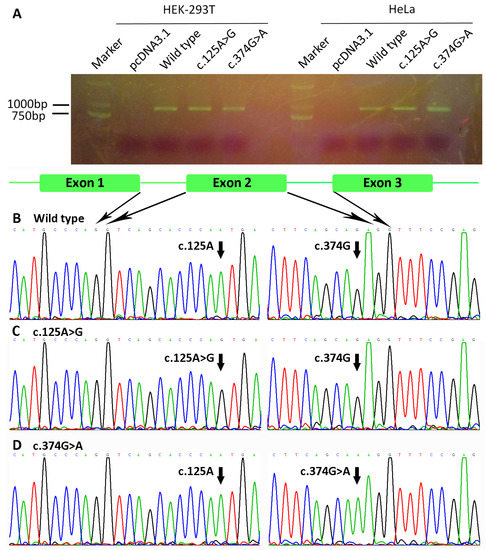
Figure 8.
Minigene splicing assay of Wnt family member 10A (WNT10A) variants c.374G>A and c.125A>G. (A) Agarose gel electrophoresis of PCR products. No abnormal splicing product was found in either HeLa or HEK-293T cells. (B–D) Sequencing results of (B) the wild-type minigene, (C) the c.125A>G minigene, and (D) the c.374G>A minigene. The minigenes containing variant c.374G>A or c.125A>G spliced normally.
3.5. Sequencing Results of PAX9, MSX1, EDAR, EDARADD, AXIN2, LRP6, and WNT10B in NSTA Families 1 and 3
To rule out the possibility of mutations in other NSTA candidate genes, we sequenced PAX9, MSX1, EDAR, EDARADD, AXIN2, LRP6, and WNT10B genes of the probands in NSTA Families 1 and 3. No pathogenic mutation was found.
4. Discussion
This article represented a comprehensive genetic study of XLHED and NSTA. Sanger sequencing of nine candidate genes and whole exome sequencing were performed, and the genetic defects of six families with XLHED or NSTA were identified. EDA mutation c.1051G>T (p.Val351Phe) and EDAR mutation c.73C>T (p.Arg25*) were novel. Bioinformatics analyses and structural modeling were used to evaluate the pathogenicity of the mutations. A minigene splicing assay was performed, and the results showed that WNT10A variants c.374G>A and c.125A>G did not affect the splicing of mRNA.
Mues et al. revealed that NSTA-related EDA mutations only impaired the receptor binding capability of mutant EDA proteins, while STA-related EDA mutations abolish the receptor binding capability of mutant EDA proteins [44]. This supports that theory that the development of the human dentition requires the highest level of EDA-receptor signaling, whereas other ectodermal appendages have less stringent requirements for EDA-receptor signaling [44]. In a previous study, we identified the EDA mutation c.776C>A (p.Ala259Glu) in a patient with XLHED [29]. Interestingly, the same mutation was also identified in patients with NSTA [45]. We proposed that that XLHED and EDA-related NTA are the same disease with different degrees of severity [29]. In this study, we found the heterozygous WNT10A mutation c.742C>T (p.Arg248*) in NSTA Family 2. This mutation in homozygous status also was reported in a patient with STA (HED) [27]. Vink et al. reported compound heterozygous WNT10A mutations c.682T>A (p.Phe228Ile) and c.321C>A (p.Cys107*) in a patient with STA (OODD), while the patient’s mother carried heterozygous WNT10A mutations c.682T>A (p.Phe228Ile) and suffered from NSTA [33]. The data indicated that WNT10A-related STA (HED or OODD) and WNT10A-related NSTA are the same disease with phenotypic variability. Based on the data of our group and the results of other researchers, we think some cases of STA and NSTA, which are caused by mutations of the same gene, are examples of the same disease with phenotypic variability. We predict that there will be more cases of the same mutations found in both patients with STA and NSTA.
In most of the previous studies, EDAR mutations were found to be the cause of hypohidrotic ectodermal dysplasia [27,46,47]. There have only been two works reporting EDAR mutations in patients with NSTA [12,48]. However, all of the EDAR mutations in these two studies were missense mutations. In our study, a novel nonsense EDAR mutation c.73C>T (p.Arg25*) was identified in NSTA Family 4. This mutation truncated 424 amino acids of the EDAR protein. The mutated protein lost its transmembrane domain and death domain. Therefore, the mutated protein is not expected to have any function. This is the first study of a nonsense EDAR mutation leading to the onset of NSTA.
There are three functional domains of the EDA protein, namely a furin protease recognition sequence, a collagen domain, and a C-terminal TNF homology domain [49]. The TNF homology domain forms homotrimers, which can bind with EDAR [50]. In this study, the novel mutation c.1051G>T (p.Val351Phe) was located on the surface of monomers. Structural modeling results showed that the mutation probably hampers the monomer–monomer interaction. The trimer may become globally instable. [51] Additionally, because the monomer–monomer interface is the core region for EDA–EDAR contact and signal transduction, the mutated EDA protein may lose its specificity to EDAR. [51] Bioinformatics analyses results, including software prediction, amino acid conservation analysis, and a public population genetic database, also indicated the deleterious nature of the mutation.
In NSTA Families 1 and 3, apart from the mutations in EDA (c.491A>C) and WNT10A (c.511C>T), we also found WNT10A variants c.374G>A and c.125A>G. The WNT10A mutation c.511C>T (p.Arg171Cys) was a pathogenic mutation reported in patients with tooth agenesis [52]. The EDA mutation c.491A>C (p.Glu164Ala) in NSTA Family 3 was an established pathogenic mutation, which is responsible for hypodontia and X-linked hypohidrotic ectodermal dysplasia [53]. Results of prediction using some software (SIFT and PolyPhen 2) did not support that the two WNT10A variants (c.374G>A and c.125A>G) were pathogenic. However, the Mutation Taster and Human Splicing Finder predicted that the two variants may affect the splicing of mRNA. Hence, minigene splicing experiments were performed. The results showed that the minigenes containing WNT10A variants c.374G>A or c.125A>G can splice normally. Hence, we consider that the two WNT10A variants were not pathogenic and did not play a role in the onset of the disease. The patient of NSTA Family 1 is caused by WNT10A mutation c.511C>T (p.Arg171Cys), and the patient of NSTA Family 3 is caused by EDA mutation c.491A>C (p.Glu164Ala).
There are 10 known causal genes of NSTA (WNT10A, EDA, PAX9, MSX1, AXIN2, EDAR, EDARADD, WNT10B, LRP6, and LTBP3) [10,12,17,20]. Screening all of the candidate genes for pathogenic mutations by Sanger sequencing would imply high financial costs and time requirements. Hence, we performed the experiments in a more efficient and economical way. In the first step, we screened for two of the most frequently mutated genes (WNT10A and EDA). If no pathogenic mutation was found, then we screened for two additional frequently mutated genes (PAX9 and MSX1). If still no pathogenic mutation was found, we applied whole exome sequencing to detect mutations of other six candidate genes. This strategy worked in the four probands. Two of the probands had mutations in the WNT10A gene, one of the probands had a mutation in the EDA gene, and the last proband had a mutation in the EDAR gene. To verify our findings and rule out the possibility of mutations in undetected genes, we screened seven more known candidate genes (PAX9, MSX1, AXIN2, EDAR, EDARADD, WNT10B, and LRP6) in NSTA Families 1 and 3. We did not find any pathogenic mutation in the exons and flanking regions. We did not check for mutations in LTBP3 because patients with LTBP3 mutations are of short stature [54], and the patients in this study were of normal stature. We did not screen the seven genes in NSTA Family 2 because the patient had a nonsense WNT10A mutation c.742C>T (p.Arg248*), which is pathogenic through the truncation of the C-terminal of the WNT10A protein.
5. Conclusions
In this study, we identified three EDA, one EDAR and two WNT10A mutations in two patients with X-linked hypohidrotic ectodermal dysplasia (XLHED) and four patients with non-syndromic tooth agenesis (NSTA). EDA mutation c.1051G>T (p.Val351Phe) and EDAR mutation c.73C>T (p.Arg25*) were novel. For the first time, these two novel mutations are hereby proven to be pathogenic. We have here reported the first nonsense EDAR mutation leading to the onset of NSTA. These results expand the mutational spectrum of WNT10A and EDAR genes and contribute to the practice of precision medicine.
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/8/10/259/s1. Table S1. Primers amplifying coding exons and flanking intronic sequences of PAX9; Table S2. Primers amplifying coding exons and flanking intronic sequences of MSX1; Table S3. Primers amplifying coding exons and flanking intronic sequences of AXIN2; Table S4. Primers amplifying coding exons and flanking intronic sequences of LRP6; Table S5. Primers amplifying coding exons and flanking intronic sequences of WNT10B.
Acknowledgments
This project was funded by the National Natural Sciences Foundation of China (81472526), the Natural Science Foundation of Guangdong Province (2014A030313126) and the Science and Technology Planning Project of Guangdong Province (2016A020215094 and 2016A020216007).
Author Contributions
D.Y. and W.Z. conceived and designed the experiments; B.Z., Q.Z., H.L., and J.L. performed the experiments; B.Z., S.L., and L.M. analyzed the data; D.Y. and W.Z. contributed reagents and materials; B.Z. and W.Z. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rakhshan, V.; Rakhshan, H. Meta-analysis of congenitally missing teeth in the permanent dentition: Prevalence, variations across ethnicities, regions and time. Int. Orthod. 2015, 13, 261–273. [Google Scholar] [CrossRef] [PubMed]
- De Coster, P.J.; Marks, L.A.; Martens, L.C.; Huysseune, A. Dental agenesis: Genetic and clinical perspectives. J. Oral Pathol. Med. 2009, 38, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Qin, C.; Yue, H.; He, H.; Bian, Z. A novel initiation codon mutation of PAX9 in a family with oligodontia. Arch. Oral Biol. 2016, 61, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Pagnan, N.A.; Visinoni, A.F. Update on ectodermal dysplasias clinical classification. Am. J. Med. Genet. A 2014, 164A, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chu, E.Y.; Watt, B.; Zhang, Y.; Gallant, N.M.; Andl, T.; Yang, S.H.; Lu, M.; Piccolo, S.; Schmidt-Ullrich, R.; et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 2008, 313, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, T.; Zheng, L.; Shitaku, Y.; Saito, M.; Tsubakimoto, T.; Takada, K.; Takano-Yamamoto, T.; Thesleff, I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation 2007, 75, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, P.; Liu, Y.; Zhang, X.; Fu, J.; Yu, H.M.I.; Qiu, M.; Chen, Y.; Hsu, W.; Zhang, Z. Intra-epithelial Requirement of Canonical Wnt Signaling for Tooth Morphogenesis. J. Biol. Chem. 2013, 288, 12080–12089. [Google Scholar] [CrossRef] [PubMed]
- van den Boogaard, M.; Creton, M.; Bronkhorst, Y.; van der Hout, A.; Hennekam, E.; Lindhout, D.; Cune, M.; van Amstel, H.K.P. Mutations in WNT10A are present in more than half of isolated hypodontia cases. J. Med. Genet. 2012, 49, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Mostowska, A.; Biedziak, B.; Zadurska, M.; Dunin-Wilczynska, I.; Lianeri, M.; Jagodzinski, P.P. Nucleotide variants of genes encoding components of the Wnt signalling pathway and the risk of non-syndromic tooth agenesis. Clin. Genet. 2013, 84, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Arzoo, P.S.; Klar, J.; Bergendal, B.; Norderyd, J.; Dahl, N. WNT10A mutations account for (1/4) of population-based isolated oligodontia and show phenotypic correlations. Am. J. Med. Genet. A 2014, 164A, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, C.; Jung, S.; Niederreither, K.; Prasad, M.; Hadj-Rabia, S.; Philip, N.; Mallet, A.; Consolino, E.; Sfeir, E.; Noueiri, B.; et al. Dental and extra-oral clinical features in 41 patients with, WNT10A gene mutations: A multicentric genotype-phenotype study. Clin. Genet. 2017. [Google Scholar] [CrossRef] [PubMed]
- Arte, S.; Parmanen, S.; Pirinen, S.; Alaluusua, S.; Nieminen, P. Candidate Gene Analysis of Tooth Agenesis Identifies Novel Mutations in Six Genes and Suggests Significant Role for WNT and EDA Signaling and Allele Combinations. PLoS ONE 2013, 8, e73705. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Han, D.; Feng, H.; Qu, H.; Song, S.; Bai, B.; Zhang, Z. Involvement of and interaction between WNT10A and EDA mutations in tooth agenesis cases in the Chinese population. PLoS ONE 2013, 8, e80393. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.S.; Headon, D.J.; Schneider, P.; Ferguson, B.M.; Overbeek, P.; Tschopp, J.; Sharpe, P.T. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development 2000, 127, 4691–4700. [Google Scholar] [PubMed]
- Peters, H.; Neubueser, A.; Kratochwil, K.; Balling, R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Satokata, I.; Maas, R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 1994, 6, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Bian, Z. The Gene Network Underlying Hypodontia. J. Dent. Res. 2015, 94, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Ruf, S.; Klimas, D.; Hoenemann, M.; Jabir, S. Genetic background of nonsyndromic oligodontia: A systematic review and meta-analysis. J. Orofac. Orthop. 2013, 74, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Bergendal, B.; Klar, J.; Stecksen-Blicks, C.; Norderyd, J.; Dahl, N. Isolated oligodontia associated with mutations in EDARADD, AXIN2, MSX1 and PAX9 genes. Am. J. Med. Genet. A 2011, 155A, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Yang, W.; Han, D.; Wang, X.; Guo, S.; Li, J.; Li, F.; Zhang, X.; Wong, S.; Bai, B.; et al. Mutations in WNT10B Are Identified in Individuals with Oligodontia. Am. J. Hum. Genet. 2016, 99, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gaczkowska, A.; Abdalla, E.M.; Dowidar, K.M.; Elhady, G.M.; Jagodzinski, P.P.; Mostowska, A. De novo EDA mutations: Variable expression in two Egyptian families. Arch. Oral Biol. 2016, 68, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Visinoni, A.F.; Lisboa-Costa, T.; Pagnan, N.A.; Chautard-Freire-Maia, E.A. Ectodermal dysplasias: Clinical and molecular review. Am. J. Med. Genet. A 2009, 149A, 1980–2002. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xie, L. Novel nonsense mutation of the EDA gene in a Chinese family with X-linked hypohidrotic ectodermal dysplasia. J. Dermatol. 2014, 41, 1014–1016. [Google Scholar] [PubMed]
- Zeng, B.; Xiao, X.; Li, S.; Lu, H.; Lu, J.; Zhu, L.; Yu, D.; Zhao, W. Eight Mutations of Three Genes (EDA, EDAR, and WNT10A) Identified in Seven Hypohidrotic Ectodermal Dysplasia Patients. Genes 2016, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Monreal, A.W.; Ferguson, B.M.; Headon, D.J.; Street, S.L.; Overbeek, P.A.; Zonana, J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat. Genet. 1999, 22, 366–369. [Google Scholar] [PubMed]
- Headon, D.J.; Emmal, S.A.; Ferguson, B.M.; Tucker, A.S.; Justice, M.J.; Sharpe, P.T.; Zonana, J.; Overbeek, P.A. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature 2001, 414, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Cluzeau, C.; Hadj-Rabia, S.; Jambou, M.; Mansour, S.; Guigue, P.; Masmoudi, S.; Bal, E.; Chassaing, N.; Vincent, M.C.; Viot, G.; et al. Only four genes (EDA1, EDAR, EDARADD, and, WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum. Mutat. 2011, 32, 70–72. [Google Scholar] [CrossRef] [PubMed]
- Adaimy, L.; Chouery, E.; Megarbane, H.; Mroueh, S.; Delague, V.; Nicolas, E.; Belguith, H.; de Mazancourt, P.; Megarbane, A. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: The odonto-onycho-dermal dysplasia. Am. J. Hum. Genet. 2007, 81, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Lu, H.; Xiao, X.; Zhou, L.; Lu, J.; Zhu, L.; Yu, D.; Zhao, W. Novel EDA mutation in X-linked hypohidrotic ectodermal dysplasia and genotype-phenotype correlation. Oral Dis. 2015, 21, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.E.; Ko, J.; Shin, T.J.; Hyun, H.K.; Lee, S.H.; Kim, J.W. Oligodontia and curly hair occur with ectodysplasin-a mutations. J. Dent. Res. 2014, 93, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Guazzarotti, L.; Tadini, G.; Mancini, G.E.; Giglio, S.; Willoughby, C.E.; Callea, M.; Sani, I.; Nannini, P.; Mameli, C.; Tenconi, A.A.; et al. Phenotypic heterogeneity and mutational spectrum in a cohort of 45 Italian males subjects with, X-linked ectodermal dysplasia. Clin. Genet. 2015, 87, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Bohring, A.; Stamm, T.; Spaich, C.; Haase, C.; Spree, K.; Hehr, U.; Hoffmann, M.; Ledig, S.; Sel, S.; Wieacker, P.; et al. WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am. J. Hum. Genet. 2009, 85, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Vink, C.P.; Ockeloen, C.W.; Ten, K.S.; Koolen, D.A.; Ploos, V.A.J.; Kuijpers-Jagtman, A.M.; van Heumen, C.C.; Kleefstra, T.; Carels, C.E. Variability in dentofacial phenotypes in four families with WNT10A mutations. Eur. J. Hum. Genet. 2014, 22, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e603. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Desmet, F.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.C.; Biancalana, V.; Ginisty, D.; Mandel, J.L.; Calvas, P. Mutational spectrum of the ED1 gene in X-linked hypohidrotic ectodermal dysplasia. Eur. J. Hum. Genet. 2001, 9, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Mues, G.; Tardivel, A.; Willen, L.; Kapadia, H.; Seaman, R.; Frazier-Bowers, S.; Schneider, P.; D’Souza, R.N. Functional analysis of Ectodysplasin-A mutations causing selective tooth agenesis. Eur. J. Hum. Genet. 2010, 18, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Han, D.; Qu, H.; Gong, Y.; Wu, H.; Zhang, X.; Zhong, N.; Feng, H. EDA gene mutations underlie non-syndromic oligodontia. J. Dent. Res. 2009, 88, 126–131. [Google Scholar] [CrossRef] [PubMed]
- van der Hout, A.H.; Oudesluijs, G.G.; Venema, A.; Verheij, J.B.; Mol, B.G.; Rump, P.; Brunner, H.G.; Vos, Y.J.; van Essen, A.J. Mutation screening of the Ectodysplasin-A receptor gene EDAR in hypohidrotic ectodermal dysplasia. Eur. J. Hum. Genet. 2008, 16, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Trzeciak, W.H.; Koczorowski, R. Molecular basis of hypohidrotic ectodermal dysplasia: An update. J. Appl. Genet. 2016, 57, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Hosomichi, K.; Yano, K.; Kim, Y.; Nakaoka, H.; Kimura, R.; Otsuka, H.; Nonaka, N.; Haga, S.; Takahashi, M.; et al. Comprehensive genetic exploration of selective tooth agenesis of mandibular incisors by exome sequencing. Hum. Genome Var. 2017, 4, 17005. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Street, S.L.; Gaide, O.; Hertig, S.; Tardivel, A.; Tschopp, J.; Runkel, L.; Alevizopoulos, K.; Ferguson, B.M.; Zonana, J. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J. Biol. Chem. 2001, 276, 18819–18827. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, J.L.; Schneider, P.; Tschopp, J. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 2002, 27, 19–26. [Google Scholar] [CrossRef]
- Hymowitz, S.G.; Compaan, D.M.; Yan, M.; Wallweber, H.J.; Dixit, V.M.; Starovasnik, M.A.; de Vos, A.M. The crystal structures of EDA-A1 and EDA-A2: Splice variants with distinct receptor specificity. Structure 2003, 11, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Mues, G.; Bonds, J.; Xiang, L.; Vieira, A.R.; Seymen, F.; Klein, O.; D’Souza, R.N. The WNT10A gene in ectodermal dysplasias and selective tooth agenesis. Am. J. Med. Genet. A 2014, 164A, 2455–2460. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Ye, X.; Shi, L.; Yin, W.; Hua, B.; Song, G.; Shi, B.; Bian, Z. Mutations in the EDA gene are responsible for X-linked hypohidrotic ectodermal dysplasia and hypodontia in Chinese kindreds. Eur. J. Oral Sci. 2008, 116, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.; Windpassinger, C.; Vitcu, I.; Orlic, M.; Rafiq, M.A.; Khalid, M.; Malik, M.N.; Ayub, M.; Alman, B.; Vincent, J.B. Oligodontia Is Caused by Mutation in LTBP3, the Gene Encoding Latent TGF-bet Binding Protein 3. Am. J. Hum. Genet. 2009, 84, 519–523. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).