Abstract
The contribution of chromatin dynamics to the regulation of human disease-associated loci such as the cystic fibrosis transmembrane conductance regulator (CFTR) gene has been the focus of intensive experimentation for many years. Recent technological advances in the analysis of transcriptional mechanisms across the entire human genome have greatly facilitated these studies. In this review we describe the complex machinery of tissue-specific regulation of CFTR expression, and put earlier observations in context by incorporating them into datasets generated by the most recent genomics methods. Though the gene promoter is required for CFTR expression, cell-type specific regulatory elements are located elsewhere in the gene and in flanking intergenic regions. Probably within its own topological domain established by the architectural proteins CTCF and cohesin, the CFTR locus utilizes chromatin dynamics to remodel nucleosomes, recruit cell-selective transcription factors, and activate intronic enhancers. These cis-acting elements are then brought to the gene promoter by chromatin looping mechanisms, which establish long-range interactions across the locus. Despite its complexity, the CFTR locus provides a paradigm for elucidating the critical role of chromatin dynamics in the transcription of individual human genes.
1. Introduction
One of the most intensively studied genes associated with human disease is the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which encompasses 189 kb at chromosome 7q31.2. It encodes a 1480 amino acid protein, which primarily functions as a cAMP-dependent chloride ion channel [1,2,3] belonging to the ATP-binding cassette (ABC) transporter superfamily. Mutations in CFTR cause the common, autosomal recessive disease cystic fibrosis (CF) [4,5,6], which is characterized by high salt levels in the sweat and the accumulation of thick, sticky mucus, which is associated with inflammation, tissue fibrosis, and impaired lung, intestinal, and pancreatic function. The CFTR gene shows tight, tissue-specific regulation of expression. The CFTR protein is abundant in the specialized epithelial cells of tissues associated with CF pathology such as the intestine, pancreas, sweat gland, and genital ducts, though levels are much lower in the respiratory epithelium [7,8,9,10]. It is also found in some other organ and cell sites. Within the intestinal epithelium there is a strong gradient of expression along the crypt (high) to the villous (low) axis and from the proximal to distal intestine [8]. CFTR is expressed from early in gestation [11] and shows temporal regulation in the lung, with the fetal lung having higher levels of transcript than adult tissue [12,13,14].
This complex pattern of coordinated tissue-specific and temporal regulation led to intensive investigation of the mechanisms controlling CFTR gene expression. These studies, which were among the first to describe critical regulatory elements in non-coding regions of the genome [15], provided a paradigm for understanding gene regulation genome-wide. The subsequent dramatic technological and conceptual advances made by the Encyclopedia of DNA elements (ENCODE) consortium [16] provided new tools to further delineate the mechanisms of CFTR regulation, particularly in differentiated primary epithelial cell types. Early work on the transcriptional regulation of CFTR focused on the promoter, which was classified as a “house-keeping” gene-like promoter. Analysis of a ~4 kb region upstream of the first exon showed a high GC content, no TATA box, and multiple transcription start sites (TSS) [17,18,19]. Utilization of different TSS may be involved in both developmental and tissue-specific control of the gene, for example, with different start sites used in the fetal and adult lung [20]. Moreover, CFTR gene promoter activity may be regulated by the recruitment of alternative 5' exons and local RNA secondary structure [21,22,23]. Initial studies also showed that the promoter contained binding sites for the transcription factors Sp1 and AP-1 [17], a cAMP response element (CRE), a CCAAT box [24,25,26], and a NF-κB binding site [27,28]. However, these analyses did not identify the mechanisms by which tissue-specific CFTR expression is achieved. The lack of cell-type specific control elements in the promoter (defined here as ~2kb 5' to the translational start site) and the large size of the gene, with multiple long introns, led to the hypothesis that critical cis-regulatory elements might exist within the gene body itself or in distal upstream or downstream regions.
2. Identification of cis-Regulatory Elements in the CFTR Locus
To identify open chromatin regions and associated regulatory elements; within and flanking the CFTR gene, DNase I hypersensitive site (DHS) mapping was performed. The first studies used Southern blotting with gene specific probes, but subsequent methodological advances enabled the whole CFTR locus to be evaluated by DNase-chip [29] and later DNase-seq [30]. Nucleosomes can prevent transcription factors from accessing their binding sites within cis-regulatory elements, therefore nucleosome-depleted regions, which are hypersensitive to digestion by DNase I, represent potentially active regulatory elements [31]. Initial analyses on CFTR by Southern blotting revealed DHS at −79.5 kb and −20.9 kb from the translational start site of CFTR [32], within several introns of the gene [15,33], and in the 3' intergenic region [34] (Figure 1). The intronic sites, particularly those within introns 1, 10, 17a, and 20, showed some cell-type restriction, suggesting a role in tissue-specific regulation of CFTR expression. Analyses of these elements, primarily using in vitro DNase I footprinting, electromobility shift assays (EMSAs), and luciferase reporter gene assays, showed that they bound various transcription factors and were likely essential for CFTR expression [15,34,35,36,37,38]. Moreover, the functional importance of these intronic sites was shown by deletion of the DHS in intron 1, which was identified as the first CFTR intronic cis-element [15], from a yeast artificial chromosome (YAC) encompassing the CFTR locus. Loss of this region from the YAC-encoded CFTR gene reduced its expression in transgenic mouse intestine by ~60% [39]. However, this deletion had no effect on CFTR expression from the YAC in transgenic mouse lung tissue, providing more direct evidence for the importance of this intronic element in tissue-specific regulation of CFTR expression. Further characterization of the intestinal-specific DHS in intron 1 showed that it contained hepatocyte nuclear factor 1 homebox A (HNF1α) binding sites and depletion of HNF1α caused a decrease in CFTR mRNA levels [40]. Subsequently, the importance of HNF1 binding at this site was confirmed by chromatin immunoprecipitation (ChIP) [41].
3. Characterization of Enhancers at the CFTR Locus
The realization and more general acceptance in the late 1990s that far from being “junk” DNA, introns contained many of the critical regulatory elements coordinating gene expression across the genome validated the earlier, laborious work on the CFTR locus. Genome-wide analyses using new methods developed by the ENCODE consortium revealed many functions for the intronic and intergenic sequences of the genome that act as cis-regulatory elements [42]. One functional class that has received much attention is enhancers, which are often located distal to the genes they regulate, utilize chromatin remodelers, and bind to transcriptional activators or co-activators to initiate gene expression. Enhancers frequently show tissue-specificity and are readily identified since they are depleted of nucleosomes, constitute open chromatin regions, and carry specific histone modifications [43].
In addition to the enhancer in the CFTR intron 1 DHS described above and a weak enhancer in intron 20 [35], DNase-chip identified several novel DHS in cells expressing different levels of CFTR mRNA [44] (Figure 1). Fibroblasts, which do not express CFTR lacked intronic DHS, while Caco2 (colorectal adenocarcinoma) cells expressing abundant CFTR showed prominent DHS within introns 10 and 11. The novel site in intron 11 was later shown to encompass a very strong enhancer [44] that appears critical for the high levels of CFTR expression in the intestinal epithelium. The DNase-chip analysis also showed cell-type selectivity of the DHS at the CFTR locus, since sites that were evident in CFTR-expressing airway cells were absent in intestinal cells and vice-versa [44]. Many of the intronic DHS in primary cells have not yet been studied intensively and may contribute further insights into the cell- specific fine-tuning of the locus.
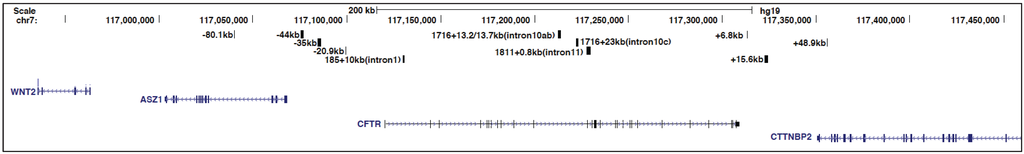
Figure 1.
Schematic to show to the genomic location of the cystic fibrosis transmembrane conductance regulator (CFTR) locus and some of its key cis-regulatory elements discussed in this review. From the University of California, Santa Cruz (UCSC) genome browser.
The presence or absence of DHS may be regulated through nucleosome positioning and remodeling [45]. Using a new method of nucleosome mapping, based on hybridization of purified nucleosomes to a bacterial artificial chromosome (BAC) encompassing the whole CFTR locus, we mapped nucleosomes in several cell types [46]. The intestinal-selective enhancer at the intron 1 DHS was nucleosome-depleted only in Caco2 cells and the HNF1 binding site, which drives this enhancer element, was occluded by nucleosomes in CFTR-expressing airway cells and in skin fibroblasts. Similar nucleosome-depleted sequences were observed at another intestinal-specific enhancer in intron 11 and an airway-selective DHS at −35 kb upstream of the translational start site [46]. However, certain ubiquitous DHS such as those at −20.9 kb 5' of the translational start site and +6.8 kb downstream of the last coding base showed a nucleosome-depleted core region across multiple cell types, suggesting occupancy of a common factor.
Enhancer sequences are also associated with specific histone modifications and occupancy by the factors that establish those marks. For example, active enhancers are often associated with acetylation at lysine 27 of histone H3 (H3K27ac) [47,48,49], which is established by p300. ChIP for p300 across the CFTR locus in Caco2 cells showed enrichment of the factor at multiple DHS identified by DNase-chip in these cells [44]. Enhancers are also often marked by mono- or di-methylation on lysine 4 of histone H3 (H3K4me1/me2) [47,49]. H3K4me1 was previously shown to be enriched at regions corresponding to cell-type specific DHS and enhancer elements within the CFTR locus [50].
4. Identification of Transcription Factors Regulating CFTR
Although the hallmark features discussed above help to identify putative enhancers, the most important characteristic of these regions is their ability to recruit transcription factors and co-activators that promote gene expression. Studies on post-confluent Caco2 cells, a model of intestinal CFTR regulation, identified a network of relevant transcription factors interacting with cis-regulatory elements at the locus. These factors included, HNF1, forkhead box protein A (FOXA), and caudal type homeobox 2 (CDX2) [40,44,51]. In Caco2 cells HNF1α was enriched at the intron 1 and 11 enhancers, and at lower levels at a DHS in intron 10 [44]. Subsequently, using in silico predictions, in vitro DNase I footprinting, EMSAs, and ChIP, the pioneer transcription factors, FOXA1 and FOXA2, were shown to occupy DHS elements in introns 10 and 11, while CDX2 was found at multiple intergenic and intronic DHS across the CFTR locus [51]. siRNA-mediated depletion of CDX2 or both FOXA1 and FOXA2 together led to 25% and 40% reductions in CFTR mRNA levels, respectively [51,52]. Binding of HNF1 and CDX2 was shown to be dependent on the presence of FOXA1/A2 [52] as was observed for other FOXA1/A2 co-factors in genome-wide studies [53,54]. Interestingly, depletion of the FOXA factors altered enhancer histone modification profiles (H3K27ac and H3K4me2) at specific sites across the CFTR locus and led to a decrease in hypersensitivity of a DHS in intron 10 [52]. These data suggest that the transcription factors involved in intestinal regulation of CFTR function in a coordinated network, in which the FOXA proteins have a pivotal role.
The DHS and nucleosome positioning data suggest that CFTR expression is regulated by different mechanisms in airway and intestinal cells. This is achieved both by utilization of airway-selective cis-regulatory elements and by recruitment of distinct transcription factors [44,46]. Analysis of CFTR regulation in airway cells revealed DHS at −44 kb and −35 kb upstream of the locus, and also in introns 18, 19, and 23 [55] (Figure 1). The DHS at −35 kb binds to the interferon regulatory factors 1 and 2 (IRF1/2) and nuclear factor Y (NF-Y) [56]. Constitutive occupancy of IRF2 at this site represses CFTR expression, but following an interferon-mediated increase in intracellular IRF1 levels, IRF2 is replaced by IRF1, which activates gene expression. NF-Y has an important role in recruiting histone-modifying enzymes to this site, where it interacts with the transcriptional repressor SIN3A and the SET domain containing lysine methyl-transferase 7 (SETD7) [56]. The DHS −35 kb element is functionally linked to regulatory sequences within DHS −44 kb for modulating CFTR expression in response to environmental stresses in the airway. DHS −44 kb contains an antioxidant response element (ARE) and is usually bound by the repressors BTB and CNC homology 1, basic leucine zipper transcription factor 1 (Bach1) and v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K (MafK) [57]. However, upon antioxidant treatment, Bach1/MafK heterodimers are replaced by the transcriptional activator Nrf2, thus enhancing CFTR expression. Nucleosome occupancy profiles combined with in silico predictions also revealed binding sites for nuclear hormone receptors within multiple airway DHS [46]. These included the glucocorticoid receptor (GR), binding of which can be induced by dexamethasone treatment, suggesting that CFTR may also be subject to hormonal regulation in some cell types.
The mechanism(s) responsible for the marked divergence in CFTR expression levels within airway (low) and intestinal (high) epithelial cells [44] are of substantial clinical relevance, since they might provide clues to new therapies based on altering gene expression levels. It is probable that not only are differences in cell-type-selective enhancers relevant, together with the recruitment of divergent activating transcription factors, but that a repressive mechanism modulates basal CFTR transcription in the airway. In this context, recent work suggesting that an airway-selective regulatory mechanism may involve a repurposing of the transcription factors FOXA1/A2 and CEBPα, which activate CFTR in the intestine to function as repressors in airway cells [58] is of interest.
5. Insulator Elements at the CFTR Locus
In addition to the DHS identified at the CFTR locus that encompass enhancers or tissue-specific regulatory elements, multiple regions of open chromatin are either ubiquitous or present in many cell types [44]. Among these features are DHS flanking the locus on the 5' side, at −79.5 kb and −20.9 kb, and others 3' at +6.8 kb, +15.6 kb, and +48.9 kb (Figure 1). Previous experiments established that the −20.9 kb, +6.8 kb, and +15.6 kb sites marked sequences with enhancer-blocking insulator activity in K562 cell based assays [50,59]. Insulators elements were initially identified based on their ability to prevent inappropriate interactions between enhancers and promoters or to demarcate distinct chromatin domains [60]. These cis-regulatory regions are especially important at the CFTR locus where the flanking genes, ankyrin repeat, SAM and basic leucine zipper domain containing 1(ASZ1) on the 5' side [61] and cortactin binding protein 2 (CTTNBP2) on the 3' side [62], have very different expression patterns from the CFTR locus. To ensure correct regulation of CFTR, it could be advantageous to create an isolated chromatin domain around the locus to prevent inappropriate interactions with regulatory elements of the neighboring genes. Insulator sites are often bound by CCCTC-binding factor (CTCF) and are readily identified genome-wide by ChIP-seq for this factor [63]. CTCF is a highly conserved DNA binding protein with 11 zinc fingers that binds to variations of the CCCTC motif [64]. It was initially identified as an activator and repressor and later recognized as the vertebrate insulator protein [65]. Besides its insulator function, CTCF is involved in many different nuclear functions including intra- and inter-chromosomal interactions [66] leading to its reclassification as a genome architectural protein [67].
The CFTR DHS elements at −20.9 kb and +6.8 kb contained binding motifs for CTCF, were occupied by CTCF as measured by ChIP, and their insulator function was shown to be dependent on CTCF [50,59]. Interestingly, although the +15.6 kb site [34] displayed insulator activity, this was independent of CTCF and might involve nuclear hormone receptors [50]. CTCF performs its functions at approximately 60%–70% sites across the genome in concert with the cohesin complex [68,69,70]. The core cohesin complex is composed of several proteins including structural maintenance of chromosomes 1 (SMC1), SMC3, SCC1 (RAD21), and SA1/SA2, which form a ring-like structure to encircle chromatin [71,72]. ChIP-seq data from the ENCODE project confirmed that the DHS at −20.9 kb and +6.8 kb were enriched for CTCF and cohesin components [42]. Also, our ChIP data on the CFTR locus identified CTCF/RAD21 occupancy at these two sites and at the ubiquitous DHS at +48.9 kb, within the last intron of the flanking gene [44,50,59]. Additional CTCF/cohesin binding sites were also found at −80.1 kb to the translational start site, which corresponds to the −79.5 kb DHS we reported previously based on Southern blotting [32]. The −80.1 kb element was shown to interact with the CFTR promoter [73] and is also in close physical association with a CTCF binding site at +83.7 kb from the 3' end of CFTR [42,74]. These data show CFTR as a paradigm for the multiple mechanisms of transcriptional regulation that are integrated in the control of a single human gene: at the promoter, at tissue-specific enhancer elements, and at CTCF/cohesin bound sites that may be acting as insulators.
6. Higher-Order Organization of the CFTR Locus
The data discussed in the previous sections of this review convincingly demonstrate that multiple cis-regulatory elements are active at the CFTR locus, many of them located at a great distance (>100 kb) from the gene promoter. A probable mechanism for these distal elements to regulate CFTR is through the establishment of long-range chromatin loops with the promoter and/or with each other. Chromatin looping between promoters and DHS harboring regulatory elements is observed at many different loci, and is particularly well-studied at the β-globin locus [75,76]. The spatial organization of the genome and higher order chromatin structure, whereby chromatin loops can bring together distinct regulatory elements, is critical for transcription and gene regulation [77,78]. Recent mapping of associations between TSS and regulatory elements revealed that TSS generally interact with regions ~120 kb away and only 7% of observed looping of regulatory elements occurs with the nearest gene [79].
Long-range chromatin interactions can be measured using several different chromosome conformation capture technologies: 3C [80], 4C-seq [81], 5C [82], HiC [83], and ChIA-PET [84]. One of the original methods, 3C, measures associations between a fixed or bait site and many other defined sites across a locus using PCR or quantitative PCR [80]. This technique was used to investigate interactions across the CFTR locus in multiple cell types [44,73]. Interactions were detected between the promoter bait and DHS marking cis-regulatory elements within introns and flanking the locus, only in cells expressing CFTR. No significant associations were observed between the promoter and the same sites in cell types where the locus is silent, suggesting that looping is dependent on the DHS being present and the elements such as enhancers being actively bound by transcription factors [44,55,73]. Furthermore, many of the interactions between different regulatory elements were shown to be reciprocal using multiple bait regions in the 3C experiments [73]. In airway cells, where distinct regulatory elements are utilized, interactions were observed between the CFTR promoter, DHS −44 kb and DHS −35 kb, though low or no significant interactions were seen with the intestinal-selective elements such as the enhancer in intron 11 [44,55,74].
An effort to reveal the mechanisms that mediated the formation of these looped chromatin structures at the CFTR locus focused on CTCF and the cohesin complex, which are known to be recruited to the gene [44,50,59]. The function(s) of these factors were examined across the 7q31.2 genomic region encompassing the CFTR locus using a siRNA-mediated depletion approach. Loss of CTCF had a significant impact on the long-range associations between CTCF/cohesin binding sites flanking the locus, but did not alter the interaction of the promoter with intronic cis-elements [74]. In contrast, depletion of RAD21 reduced all the interactions across the locus including those between the promoter, enhancers, and other cis-regulatory elements including CTCF/cohesin bound regions, suggesting cohesin was primarily stabilizing the different sets of chromatin loops [74]. These data suggested that CTCF and cohesin performed distinct functions at the CFTR locus. Recent genome-wide studies confirmed our observations [85] and provided evidence for a CTCF-independent role for cohesin in enhancer-promoter interactions [86]. Specifically, cohesin depletion caused a decrease in intra-chromosomal interactions, while loss of CTCF increased associations between topologically associated domains (TADs) [85], which are large, mega-based sized domains that under normal conditions show lower interaction frequencies with each other [67]. Hence, at the CFTR locus, where CTCF mediates the loops between flanking CTCF/cohesin binding sites, those regions may mark the boundaries of a TAD that isolates the locus for appropriate regulation. Our recent 4C-seq data generated across the CFTR locus support this hypothesis (Yang et al., unpublished). The cohesin complex stabilizes interactions between TSS and cis-regulatory elements such as enhancers at many loci including the CFTR locus. However, despite the changes in the three-dimensional conformation of the locus after depletion of CTCF and/or cohesin, CFTR mRNA and protein levels increased [74]. Further analysis revealed that the increase in expression was associated with alterations in the chromatin landscape, transcription factor occupancy, and nuclear positioning of the CFTR alleles.
The role of the cohesin complex in stabilizing associations between the CFTR promoter and cis-regulatory elements/enhancers implicated transcription factors binding to these regions in the looping mechanism. Since the FOXA1/A2 pioneer factors were identified as the pivotal proteins in the transcriptional complex regulating CFTR intestinal enhancers [52], their role in the three-dimensional organization of CFTR was examined. Combined depletion of both FOXA1 and FOXA2 significantly decreased interactions between the promoter and important intestinal cis-regulatory elements located in introns 10 and 11 [52]. Although there was evidence for an indirect role of the FOXA family members in chromatin looping genome-wide at estrogen receptor binding sites [53,84], the CFTR data may be the first evidence of a direct role of FOXA1/A2 in chromatin organization. Recent work also identified a role for the chromatin remodeler, chromodomain helicase DNA-binding 6 (CHD6), in regulating CFTR expression. CHD6 binds to several cis-regulatory elements at the CFTR locus, including the DHS at intron 1, and interacts with members of the facilitates chromatin transcription (FACT) complex and other transcription factors such as CDX2 [87]. shRNA-mediated depletion of CHD6 decreased CFTR mRNA levels and reduced long-range chromatin looping across the locus, indicating its importance both as an activating factor and a chromatin organizer at the locus. However, in these experiments depletion of CTCF in CF-PAC cells reduced CFTR transcript levels in contrast to the increase we observed in Caco2 cells. This suggests there may be some cell specificity in the role of architectural proteins in organizing the active locus or that the relative abundance of CFTR transcripts in different cell types may be important.
7. Conclusions
In summary, though the CFTR promoter is necessary to drive basal levels of gene expression, cis-elements located elsewhere in the locus are required to confer tissue specificity and control abundance of the transcript. These elements interact directly with the promoter via chromosome looping and perhaps other mechanisms, and recruit diverse cell-selective transcription factors.
The extensive studies carried out over many years, which are discussed in this review, have provided substantial insights into the temporal and tissue-specific regulation of CFTR expression, though our understanding remains incomplete. They also illustrate the critical importance of investigating the locus in differentiated cell types from many lineages, which utilize different transcriptional networks. More recently, the integration of data on cis-regulatory elements, their trans-interacting factors, and the contribution of higher order chromatin structure have illustrated the important role of chromatin dynamics in the regulatory mechanism. Many of the insights gained from CFTR are equally relevant to other disease-associated human genes and coincide with observations from multiple, elegant genome-wide analyses. Future work will identify the specifics of critical cell-type selective transcription factors and regulatory elements in primary cells from the airway, intestinal organoids, sweat gland ducts, and the epididymis, where CFTR expression is required for normal epithelial function. Moreover, the ultimate proof for the importance of these cis-regulatory elements in vivo will require deleting the elements individually and in combination using systems such as CRISPR-Cas9 [88,89] to assess their affect on CFTR expression, its chromatin landscape, and higher order organization. Use of the same technology to modify transcription factor binding sites within individual elements may also identify the proteins critical for maintaining appropriate, tissue-specific levels of CFTR expression (see many online tools for recent CRISPR applications). Finally, it will be important to incorporate our understanding of post-transcriptional mechanisms of CFTR regulation, for example by microRNAs (miRNAs), with knowledge of the critical role of chromatin dynamics at the locus. Though not a focus of this review, CFTR is also subject to direct repression via its 3' UTR by several miRs including miR-145 and miR-494 [90,91,92]. CFTR may also be indirectly regulated by miR-138, which inhibits SIN3A [93], a transcriptional repressor that regulates the gene and alters its chromatin dynamics [56]. Integrating these diverse regulatory mechanisms may ultimately facilitate approaches to manipulate CFTR gene expression for therapeutic benefit.
Acknowledgments
Funding sources: work in the Harris lab is funded in part by the Cystic Fibrosis Foundation and the National Institutes of Health (PI: AH) R01 HL094585 and HD068901.
Author Contributions
Both authors contributed to writing the review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anderson, M.P.; Gregory, R.J.; Thompson, S.; Souza, D.W.; Paul, S.; Mulligan, R.C.; Smith, A.E.; Welsh, M.J. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 1991, 253, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.H.; Rich, D.P.; Marshall, J.; Gregory, R.J.; Welsh, M.J.; Smith, A.E. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell 1991, 66, 1027–1036. [Google Scholar] [CrossRef]
- Anderson, M.P.; Berger, H.A.; Rich, D.P.; Gregory, R.J.; Smith, A.E.; Welsh, M.J. Nucleoside triphosphates are required to open the CFTR chloride channel. Cell 1991, 67, 775–784. [Google Scholar] [CrossRef]
- Rommens, J.M.; Iannuzzi, M.C.; Kerem, B.; Drumm, M.L.; Melmer, G.; Dean, M.; Rozmahel, R.; Cole, J.L.; Kennedy, D.; Hidaka, N.; et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science 1989, 245, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Kerem, B.; Rommens, J.M.; Buchanan, J.A.; Markiewicz, D.; Cox, T.K.; Chakravarti, A.; Buchwald, M.; Tsui, L.C. Identification of the cystic fibrosis gene: Genetic analysis. Science 1989, 245, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Rozmahel, R.; Grzelczak, Z.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.L. Al Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Crawford, I.; Maloney, P.C.; Zeitlin, P.L.; Guggino, W.B.; Hyde, S.C.; Turley, H.; Gatter, K.C.; Harris, A.; Higgins, C.F. Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc. Natl. Acad. Sci. USA 1991, 88, 9262–9266. [Google Scholar] [CrossRef] [PubMed]
- Trezise, A.E.; Buchwald, M. In vivo cell-specific expression of the cystic fibrosis transmembrane conductance regulator. Nature 1991, 353, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, B.C.; Chu, C.S.; Paakko, P.K.; Banks, T.C.; Yoshimura, K.; Ferrans, V.J.; Chernick, M.S.; Crystal, R.G. Expression of the cystic fibrosis transmembrane conductance regulator gene in the respiratory tract of normal individuals and individuals with cystic fibrosis. Proc. Natl. Acad. Sci. USA 1991, 88, 6565–6569. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, J.F.; Yankaskas, J.R.; Ernst, S.A.; Yang, Y.; Marino, C.R.; Boucher, R.C.; Cohn, J.A.; Wilson, J.M. Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat. Genet. 1992, 2, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.; Chalkley, G.; Goodman, S.; Coleman, L. Expression of the cystic fibrosis gene in human development. Dev. Camb. Engl. 1991, 113, 305–310. [Google Scholar]
- Trezise, A.E.; Chambers, J.A.; Wardle, C.J.; Gould, S.; Harris, A. Expression of the cystic fibrosis gene in human foetal tissues. Hum. Mol. Genet. 1993, 2, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, J.F.; Zepeda, M.; Cohn, J.A.; Yankaskas, J.R.; Wilson, J.M. Expression of the cystic fibrosis gene in adult human lung. J. Clin. Investig. 1994, 93, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Broackes-Carter, F.C.; Mouchel, N.; Gill, D.; Hyde, S.; Bassett, J.; Harris, A. Temporal regulation of CFTR expression during ovine lung development: Implications for CF gene therapy. Hum. Mol. Genet. 2002, 11, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.N.; Barth, M.L.; McDowell, T.L.; Moulin, D.S.; Nuthall, H.N.; Hollingsworth, M.A.; Harris, A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996, 271, 9947–9954. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Nakamura, H.; Trapnell, B.C.; Dalemans, W.; Pavirani, A.; Lecocq, J.P.; Crystal, R.G. The cystic fibrosis gene has a “housekeeping”-type promoter and is expressed at low levels in cells of epithelial origin. J. Biol. Chem. 1991, 266, 9140–9144. [Google Scholar] [PubMed]
- Chou, J.L.; Rozmahel, R.; Tsui, L.C. Characterization of the promoter region of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1991, 266, 24471–24476. [Google Scholar] [PubMed]
- Koh, J.; Sferra, T.J.; Collins, F.S. Characterization of the cystic fibrosis transmembrane conductance regulator promoter region. Chromatin context and tissue-specificity. J. Biol. Chem. 1993, 268, 15912–15921. [Google Scholar] [PubMed]
- White, N.L.; Higgins, C.F.; Trezise, A.E. Tissue-specific in vivo transcription start sites of the human and murine cystic fibrosis genes. Hum. Mol. Genet. 1998, 7, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Mouchel, N.; Broackes-Carter, F.; Harris, A. Alternative 5' exons of the CFTR gene show developmental regulation. Hum. Mol. Genet. 2003, 12, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.A.; Costa, F.F.; Bischof, J.M.; Williams, S.H.; Soares, M.B.; Harris, A. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am. J. Respir. Cell Mol. Biol. 2010, 43, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Lukowski, S.W.; Rothnagel, J.A.; Trezise, A.E.O. CFTR mRNA expression is regulated by an upstream open reading frame and RNA secondary structure in its 5' untranslated region. Hum. Mol. Genet. 2015, 24, 899–912. [Google Scholar] [CrossRef] [PubMed]
- McDonald, R.A.; Matthews, R.P.; Idzerda, R.L.; McKnight, G.S. Basal expression of the cystic fibrosis transmembrane conductance regulator gene is dependent on protein kinase A activity. Proc. Natl. Acad. Sci. USA 1995, 92, 7560–7564. [Google Scholar] [CrossRef] [PubMed]
- Pittman, N.; Shue, G.; LeLeiko, N.S.; Walsh, M.J. Transcription of cystic fibrosis transmembrane conductance regulator requires a CCAAT-like element for both basal and cAMP-mediated regulation. J. Biol. Chem. 1995, 270, 28848–28857. [Google Scholar] [CrossRef] [PubMed]
- Matthews, R.P.; McKnight, G.S. Characterization of the cAMP response element of the cystic fibrosis transmembrane conductance regulator gene promoter. J. Biol. Chem. 1996, 271, 31869–31877. [Google Scholar] [CrossRef] [PubMed]
- Cafferata, E.G.; Guerrico, A.M.; Pivetta, O.H.; Santa-Coloma, T.A. NF-kappaB activation is involved in regulation of cystic fibrosis transmembrane conductance regulator (CFTR) by interleukin-1beta. J. Biol. Chem. 2001, 276, 15441–15444. [Google Scholar] [CrossRef] [PubMed]
- Brouillard, F.; Bouthier, M.; Leclerc, T.; Clement, A.; Baudouin-Legros, M.; Edelman, A. NF-κB mediates up-regulation of CFTR gene expression in calu-3 cells by interleukin-1β. J. Biol. Chem. 2001, 276, 9486–9491. [Google Scholar] [CrossRef] [PubMed]
- Crawford, G.E.; Davis, S.; Scacheri, P.C.; Renaud, G.; Halawi, M.J.; Erdos, M.R.; Green, R.; Meltzer, P.S.; Wolfsberg, T.G.; Collins, F.S. DNase-chip: A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods 2006, 3, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Crawford, G.E. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010. [Google Scholar] [CrossRef] [PubMed]
- Gross, D.S.; Garrard, W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988, 57, 159–197. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.N.; Wardle, C.J.; Harris, A. Characterization of DNASE I hypersensitive sites in the 120 kb 5' to the CFTR gene. Biochem. Biophys. Res. Commun. 1995, 211, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Nuthall, H.N.; Majetti, M.E.; Harris, A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics 2000, 64, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Nuthall, H.N.; Moulin, D.S.; Huxley, C.; Harris, A. Analysis of DNase-I-hypersensitive sites at the 3' end of the cystic fibrosis transmembrane conductance regulator gene (CFTR). Biochem. J. 1999, 341, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Phylactides, M.; Rowntree, R.; Nuthall, H.; Ussery, D.; Wheeler, A.; Harris, A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur. J. Biochem. 2002, 269, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Moulin, D.S.; Manson, A.L.; Nuthall, H.N.; Smith, D.J.; Huxley, C.; Harris, A. In vivo analysis of DNase I hypersensitive sites in the human CFTR gene. Mol. Med. 1999, 5, 211–223. [Google Scholar] [PubMed]
- Nuthall, H.N.; Vassaux, G.; Huxley, C.; Harris, A. Analysis of a DNase I hypersensitive site located −20.9 kb upstream of the CFTR gene. Eur. J. Biochem. 1999, 266, 431–443. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, V.A.; Ott, C.J.; Phylactides, M.; Harris, A. Interaction of intestinal and pancreatic transcription factors in the regulation of CFTR gene expression. Biochim. Biophys. Acta 2009, 1789, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Rowntree, R.K.; Vassaux, G.; McDowell, T.L.; Howe, S.; McGuigan, A.; Phylactides, M.; Huxley, C.; Harris, A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001, 10, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Mouchel, N.; Henstra, S.A.; McCarthy, V.A.; Williams, S.H.; Phylactides, M.; Harris, A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem. J. 2004, 378, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Ott, C.J.; Suszko, M.; Blackledge, N.P.; Wright, J.E.; Crawford, G.E.; Harris, A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell. Mol. Med. 2009, 13, 680–692. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Zentner, G.E.; Scacheri, P.C. The Chromatin Fingerprint of Gene Enhancer Elements. J. Biol. Chem. 2012, 287, 30888–30896. [Google Scholar] [CrossRef] [PubMed]
- Ott, C.J.; Blackledge, N.P.; Kerschner, J.L.; Leir, S.-H.; Crawford, G.E.; Cotton, C.U.; Harris, A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. USA 2009, 106, 19934–19939. [Google Scholar] [CrossRef] [PubMed]
- Valouev, A.; Johnson, S.M.; Boyd, S.D.; Smith, C.L.; Fire, A.Z.; Sidow, A. Determinants of nucleosome organization in primary human cells. Nature 2011, 474, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Yigit, E.; Bischof, J.M.; Zhang, Z.; Ott, C.J.; Kerschner, J.L.; Leir, S.-H.; Buitrago-Delgado, E.; Zhang, Q.; Wang, J.-P.Z.; Widom, J.; et al. Nucleosome mapping across the CFTR locus identifies novel regulatory factors. Nucleic Acids Res. 2013, 41, 2857–2868. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011, 470, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Carter, E.J.; Evans, J.R.; Lawson, V.; Rowntree, R.K.; Harris, A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007, 408, 267–275. [Google Scholar] [PubMed]
- Kerschner, J.L.; Harris, A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem. J. 2012, 446, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Kerschner, J.L.; Gosalia, N.; Leir, S.-H.; Harris, A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics 2014, 9, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.S.; Liu, X.S.; Brodsky, A.S.; Li, W.; Meyer, C.A.; Szary, A.J.; Eeckhoute, J.; Shao, W.; Hestermann, E.V.; Geistlinger, T.R.; et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005, 122, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Lupien, M.; Eeckhoute, J.; Meyer, C.A.; Wang, Q.; Zhang, Y.; Li, W.; Carroll, J.S.; Liu, X.S.; Brown, M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008, 132, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ott, C.J.; Lewandowska, M.A.; Leir, S.-H.; Harris, A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012, 16, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Leir, S.-H.; Harris, A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol. Cell. Biol. 2013, 33, 2843–2853. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Leir, S.-H.; Harris, A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am. J. Respir. Cell Mol. Biol. 2015, 52, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Viart, V.; Bergougnoux, A.; Bonini, J.; Varilh, J.; Chiron, R.; Tabary, O.; Molinari, N.; Claustres, M.; Taulan-Cadars, M. Transcription factors and miRNAs that regulate fetal to adult CFTR expression change are new targets for cystic fibrosis. Eur. Respir. J. 2015, 45, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Ott, C.J.; Gillen, A.E.; Harris, A. An insulator element 3' to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009, 37, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Cremins, J.E.; Corces, V.G. Chromatin insulators: Linking genome organization to cellular function. Mol. Cell 2013, 50, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Rajkovic, A.; Viveiros, M.M.; Burns, K.H.; Eppig, J.J.; Matzuk, M.M. Identification of Gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-alpha motif, and a basic leucine zipper. Mol. Endocrinol. 2002, 16, 1168–1184. [Google Scholar] [PubMed]
- Cheung, J.; Petek, E.; Nakabayashi, K.; Tsui, L.C.; Vincent, J.B.; Scherer, S.W. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics 2001, 78, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Shulha, H.P.; Lin, J.M.; Vales, T.R.; Fu, Y.; Bodine, D.M.; McKay, R.D.G.; Chenoweth, J.G.; Tesar, P.J.; Furey, T.S.; et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007, 3, e136. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, R.; Renkawitz, R.; Lobanenkov, V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001, 17, 520–527. [Google Scholar] [CrossRef]
- Bell, A.C.; West, A.G.; Felsenfeld, G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999, 98, 387–396. [Google Scholar] [CrossRef]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.-T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Wendt, K.S.; Yoshida, K.; Itoh, T.; Bando, M.; Koch, B.; Schirghuber, E.; Tsutsumi, S.; Nagae, G.; Ishihara, K.; Mishiro, T.; et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008, 451, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Rubio, E.D.; Reiss, D.J.; Welcsh, P.L.; Disteche, C.M.; Filippova, G.N.; Baliga, N.S.; Aebersold, R.; Ranish, J.A.; Krumm, A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. USA 2008, 105, 8309–8314. [Google Scholar] [CrossRef] [PubMed]
- Parelho, V.; Hadjur, S.; Spivakov, M.; Leleu, M.; Sauer, S.; Gregson, H.C.; Jarmuz, A.; Canzonetta, C.; Webster, Z.; Nesterova, T.; et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008, 132, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Haering, C.H. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef] [PubMed]
- Horsfield, J.A.; Print, C.G.; Mönnich, M. Diverse developmental disorders from the one ring: Distinct molecular pathways underlie the cohesinopathies. Front. Genet. 2012, 3, 171. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, N.; Smith, E.M.; Tabuchi, T.M.; Koch, C.M.; Dunham, I.; Stamatoyannopoulos, J.A.; Dekker, J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010, 38, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Gosalia, N.; Neems, D.; Kerschner, J.L.; Kosak, S.T.; Harris, A. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014, 42, 9612–9622. [Google Scholar] [CrossRef] [PubMed]
- Tolhuis, B.; Palstra, R.J.; Splinter, E.; Grosveld, F.; de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 2002, 10, 1453–1465. [Google Scholar] [CrossRef]
- Ragoczy, T.; Telling, A.; Sawado, T.; Groudine, M.; Kosak, S.T. A genetic analysis of chromosome territory looping: Diverse roles for distal regulatory elements. Chromosome Res. 2003, 11, 513–525. [Google Scholar] [CrossRef] [PubMed]
- De Laat, W.; Grosveld, F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003, 11, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.-T.; Corces, V.G. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011, 12, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The long-range interaction landscape of gene promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing chromosome conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Splinter, E.; de Wit, E.; van de Werken, H.J.G.; Klous, P.; de Laat, W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods 2012, 58, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Fullwood, M.J.; Liu, M.H.; Pan, Y.F.; Liu, J.; Xu, H.; Mohamed, Y.B.; Orlov, Y.L.; Velkov, S.; Ho, A.; Mei, P.H.; et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 2009, 462, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Zuin, J.; Dixon, J.R.; van der Reijden, M.I.J.A.; Ye, Z.; Kolovos, P.; Brouwer, R.W.W.; van de Corput, M.P.C.; van de Werken, H.J.G.; Knoch, T.A.; van IJcken, W.F.J.; et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Chen, C.; Teng, L.; Tan, K. Global view of enhancer-promoter interactome in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, E2191–E2199. [Google Scholar] [CrossRef] [PubMed]
- Sancho, A.; Li, S.; Paul, T.; Zhang, F.; Aguilo, F.; Vashisht, A.; Balasubramaniyan, N.; Leleiko, N.S.; Suchy, F.J.; Wohlschlegel, J.A.; et al. CHD6 regulates the topological arrangement of the CFTR locus. Hum. Mol. Genet. 2015, 24, 2724–2732. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; Zhang, F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Gillen, A.E.; Gosalia, N.; Leir, S.-H.; Harris, A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 2011, 438, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Megiorni, F.; Cialfi, S.; Dominici, C.; Quattrucci, S.; Pizzuti, A. Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS ONE 2011, 6, e26601. [Google Scholar] [CrossRef] [PubMed]
- Hassan, F.; Nuovo, G.J.; Crawford, M.; Boyaka, P.N.; Kirkby, S.; Nana-Sinkam, S.P.; Cormet-Boyaka, E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE 2012, 7, e50837. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Karp, P.H.; Jiang, P.; Ostedgaard, L.S.; Walz, A.E.; Fisher, J.T.; Keshavjee, S.; Lennox, K.A.; Jacobi, A.M.; Rose, S.D.; et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. USA 2012, 109, 13362–13367. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).