Abstract
Pericentral retinitis pigmentosa (RP) is an atypical form of RP that affects the near-peripheral retina first and tends to spare the far periphery. This study was performed to further define the genetic basis of this phenotype. We identified a cohort of 43 probands with pericentral RP based on a comprehensive analysis of their retinal phenotype. Genetic analyses of DNA samples from these patients were performed using panel-based next-generation sequencing, copy number variations, and whole exome sequencing (WES). Mutations provisionally responsible for disease were found in 19 of the 43 families (44%) analyzed. These include mutations in RHO (five patients), USH2A (four patients), and PDE6B (two patients). Of 28 putatively pathogenic alleles, 15 (54%) have been previously identified in patients with more common forms of typical RP, while the remaining 13 mutations (46%) were novel. Burden testing of WES data successfully identified HGSNAT as a cause of pericentral RP in at least two patients, suggesting it is also a relatively common cause of pericentral RP. While additional sequencing might uncover new genes specifically associated with pericentral RP, the current results suggest that genetically pericentral RP is not a separate clinical entity, but rather is part of the spectrum of mild RP phenotypes.
1. Introduction
Retinitis pigmentosa (RP) is well known for being a genetically heterogeneous disease, with mutations in at least 89 different genes known to cause nonsyndromic RP alone [1]. As mutations in so many different genes can cause essentially the same phenotype, this makes RP one of the most genetically heterogeneous diseases in humans. However, while RP is usually characterized by typical “bone-spicule” pigmentation and photoreceptor degeneration beginning in the mid-peripheral retina [2,3,4], it would be an oversimplification to say that all RP phenotypes are the same; several subtypes of RP have been clinically defined, including pericentral RP, sector RP, pigmented paravenous RP, and RP without pigment [5,6,7,8]. It remains to be determined to what extent these clinical subtypes stem from different genetic causes, or whether they are, for example, a reflection of variable expressivity of phenotypes due to mutations in the same set of genes [3]. This study aimed to address this question by expanding the search for genetic causes of a particular subtype of RP—pericentral RP. We further hypothesized that by studying a cohort of patients with an atypical phenotype, it would increase the possibility of uncovering new biological pathways or genes involved in RP.
Pericentral RP has been described as a subtype of RP in which, instead of the pathology starting in the mid-periphery like typical RP, the disease starts in the near periphery closer to the vascular arcades and tends to spare the far periphery. As this is a clinically defined phenotype, the definition of pericentral RP and even the name of the condition can vary somewhat between authors. Similar phenotypes have been described as: pericentral pigmentary retinopathy [9,10,11], pericentral pigmentary retinal degeneration [12,13,14,15], pericentral retinal dystrophy [16], peripapillary retinal degeneration [14,17], perivascular retinal pigment epithelium atrophy [18], pericentral retinal degeneration [19], RP with perimacular or paramacular pattern [2], and pericentral RP [20,21,22]. A natural history study from our institution defined the phenotype as bone-spicule pigmentation or atrophy in the near mid-periphery corresponding to an annular scotoma from 5–30 degrees, a normal or nearly normal dark adaptation threshold, and subnormal but readily detectable full-field electroretinogram (ERG) responses [22].
The genetic causes of pericentral RP have not been fully defined to date. The pericentral RP phenotype has been found in both dominant and recessive pedigrees [10,16,18,19]. Three families with pericentral phenotypes were found to have mutations in rhodopsin (RHO) [23]. Further studies, specifically of the phenotypes of patients with RHO-associated RP, revealed one subset with pericentral defects [24]. Mutations in the TOPORS gene have been reported to cause a pericentral RP-like phenotype in two pedigrees [18,25]. In those studies, the phenotype was termed “pericentral retinal dystrophy” or “RP with perivascular retinal pigment epithelium atrophy” [18,25]. Another study reported two patients with TOPORS mutations that caused typical autosomal dominant RP without the presence of a perivascular cuff of retinal pigment epithelium atrophy [26]. Further putative mutations in TOPORS in a panel of RP patients have been reported [27]. The largest study of pericentral RP genetics to date identified the molecular cause of disease in 14 of 28 pericentral cases [19]. The most commonly identified genes among the 14 solved cases were ABCA4 with 5 cases and CERKL with 2 cases. The authors concluded that there was molecular heterogeneity in the pericentral phenotype, making it an uncommon phenotype composed of many different genotypes. They noted the genes involved have also been associated with other phenotypes, such as maculopathies and typical RP.
One non-genetic cause of a pericentral RP-like phenotype is a form of hydroxychloroquine (Plaquenil) toxicity; the pericentral pattern of pathology (as opposed to the usual perifoveal type) is more common in Asian patients [28,29]. None of the patients described in this study are known to have a history of hydroxychloroquine use.
We performed panel-based sequencing and whole exome sequencing (WES) of a larger cohort of patients with pericentral RP to better define the genetic causes of this phenotype. In addition to extending the observation that the genetics of pericentral RP show a diversity of genotypes, the results obtained from this study indicate that the genetic causes of pericentral RP are similar to those that can cause mild versions of typical RP. Using WES, HGSNAT was also identified as a recurring cause of pericentral RP.
2. Materials and Methods
2.1. Cohort
We identified a cohort of 45 probands with pericentral RP based on a comprehensive analysis of their retinal phenotype. They underwent a comprehensive evaluation in the Electroretinography Service at the Massachusetts Eye and Ear Infirmary, including best corrected visual acuity, Goldmann visual field tests with V4e and I4e test lights, final dark-adapted threshold with an 11 degree test light in the Goldmann-Weekers dark adaptometer, and full-field ERG. ERG responses included a white light mixed response measured at 0.5 Hz and a cone flicker response measured at 30 Hz, as previously described [30]. Inclusion in the cohort was determined by the clinical diagnosis of an experienced physician (E.L. Berson, Massachusetts Eye and Ear Infirmary, Massachusetts, MA, USA) based on the above testing. In summary, factors included bone-spicule pigmentation or atrophy in the near mid-periphery (5–30 degrees) corresponding to an annular scotoma, a normal or nearly normal dark adaptation threshold, subnormal but readily detectable full-field ERG responses, and a healthy-appearing anterior retina [22]. Not all findings were present in every patient. The phenotype has been described in depth in previous studies [19,22]. For further references please see the introduction.
Blood samples were collected for leukocyte DNA. DNA samples were collected from 45 probands, along with affected and unaffected relatives when available, for a total of 61 samples. To the best of our knowledge, none of the probands were related. The study protocol adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Board (Human Studies Committee Protocol 11-057H, approved 2012-present) of the Massachusetts Eye and Ear Infirmary.
2.2. Panel-Based Exon Sequencing
A custom SureSelect targeted enrichment kit (Agilent, Santa Clara, CA, USA) was designed to capture and analyze the coding regions and untranslated regions (UTRs) of 196 genes known to cause inherited eye disease [31]. Later versions tested 226 genes. Libraries were generated using standard methods [31]. This Genetic Eye Disease (GEDi) panel was analyzed with a MiSeq platform (Illumina, San Diego, CA, USA) using 2 × 121 bp reads, multiplexing 9 to 12 samples per run.
Panel-based sequencing data was analyzed as previously described [32,33]. Briefly, sequences were aligned with BWA [34], and SAMtools [35] was used for duplicate removal and variant detection. Variants were annotated and filtered using internal data as well as publicly-available sources. Variant types that were considered included nonsynonymous changes (i.e., protein sequence altering changes), splice-site changes, or variants previously described as disease-causing in the Human Gene Mutation Database (HGMD) [36] or Clinvar [37]. An in-house, web-based variant browsing tool aided browsing and collation of results. Initial allele frequency cutoffs were set at 1/10,000 for dominant disease and 1/700 for recessive disease based on similar values for the most common alleles that cause RP. Standardized variant names were validated using the online tool Mutalyzer (https://mutalyzer.nl/).
2.3. Sanger Validation
Mutations detected by next-generation sequencing (NGS) were sequenced using Sanger sequencing. Genomic regions of interest were PCR amplified with optimization of cycling temperatures, purified (ExoSap-IT, Affymetrix, Santa Clara, CA, USA), and sequenced (BigDye Terminator v3.1, ABI 3730xl, Life Technologies, Grand Island, NY, USA).
2.4. Copy Number Variation (CNV) Analysis
Nine patients without disease-causing mutations identified using targeted exon sequencing then underwent CNV analysis using Omni2.5 chips (Illumina) according to manufacturer instructions. Regions with potential deletions or duplications were identified using CNV Workshop [38].
2.5. Whole Exome Sequencing and Burden Test
Targeted enrichment was performed using the SureSelect XT Human All Exon + UTR v5 baits (Agilent). Libraries were sequenced on an Illumina HiSeq platform using 2 × 101 bp reads in a 16-sample multiplex. Variants were identified using the pipeline described above for panel-based sequencing. Variants of interest were also validated using a pipeline based on BWA alignments and Genome Analysis Toolkit (GATK) joint variant calling, following Broad Institute best practices [39]. An automated gene “burden” analysis [40] was conducted to detect genes where predicted loss-of-function mutations are overrepresented in the unsolved pericentral cohort compared to all other exomes run at our institution. The pericentral cohort consists of 16 samples that were not initially solved using panel-based sequencing (except #24, which had both panel and exome sequencing). The test looks for overrepresentation, on a gene-by-gene basis, of damaging variants in the 16 unsolved samples, as compared to the remainder of the WES sample cohort. A variant was considered “damaging” if it was an exonic, non-synonymous variant which is: either a frameshift insertion or deletion (i.e., length not divisible by 3), a stop-gain mutation, a splice variant at ±1 or ±2 locations, or predicted as damaging by either PolyPhen [41] or SIFT [42]. Synonymous and UTR variants were not included. Variants seen >20 times in the entire exome dataset were discarded as too common to be a cause of a rare Mendelian disease. The number of variants meeting these conditions was counted on an allelic basis in each sample (e.g., autosomal homozygous calls counted as 2, heterozygotes as 1). First, a recessive model was applied, which required a damaging homozygous or compound heterozygous variant for a sample to be counted. Separately, a dominant model was applied where only one damaging heterozygous or homozygous variant had to be present to be counted. Then, for each gene in the dataset, a right-sided Fisher test was computed to assess the overrepresentation of samples with damaging variants in the pericentral cohort versus all other exomes. All genes were then ranked by their resultant Fisher scores to identify genes whose damaging variants have maximal over-representation in the pericentral cohort.
3. Results
3.1. Cohort
Baseline clinical characteristics are shown in Table 1. An example of how the pericentral RP phenotype differs from typical RP is shown in Figure 1.

Table 1.
Baseline clinical characteristics of cohort probands. Further details regarding genes (last column) are provided in Table 2.
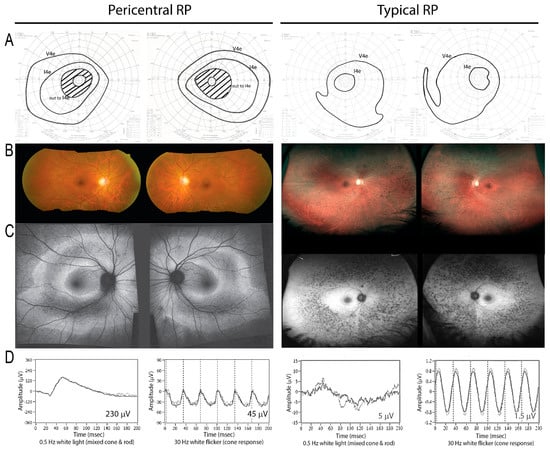
Figure 1.
On the left, a patient (Family #14) with a heterozygous RHO mutation demonstrates a pericentral phenotype, with pericentral scotomas but preserved peripheral field to the I4e stimulus on visual field testing (A). There is retinal pigment epitheium (RPE) atrophy in the near periphery as shown by fundus photo (B) and autofluorescence imaging (C), as well as relatively preserved electroretinograms (ERGs) (D). In contrast, in typical retinitis pigmentosa (RP) (right), the peripheral response to I4e is lost (A), and the affected area is located farther away from the macula in the mid-periphery (right, B,C; note lower magnification). The ERG is more severely affected (D; note different scales; see numerical values, inset).
3.2. Panel-Based Sequencing Results
The panel-based NGS approach (GEDi panel [31]) provided a mean depth of coverage of the targeted sequences of >100-fold, and >97% of the targeted sequences were covered with ≥10× depth. Out of 43 families, 44% (19 families) were solved genetically by identifying mutations provisionally responsible for disease (Table 2). Within those 19 solved families, there were: 7 dominant, 1 X-linked carrier, 2 homozygous recessive, and 9 compound heterozygous recessive. Therefore, there were a total of 28 (7 + 1 + 2 + 18) putatively pathogenic alleles. Fifteen of these 28 alleles (54%) have been previously reported in patients with typical RP or an RP-associated syndrome, while 13 alleles (46%) are novel. See supplemental Table S1 for additional annotation including genomic coordinates, pathogenicity predictions, pedigree type, references, and notes discussing the unsolved/partially solved patients with variants of unknown significance (VUS).

Table 2.
Genetic causes of disease in the pericentral RP cohort, including probands whose cause of disease was solved by panel-based sequencing (top) or whole exome sequencing (WES) (middle). Unsolved probands with VUS are shown below. Also, see Supplemental Table S1 for a more fully annotated version including genomic coordinates, pathogenicity predictions, and references.
All variants that were validated with Sanger sequencing showed the expected results (Table 2). During the course of this study, in-depth assessment of validation rates became available, showing that >10× NGS coverage of a variant provides high confidence of its existence, and the several variants in Table 2 were not Sanger sequenced, as indicated. However, segregation studies in family members were always performed whenever samples were available, which showed 100% concordance with expected results, as shown (Table 2).
Five samples had mutations in RHO, making it the most commonly identified gene associated with pericentral RP, to date, across studies (see Table 3 and Discussion). No mutations were found in TOPORS or ABCA4, two genes identified in other studies (see Discussion.)

Table 3.
Genes identified in patients with pericentral RP (this study), pericentral retinal degeneration [19], or pericentral retinal dystrophy [23,25], with numbers of probands solved by each gene.
Family #18 was the largest family available in this study, with a proband, his three affected children, and his unaffected wife. A PRPF31 3′ splice mutation (c.421-1G>A) was detected in all affected family members and was absent in the unaffected wife. Interestingly, while the proband had pericentral RP, his children with the same mutation had typical RP (n = 2) or mild RP without pericentral features (n = 1). See Figure 2.
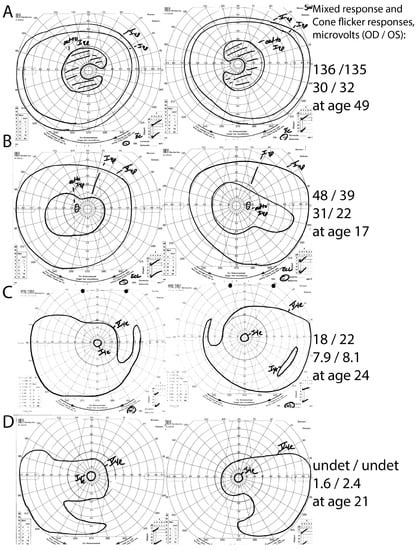
Figure 2.
Intra-familial phenotypic variation in family #18. The proband (A) had pericentral RP, with near-peripheral C-shaped scotomas with preserved peripheral I4e responses, and relatively preserved full-field ERG responses. His children, in contrast, had either atypically mild RP without pericentral features (B) or typical RP (C,D). Note the constricted peripheral I4e stimulus responses and lower ERG response amplitudes in the children (B–D) in comparison to the proband. undet = undetectable.
Nine samples unsolved by panel-based NGS were analyzing using Omni2.5 chips (Illumina). No pathogenic CNVs were identified.
3.3. Whole Exome Sequencing Results
Previous studies used a candidate gene approach or panel-based sequencing to identify genetic causes of pericentral RP. To broaden this search for novel gene that cause pericentral RP, WES was performed on unsolved probands, and their family members, when available. From the probands not solved by panel-based NGS, 16 families were selected for WES. Three families had DNA available from multiple family members, while 13 were run as single samples.
A “burden test” was used to rank genes according to which genes had more damaging mutations in the pericentral set than in a control set (see Section 2.5 and Discussion.) Sixteen unsolved pericentral RP probands were compared to a set of 1724 exomes representing all other WES samples in our database. First, a dominant model was assumed in which only one damaging allele (i.e., a heterozygote mutation) was required to be counted. In each sample, there were a large number of genes contained “damaging” variants (as defined in Section 2.5); over 100 genes were implicated in every sample (range 117–233 not shown). This large number of hits makes it difficult to find the real solution, which is mostly likely a single gene in these Mendelian diseases/families. The top hits included a number of genes with large transcripts (e.g., OBSCN 24 kb mRNA, HMCN1 18 kb mRNA) with “damaging” alleles in >100 exomes in the control set, indicating that they are not likely to cause a rare disease. A more stringent test was performed which added a filter for ExAC allele frequency and omitted variants predicted as damaging by PolyPhen or SIFT. In this case, fewer positive hits were detected from each sample (range: 4–23 genes per sample, Supplemental Table S2), but still not few enough to identify meaningful solutions. Also, the list contains an overrepresentation of single base pair insertions (not shown), suggesting additional filtering strategies are required. Until these issues can be resolved, analyses of hits from the dominant inheritance model have been deferred.
Next, a recessive model was assumed which required two predicted-damaging variants (i.e., homozygous or compound heterozygous variants) in a sample for it to contribute to the gene count. Each sample contributed only 0–15 genes (average 6) under these conditions (Supplemental Table S3). The most enriched gene was HGSNAT, which was found in 3 of 16 unsolved pericentral RP samples (#34, #15, #13) and only 2 of 1726 other samples, p = 6 × 10−6 for enrichment, p = 6 × 10−4 with Bonferroni correction. One of the other samples was actually a pericentral sample provisionally considered solved (#24) by panel-based sequencing, making the enrichment even stronger. We categorized two of these samples as solved by HGSNAT mutations (#34 and #15), and two samples (#24, #13) with VUS in HGSNAT (Table 2). The latter two samples have homozygous mutations in a known, relatively common hypomorphic allele A615T, which has been reported to be pathogenic when combined with other alleles (see Discussion). Sample #15 contained a + 1 5’ splice variant that has been reported as pathogenic [43], in combination with the common A615T variant discussed below. Family members are not available for segregation testing. Sample #34 has a novel homozygous S318N mutation predicted to be damaging.
HGSNAT mutations most commonly cause Mucopolysaccharidosis type IIIC, but review of the literature reveals nonsyndromic RP cases with what is essentially a pericentral phenotype (see Discussion). Fundus photographs of the two patients considered solved by HGSNAT mutations in this study are shown in Figure 3. Available records did not make any note of nonocular manifestations of mild HGSNAT deficiency, such as “coarse facial features, hypertrichosis, contractures, organomegaly, hearing impairment, behavioral and sleeping problems, recurrent infections, diarrhea, epilepsy or late onset of mental deterioration.” [44], p. 3745.
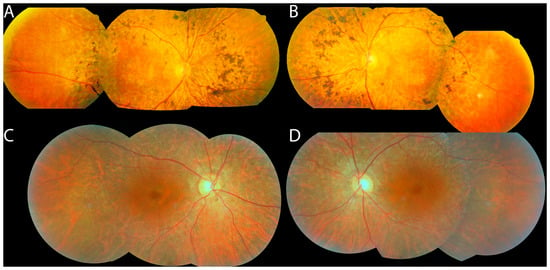
Figure 3.
Fundus photos of patients with HGSNAT mutations: #34 (A,B) and #15 (C,D). Note that the bone spicules (A,B) or RPE depigmentation (C,D) are more posteriorly located than in typical RP, and that the fundus begins to show a more normal color peripherally (especially temporally in these photos).
Additional enriched genes under the recessive model were not significant after multiple test correction.
In essence, burden testing analyses on the WES data successfully identified HGSNAT as a cause of pericentral RP, in at least two patients.
4. Discussion
4.1. What Genetics Reveals about the Causes of the Pericentral RP Phenotype
There is no specific genetic cause of pericentral RP. This study more than doubles the number of pericentral RP patients successfully genotyped. Yet there is still no single pericentral RP gene that explains most of the probands with this clinical phenotype. However, closer analysis of the genetic causes does give some flavor of what the genetic source of the phenotype is, and which other phenotypes are nearby to this phenotype in the sense that the causal genes overlap. Namely, the genetic causes of pericentral RP are similar to those that have been reported to cause other forms of mild RP. Grouping the three pericentral cohorts summarized in Table 3, RHO is the most commonly identified causative gene. RHO typically produces milder dominant disease [24,45,46,47] or sometimes minimally progressive sector RP [5,48]. We also identified one case as the X-linked RP/RP2 carrier state [49], which also tends to produce a relatively mild phenotype compared to typical RP. About half of the gene variants described in this study have been previously reported in patients with typical RP. While it is possible that, on close inspection of the phenotype, all of these variants will be mild/hypomorphic mutations at a molecular level, it seems much more likely that there is broad overlap with the phenotypes of typical RP, atypically mild RP, and pericentral RP, even within a particular genotype. This overlap is demonstrated perfectly by family #18 in Figure 2 above where the same genotype causes typical RP, atypically mild RP, and pericentral RP. Cis-acting variants or modifier genes may explain some of this intra-familial diversity (e.g., [50]) and identifying additional modifier genes is an area of active research.
There are admittedly limitations to this interpretation of pericentral RP being caused by genes that typically produce mild disease; there are multiple recessive RP genes in Table 3, and some such as USH2A typically cause a large amount of field loss [47]. Furthermore, while pericentral RP is mild (by definition) in that there is a large amount of remaining visual field function as reflected by total visual field area and ERG responses, the better peripheral retinal function is accompanied by worse central/pericentral field and sometimes worse central acuity [19,22].
The genotypes identified in this study were most similar to those reported by Grondahl et al. [23] and Selmer et al. [25], where RHO was the most commonly identified gene. In contrast, as displayed in Table 3, Matsui et al. [19] found ABCA4 as the most commonly identified gene in their “pericentral retinal degeneration” cohort. Matsui et al. carefully detail how the phenotypes of their five ABCA4 cases are different from typical Stargardt disease with foveal sparing (e.g., no flecks, larger remaining central island of vision); nonetheless, subtle differences in inclusion criteria of mild, macula-predominant cases whose phenotype falls in between that of typical Stargardt disease and typical RP probably explain the differences observed genotypes.
4.2. Whole Exome Sequencing-Based Gene Discovery and HGSNAT
WES analysis strategies for this cohort are not trivial. Any WES dataset contains a large number of nonpathogenic variants. The inclusion of a control group for comparison helped to address this problem. Furthermore, some notable successes of WES-based gene discovery analyzed diseases where the genetic cause is limited to one or two genes (e.g., Kabuki syndrome [51]), and where the phenotype is very distinctive. For any cohort of RP patients, these advantages are not expected due to the large number of genes that can cause RP, ~50% diagnostic rates, and imperfections in clinically-defined phenotypic boundaries. Therefore, we assumed there would be heterogeneity in the results, decreasing the power to detect new genes with confidence. Given those challenges of WES-based cohort analyses, it is notable that WES did provide additional information in this cohort—the identification of HGSNAT as a recurring cause of pericentral RP.
Mutations in HGSNAT usually cause Sanfilippo Syndrome/Mucopolysaccharidosis Type IIIC—a severe multisystemic lysosomal storage disease that presents in infancy or childhood and leads to mental retardation, early death and, among other problems, RP [52]. More recently, HGSNAT mutations have been identified as a cause of nonsyndromic RP [44]. Haer-Wigman et al. [44] describe multiple patients with ring scotomas and one with a pericentral field defect. The images published in Figure 3G,H of that study show autofluorescence defects restricted to the near periphery [44]. Similarly, Van Cauwenbergh et al. [53] report on a patient with compound heterozygote mutations in HGSNAT (A615T/deletion of exons 7–8). While not explicitly described as a pericentral phenotype, Figure 2A of that study shows pigment changes, atrophy, and autofluorescence changes in the near periphery [53]. Fundus photographs of the two HGSNAT probands identified in the present study are shown in Figure 3, and they have a similar pericentral phenotype. (Unfortunately, those patients are no longer available for a more detailed phenotypic analysis such as enzyme levels.) Most recently, one additional case of HGSNAT-associated RP, with compound heterozygous A615T/P283L mutations, has been reported but without detailed phenotype information [54]. In summary, while there is mention of HGSNAT-associated nonsyndromic RP as having a perimacular or pericentral phenotype [44], this study serves to merge those observations with the larger body of literature about pericentral RP per se, and also suggests that HGSNAT is one of the more common genes to cause the pericentral RP phenotype. Practical implications of this finding are that HGSNAT should be included in panel-based testing of nonsyndromic RP, and evaluation for the reported [53] deletion of exons 7–8 should be considered in pericentral cases as well.
The pathogenicity status of the HGSNAT A615T allele appears to be complicated. This is particularly relevant to probands #13 and #24 in this study with homozygous A615T variants, but without another allele or detectable deletions. Our best interpretation of the existing data is that A615T is a weak mutation in that may cause no disease or mild disease in the homozygous state, but can cause mucopolysaccharidosis and/or RP when paired with a stronger allele [44,55,56,57]. The evidence supporting this interpretation is complex and not definitive. The A615T variant was originally identified as a cause of Mucopolysaccharidosis Type IIIC (described in that study as A643T) [55]. Feldhammer el al. [56] identify A615T as having slightly reduced activity, which was interpreted as wild type level in their original paper, but has been reinterpreted as a hypomorphic allele by Haer-Wigman et al. [44]. Haer-Wigman et al. [44] report an RP pedigree with heterozygous G133A mutations in combination with homozygous A615T variants—that is, G133A in cis with A615T on one allele, and A615T alone on the other allele. This suggests A615T can cause RP when paired with a stronger allele. Biochemically, a similar pattern has been reported. Fedele et al. [57] show that a combination of W403C and A615T in cis additively leads to lower activity, but that the A615T variant alone has a negligible decrease in activity; they state that A615T homozygotes probably would not be affected. Frequency data supports non-pathogenicity of homozygous A615T as well; the allele frequency for A615T appears particularly high in Ashkenazi Jews (1.5% with 2/4993 homozygotes reported [58]), such that this mutation is likely too common to be a rare cause of RP. Additional data such as evidence of partial penetrance or of high actual HGSNAT-associated disease incidence in Ashkenazi Jews could lead to reevaluation of this conclusion, however. Conversely, it is also not certain that every instance of compound heterozygote with A615T is disease-causing. For now, Table 2 lists homozygous A615T variants as a VUS, while compound heterozygotes with another pathogenic allele are listed as provisionally disease-causing.
In conclusion, these findings confirm and extend the observation that pericentral RP can be caused by many of the same mutations that cause typical RP, in a broad spectrum of genes that are known to cause typical RP. Additionally, the genotypes in the present cohort (e.g., RHO, RP2 carrier state) suggest that pericentral RP shares genetic causes with other forms of mild RP. WES-based gene discovery analysis also allowed us to identify HGSNAT mutations as a recurring cause of pericentral RP. Out of the small number of reported cases of HGSNAT-associated RP, many appear to have pericentral features.
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/8/10/256/s1. Table S1: Genetic causes of disease in the pericentral RP cohort- fully annotated version. Table S2: Burden test results for dominant gene model. Table S3: Burden test results for recessive gene model.
Acknowledgments
This manuscript is dedicated to Eliot L. Berson M.D. (deceased), who originally conceived of the project in 2012 and contributed the patient phenotypes and DNA samples. We also thank Mark Consugar for technical support. Financial support: NEI K12 EY16335 (J.C.), a Research to Prevent Blindness Career Development Award (J.C.), NIH EY012910 (E.A.P.), grants from the Foundation Fighting Blindness (E.A.P. and J.C.), NIH P30EY014104 Core grant.
Author Contributions
J.C. and E.A.P. conceived and designed the experiments; A.L., S.H., and J.C. performed the experiments; J.C., C.W.-D., E.P., E.A.P., M.A.S., M.M., and D.N.-G. analyzed the data; M.M., J.C., M.A.S. contributed materials/analysis tools; J.C. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Daiger, S.P. RetNet, the Retinal Information Network. Available online: http://www.sph.uth.tmc.edu/RetNet/ (accessed on 24 June 2017).
- Grover, S.; Fishman, G.A.; Brown, J., Jr. Patterns of visual field progression in patients with retinitis pigmentosa. Ophthalmology 1998, 105, 1069–1075. [Google Scholar] [CrossRef]
- Chang, S.; Vaccarella, L.; Olatunji, S.; Cebulla, C.; Christoforidis, J. Diagnostic challenges in retinitis pigmentosa: Genotypic multiplicity and phenotypic variability. Curr. Genomics 2011, 12, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Gonin, J. Le scotome Annulaire Dans la Dégénérescence Pigmentaire de la Rétine; A. Maloine: Paris, France, 1901. [Google Scholar]
- Berson, E.L.; Howard, J. Temporal aspects of the electroretinogram in sector retinitis pigmentosa. Arch. Ophthalmol. 1971, 86, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Valentine, J.A. Night Blindness: Retinitis Pigmentosa sine Pigmento. Proc. R. Soc. Med. 1923, 16, 17. [Google Scholar] [PubMed]
- Choi, J.Y.; Sandberg, M.A.; Berson, E.L. Natural course of ocular function in pigmented paravenous retinochoroidal atrophy. Am. J. Ophthalmol. 2006, 141, 763–765. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.B.; Zhang, Y.X. Pigmented paravenous retinochoroidal atrophy (Review). Exp. Ther. Med. 2014, 7, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Francois, J.; De Rouck, A.; Cambie, E.; De Laey, J.J. Visual functions in pericentral and central pigmentary retinopathy. Ophthalmologica 1972, 165, 38–61. [Google Scholar] [CrossRef] [PubMed]
- Traboulsi, E.I.; O’Neill, J.F.; Maumenee, I.H. Autosomal recessive pericentral pigmentary retinopathy. Am. J. Ophthalmol. 1988, 106, 551–556. [Google Scholar] [CrossRef]
- Durlu, Y.K.; Burumcek, E.; Devranoglu, K.; Mudun, A.B.; Karacorlu, S.; Arslan, M.O. Associated ocular findings in pericentral pigmentary retinopathy. Acta Ophthalmol. Scand. 1997, 75, 101–103. [Google Scholar] [CrossRef] [PubMed]
- Hayasaka, S.; Fukuda, K.; Tsuchiya, M.; Mizuno, K. Pericentral pigmentary retinal degeneration. Jpn. J. Ophthalmol. 1985, 29, 161–169. [Google Scholar] [PubMed]
- Yamaguchi, K.; Kin-para, Y.; Tamai, M. Idiopathic central serous choroidopathy in a patient with pericentral pigmentary retinal degeneration. Ann. Ophthalmol. 1991, 23, 251–253. [Google Scholar] [PubMed]
- Noble, K.G. Peripapillary (pericentral) pigmentary retinal degeneration. Am. J. Ophthalmol. 1989, 108, 686–690. [Google Scholar] [CrossRef]
- Hotta, K.; Kondo, M.; Nakamura, M.; Hotta, J.; Terasaki, H.; Miyake, Y.; Hida, T. Negative electroretinograms in pericentral pigmentary retinal degeneration. Clin. Exp. Ophthalmol. 2006, 34, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Grondahl, J. Pericentral retinal dystrophy. Acta Ophthalmol. 1987, 65, 344–351. [Google Scholar] [CrossRef]
- Noble, K.G.; Carr, R.E. Peripapillary pigmentary retinal degeneration. Am. J. Ophthalmol. 1978, 86, 65–75. [Google Scholar] [CrossRef]
- Chakarova, C.F.; Papaioannou, M.G.; Khanna, H.; Lopez, I.; Waseem, N.; Shah, A.; Theis, T.; Friedman, J.; Maubaret, C.; Bujakowska, K.; et al. Mutations in TOPORS cause autosomal dominant retinitis pigmentosa with perivascular retinal pigment epithelium atrophy. Am. J. Hum. Genet. 2007, 81, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Matsui, R.; Cideciyan, A.V.; Schwartz, S.B.; Sumaroka, A.; Roman, A.J.; Swider, M.; Huang, W.C.; Sheplock, R.; Jacobson, S.G. Molecular Heterogeneity Within the Clinical Diagnosis of Pericentral Retinal Degeneration. Invest. Ophthalmol. Vis. Sci. 2015, 56, 6007–6018. [Google Scholar] [CrossRef] [PubMed]
- Krill, A. Central and pericentral retinitis pigmentosa. In Krill’s Hereditary Retinal and Choroidal Diseases; Archer, D.B., Ed.; Blackwell Publishing: Hagerstown, MD, USA, 1977; Volume 2, pp. 617–625. [Google Scholar]
- Szamier, R.B.; Berson, E.L. Histopathologic study of an unusual form of retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 1982, 22, 559–570. [Google Scholar] [PubMed]
- Sandberg, M.A.; Gaudio, A.R.; Berson, E.L. Disease course of patients with pericentral retinitis pigmentosa. Am. J. Ophthalmol. 2005, 140, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Grondahl, J.; Riise, R.; Heiberg, A.; Leren, T.; Christoffersen, T.; Bragadottir, R. Autosomal dominant retinitis pigmentosa in Norway: A 20-year clinical follow-up study with molecular genetic analysis. Two novel rhodopsin mutations: 1003delG and I179F. Acta Ophthalmol. Scand. 2007, 85, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; McGuigan, D.B., 3rd; Sumaroka, A.; Roman, A.J.; Gruzensky, M.L.; Sheplock, R.; Palma, J.; Schwartz, S.B.; Aleman, T.S.; Cideciyan, A.V. Complexity of the Class B Phenotype in Autosomal Dominant Retinitis Pigmentosa Due to Rhodopsin Mutations. Invest. Ophthalmol. Vis. Sci. 2016, 57, 4847–4858. [Google Scholar] [CrossRef] [PubMed]
- Selmer, K.K.; Grondahl, J.; Riise, R.; Brandal, K.; Braaten, O.; Bragadottir, R.; Undlien, D.E. Autosomal dominant pericentral retinal dystrophy caused by a novel missense mutation in the TOPORS gene. Acta Ophthalmol. 2010, 88, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Bowne, S.J.; Sullivan, L.S.; Gire, A.I.; Birch, D.G.; Hughbanks-Wheaton, D.; Heckenlively, J.R.; Daiger, S.P. Mutations in the TOPORS gene cause 1% of autosomal dominant retinitis pigmentosa. Mol. Vis. 2008, 14, 922–927. [Google Scholar] [PubMed]
- Schob, C.; Orth, U.; Gal, A.; Kindler, S.; Chakarova, C.F.; Bhattacharya, S.S.; Ruther, K. Mutations in TOPORS: A rare cause of autosomal dominant retinitis pigmentosa in continental Europe? Ophthalmic Genet. 2009, 30, 96–98. [Google Scholar] [CrossRef] [PubMed]
- Melles, R.B.; Marmor, M.F. Pericentral retinopathy and racial differences in hydroxychloroquine toxicity. Ophthalmology 2015, 122, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.A.; Marmor, M.F. ERG and other discriminators between advanced hydroxychloroquine retinopathy and retinitis pigmentosa. Doc. Ophthalmol. 2017, 134, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Berson, E.L.; Rosner, B.; Sandberg, M.A.; Hayes, K.C.; Nicholson, B.W.; Weigel-DiFranco, C.; Willett, W. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch. Ophthalmol. 1993, 111, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Consugar, M.B.; Navarro-Gomez, D.; Place, E.M.; Bujakowska, K.M.; Sousa, M.E.; Fonseca-Kelly, Z.D.; Taub, D.G.; Janessian, M.; Wang, D.Y.; Au, E.D.; et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet. Med. 2015, 17, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.J.; Zhang, Q.; Nakamaru-Ogiso, E.; Kannabiran, C.; Fonseca-Kelly, Z.; Chakarova, C.; Audo, I.; Mackay, D.S.; Zeitz, C.; Borman, A.D.; et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat. Genet. 2012, 44, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Bujakowska, K.M.; Consugar, M.; Place, E.; Harper, S.; Lena, J.; Taub, D.G.; White, J.; Navarro-Gomez, D.; Weigel DiFranco, C.; Farkas, M.H.; et al. Targeted exon sequencing in Usher syndrome type I. Invest. Ophthalmol. Vis. Sci. 2014, 55, 8488–8496. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef] [PubMed]
- Gai, X.; Perin, J.C.; Murphy, K.; O’Hara, R.; D’Arcy, M.; Wenocur, A.; Xie, H.M.; Rappaport, E.F.; Shaikh, T.H.; White, P.S. CNV Workshop: An integrated platform for high-throughput copy number variation discovery and clinical diagnostics. BMC Bioinform. 2010, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- GATK Best Practices. Available online: https://software.broadinstitute.org/gatk/best-practices/ (accessed on 1 October 2017).
- Purcell, S.M.; Moran, J.L.; Fromer, M.; Ruderfer, D.; Solovieff, N.; Roussos, P.; O’Dushlaine, C.; Chambert, K.; Bergen, S.E.; Kahler, A.; et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014, 506, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Fedele, A.O.; Filocamo, M.; Di Rocco, M.; Sersale, G.; Lubke, T.; di Natale, P.; Cosma, M.P.; Ballabio, A. Mutational analysis of the HGSNAT gene in Italian patients with mucopolysaccharidosis IIIC (Sanfilippo C syndrome). Mutation in brief #959. Online. Hum. Mutat. 2007, 28, 523. [Google Scholar] [PubMed]
- Haer-Wigman, L.; Newman, H.; Leibu, R.; Bax, N.M.; Baris, H.N.; Rizel, L.; Banin, E.; Massarweh, A.; Roosing, S.; Lefeber, D.J.; et al. Non-syndromic retinitis pigmentosa due to mutations in the mucopolysaccharidosis type IIIC gene, heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT). Hum. Mol. Genet. 2015, 24, 3742–3751. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, V.; Abrahamson, M.; Andreasson, S.; Ehinger, B.; Fex, G.; Polland, W. A mild phenotype of autosomal dominant retinitis pigmentosa is associated with the rhodopsin mutation Pro-267-Leu. Ophthalmic Genet. 1997, 18, 63–70. [Google Scholar] [PubMed]
- Berson, E.L.; Rosner, B.; Weigel-DiFranco, C.; Dryja, T.P.; Sandberg, M.A. Disease progression in patients with dominant retinitis pigmentosa and rhodopsin mutations. Invest. Ophthalmol. Vis. Sci. 2002, 43, 3027–3036. [Google Scholar] [PubMed]
- Sandberg, M.A.; Rosner, B.; Weigel-DiFranco, C.; McGee, T.L.; Dryja, T.P.; Berson, E.L. Disease course in patients with autosomal recessive retinitis pigmentosa due to the USH2A gene. Invest. Ophthalmol. Vis. Sci. 2008, 49, 5532–5539. [Google Scholar] [CrossRef] [PubMed]
- Fishman, G.A.; Stone, E.M.; Gilbert, L.D.; Sheffield, V.C. Ocular findings associated with a rhodopsin gene codon 106 mutation. Glycine-to-arginine change in autosomal dominant retinitis pigmentosa. Arch. Ophthalmol. 1992, 110, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Comander, J.; Weigel-DiFranco, C.; Sandberg, M.A.; Berson, E.L. Visual Function in Carriers of X-Linked Retinitis Pigmentosa. Ophthalmology 2015, 122, 1899–1906. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.M.; Shah, A.Z.; Venturini, G.; Rivolta, C.; Rose, G.E.; Bhattacharya, S.S. Dominant PRPF31 mutations are hypostatic to a recessive CNOT3 polymorphism in retinitis pigmentosa: A novel phenomenon of “linked trans-acting epistasis”. Ann. Hum. Genet. 2014, 78, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.B.; Bigham, A.W.; Buckingham, K.J.; Hannibal, M.C.; McMillin, M.J.; Gildersleeve, H.I.; Beck, A.E.; Tabor, H.K.; Cooper, G.M.; Mefford, H.C.; et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010, 42, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Feldhammer, M.; Durand, S.; Mrazova, L.; Boucher, R.M.; Laframboise, R.; Steinfeld, R.; Wraith, J.E.; Michelakakis, H.; van Diggelen, O.P.; Hrebicek, M.; et al. Sanfilippo syndrome type C: Mutation spectrum in the heparan sulfate acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT) gene. Hum. Mutat. 2009, 30, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Van Cauwenbergh, C.; Van Schil, K.; Cannoodt, R.; Bauwens, M.; Van Laethem, T.; De Jaegere, S.; Steyaert, W.; Sante, T.; Menten, B.; Leroy, B.P.; et al. arrEYE: A customized platform for high-resolution copy number analysis of coding and noncoding regions of known and candidate retinal dystrophy genes and retinal noncoding RNAs. Genet. Med. 2017, 19, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Carss, K.J.; Arno, G.; Erwood, M.; Stephens, J.; Sanchis-Juan, A.; Hull, S.; Megy, K.; Grozeva, D.; Dewhurst, E.; Malka, S.; et al. Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am. J. Hum. Genet. 2017, 100, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Hrebicek, M.; Mrazova, L.; Seyrantepe, V.; Durand, S.; Roslin, N.M.; Noskova, L.; Hartmannova, H.; Ivanek, R.; Cizkova, A.; Poupetova, H.; et al. Mutations in TMEM76* cause mucopolysaccharidosis IIIC (Sanfilippo C syndrome). Am. J. Hum. Genet. 2006, 79, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Feldhammer, M.; Durand, S.; Pshezhetsky, A.V. Protein misfolding as an underlying molecular defect in mucopolysaccharidosis III type C. PLoS ONE 2009, 4, e7434. [Google Scholar] [CrossRef] [PubMed]
- Fedele, A.O.; Hopwood, J.J. Functional analysis of the HGSNAT gene in patients with mucopolysaccharidosis IIIC (Sanfilippo C Syndrome). Hum. Mutat. 2010, 31, E1574–E1586. [Google Scholar] [CrossRef] [PubMed]
- gnomAD browser beta. Variant: 8:43054647 G/A. Available online: http://gnomad.broadinstitute.org/variant/8-43054647-G-A (accessed on 1 October 2017).
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).