One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation
Abstract
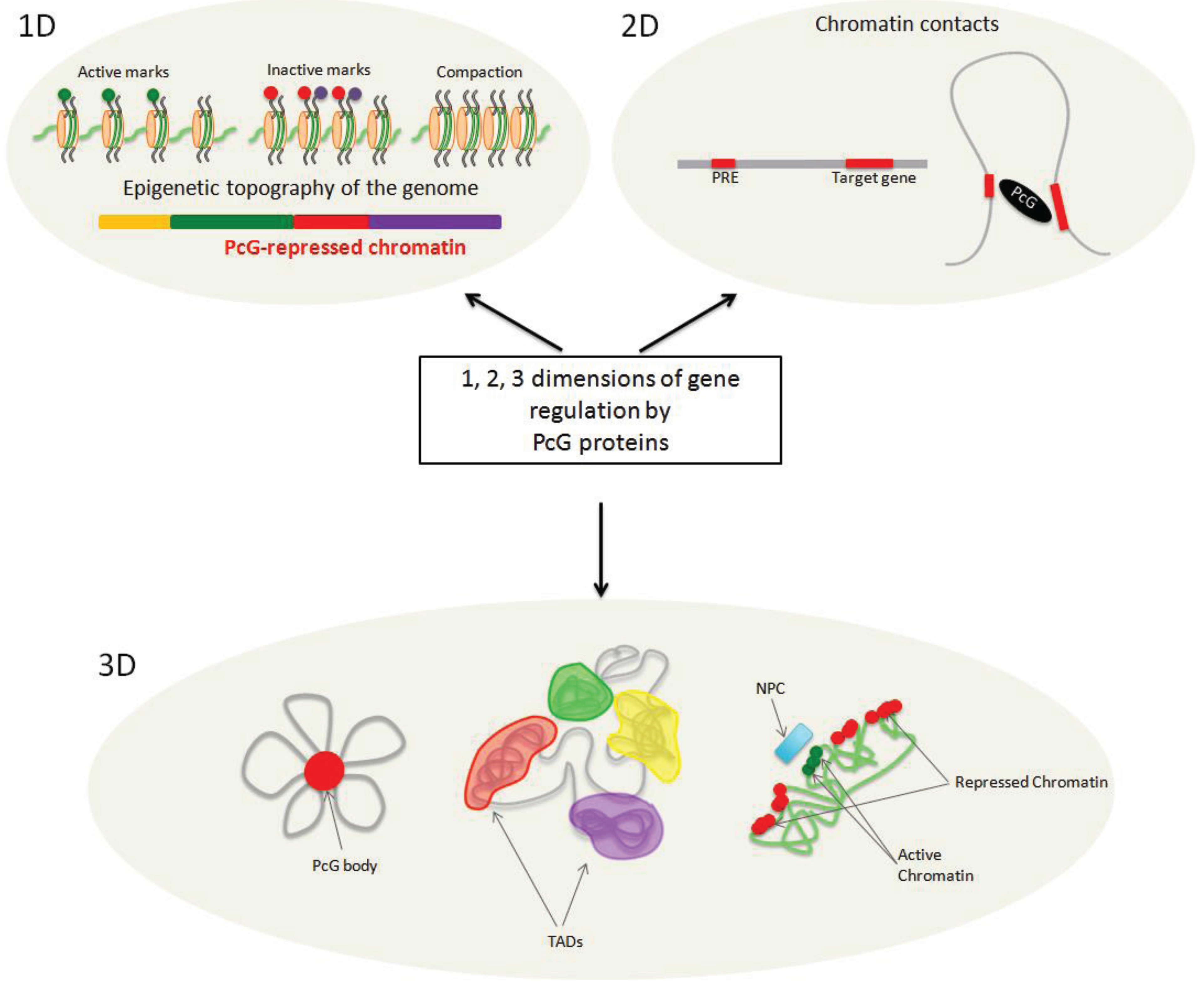
:1. Introduction
2. Hierarchy vs. Plasticity for More Flexibility in Repression?
2.1. Canonical Two-Step Mechanism of Repression
2.2. Non-Canonical Mechanisms of Repression
3. Various Ways to Hunt for Targets
4. On the Path from 1D to 3D
4.1. Never Walk Alone—Polycomb and Chromatin Domains
4.2. PcG and Chromatin Fiber Packaging
4.3. It Takes Two to Tango—Chromatin Loops Mediated by PcG Proteins
5. In the Third Dimension: Polycomb Mediates Higher Order Chromatin Organization
5.1. Polycomb and Topologically Associating Domains
5.2. Polycomb Clustering
5.3. Polycomb Regulation and Spatial Distribution
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lewis, E.B. New mutants: Reports of P. lewis. Drosoph. Inf. Serv. 1947, 21, 69. [Google Scholar]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.; Villar, C.B. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008, 18, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Pirrotta, V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013, 14, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Pu, L.; Sung, Z.R. PcG and trxG in plants—Friends or foes. Trends Genet. 2015, 31, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Wagner, D. Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 2015, 23, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, F.; Gaudin, V. Epigenetic control by plant Polycomb proteins: New perspectives and emerging roles in stress response. In From Plant Genomics to Plant Biotechnology; Palmiro Poltronieri, N.B., Corrado, F., Eds.; Woodhead Publishing, Elsevier: Cambridge, UK, 2013; pp. 31–48. [Google Scholar]
- Kleinmanns, J.A.; Schubert, D. Polycomb and Trithorax group protein-mediated control of stress responses in plants. Biol. Chem. 2014, 395, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Molitor, A.; Shen, W.H. The polycomb complex PRC1: Composition and function in plants. J. Genet. Genomics 2013, 40, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Bemer, M.; Grossniklaus, U. Dynamic regulation of Polycomb group activity during plant development. Curr. Opin. Plant Biol. 2012, 15, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Pien, S.; Grossniklaus, U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta 2007, 1769, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, V.; Libault, M.; Pouteau, S.; Juul, T.; Zhao, G.; Lefebvre, D.; Grandjean, O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 2001, 128, 4847–4858. [Google Scholar] [PubMed]
- Zhang, X.; Germann, S.; Blus, B.J.; Khorasanizadeh, S.; Gaudin, V.; Jacobsen, S.E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007, 14, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Bratzel, F.; Lopez-Torrejon, G.; Koch, M.; del Pozo, J.C.; Calonje, M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 2010, 20, 1853–1859. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Lagarou, A.; Mohd-Sarip, A.; Moshkin, Y.M.; Chalkley, G.E.; Bezstarosti, K.; Demmers, J.A.; Verrijzer, C.P. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008, 22, 2799–2810. [Google Scholar] [CrossRef] [PubMed]
- Beh, L.Y.; Colwell, L.J.; Francis, N.J. A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc. Natl. Acad. Sci. USA 2012, 109, E1063–E1071. [Google Scholar] [CrossRef] [PubMed]
- Bantignies, F.; Cavalli, G. Polycomb group proteins: Repression in 3D. Trends Genet. 2011, 27, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin compaction by a polycomb group protein complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Cavalli, G. Polycomb domain formation depends on short and long distance regulatory cues. PLoS ONE 2013, 8, e56531. [Google Scholar] [CrossRef] [PubMed]
- Ringrose, L.; Paro, R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Ann. Rev. Genet. 2004, 38, 413–443. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, E.J.; Dennis, E.S. Vernalization-Induced Trimethylation of Histone H3 Lysine 27 at FLC Is Not Maintained in Mitotically Quiescent Cells. Curr. Biol. 2007, 17, 1978–1983. [Google Scholar] [CrossRef] [PubMed]
- De Lucia, F.; Crevillen, P.; Jones, A.M.; Greb, T.; Dean, C. A PHD-Polycomb Repressive Complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 16831–16836. [Google Scholar] [CrossRef] [PubMed]
- Pinter, S.F.; Sadreyev, R.I.; Yildirim, E.; Jeon, Y.; Ohsumi, T.K.; Borowsky, M.; Lee, J.T. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012, 22, 1864–1876. [Google Scholar] [CrossRef] [PubMed]
- Dorighi, K.M.; Tamkun, J.W. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 2013, 140, 4182–4192. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Dorighi, K.M.; Tamkun, J.W. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008, 4, e1000217. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Kohler, C.; Hennig, L. Keeping the gate closed: Functions of the Polycomb repressive complex PRC2 in development. Plant J. 2015, 83, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Hu, Y.; Belmont, A.S. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 2014, 24, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Lee, A.J.; Roh, T.Y. Polycomb group protein-mediated histone modifications during cell differentiation. Epigenomics 2015, 7, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, Y.; Zhou, D.X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 2011, 1809, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Gearhart, M.D.; Corcoran, C.M.; Wamstad, J.A.; Bardwell, V.J. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 2006, 26, 6880–6889. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Calonje, M.; Sanchez, R.; Chen, L.; Sung, Z.R. EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 2008, 20, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Ishiguro, K.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; O’Loghlen, A. PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 2014, 24, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 2002, 22, 6070–6078. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 2012. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, J.C.; Gutierrez, L.; Muller, J. Histone H2A monoubiquitination and Polycomb repression: The missing pieces of the puzzle. Fly 2012, 6, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Klymenko, T.; Papp, B.; Fischle, W.; Kocher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Muller, J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Pirrotta, V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008, 20, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Sengupta, A.K.; Kubicek, S.; Mechtler, K.; Spahn, L.; Koseki, H.; Jenuwein, T.; Wutz, A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006, 25, 3110–3122. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Muller, C.W.; Vermeulen, M.; Muller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Dienstbier, M.; Hassan, R.; Schermelleh, L.; Sharif, J.; Blackledge, N.P.; de Marco, V.; Elderkin, S.; Koseki, H.; Klose, R.; et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014, 7, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sung, S. Polycomb-mediated gene silencing in Arabidopsis thaliana. Mol. Cells 2014, 37, 841–850. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Huang, H.; Xu, L. Mechanisms guiding Polycomb activities during gene silencing in Arabidopsis thaliana. Front. Plant Sci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kassis, J.A.; Brown, J.L. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 2013, 81, 83–118. [Google Scholar] [PubMed]
- Ku, M.; Koche, R.P.; Rheinbay, E.; Mendenhall, E.M.; Endoh, M.; Mikkelsen, T.S.; Presser, A.; Nusbaum, C.; Xie, X.; Chi, A.S.; et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008, 4, e1000242. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Buzas, D.M.; Ying, H.; Robertson, M.; Taylor, J.; Peacock, W.J.; Dennis, E.S.; Helliwell, C. Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genomics 2013, 14, 593. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.M.; Austin, R.S.; Blanvillain-Baufume, S.; Reback, M.A.; Monniaux, M.; Wu, M.F.; Sang, Y.; Yamaguchi, A.; Yamaguchi, N.; Parker, J.E.; et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 2011, 20, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-binding factor BPC6 recruits PRC1 component LHP1 to GAGA DNA-motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Berger, N.; Dubreucq, B.; Roudier, F.; Dubos, C.; Lepiniec, L. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 2011, 23, 4065–4078. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Yun, H.; Park, K.; Ohr, H.; Lee, O.; Kim, D.H.; Sung, S.; Choi, Y. The catalytic subunit of Arabidopsis DNA polymerase alpha ensures stable maintenance of histone modification. Development 2013, 140, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS Terminates Floral Stem Cell Maintenance in Arabidopsis by Directly Repressing WUSCHEL through Recruitment of Polycomb Group Proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Benfey, P.N. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009, 58, 1016–1027. [Google Scholar] [CrossRef] [PubMed]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Marchese, F.P.; Huarte, M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 2013, 191, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Nazer, E.; Lei, E.P. Modulation of chromatin modifying complexes by noncoding RNAs in trans. Curr. Opin. Genet. Dev. 2014, 25, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef] [PubMed]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Marquardt, S.; Lister, C.; Swiezewski, S.; Dean, C. Targeted 3' processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 2010, 327, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Latrasse, D.; Germann, S.; Houba-Herin, N.; Dubois, E.; Bui-Prodhomme, D.; Hourcade, D.; Juul-Jensen, T.; le Roux, C.; Majira, A.; Simoncello, N.; et al. Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS ONE 2011, 6, e16592. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Wang, X.; Cifuentes-Rojas, C.; Goodrich, K.J.; Gooding, A.R.; Lee, J.T.; Cech, T.R. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 2015, 57, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Wu, T.; Fu, H.; Dai, C.; Wu, H.; Liu, N.; Li, X.; Xu, M.; Zhang, Z.; Niu, T.; et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 2012, 337, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Dietrich, N.; Pasini, D.; Hansen, K.H.; Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006, 20, 1123–1136. [Google Scholar] [CrossRef] [PubMed]
- Tolhuis, B.; de Wit, E.; Muijrers, I.; Teunissen, H.; Talhout, W.; van Steensel, B.; van Lohuizen, M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006, 38, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Bouyer, D.; Roudier, F.; Heese, M.; Andersen, E.D.; Gey, D.; Nowack, M.K.; Goodrich, J.; Renou, J.P.; Grini, P.E.; Colot, V.; et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011, 7, e1002014. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Park, S.; van Nocker, S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008, 4, e1000077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sridhar, V.V.; Zhu, J.; Kapoor, A.; Zhu, J.K. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2007, 2, e1210. [Google Scholar] [CrossRef] [PubMed]
- Lafos, M.; Kroll, P.; Hohenstatt, M.L.; Thorpe, F.L.; Clarenz, O.; Schubert, D. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011, 7, e1002040. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Weber, M.; Rebhan, M.; Roloff, T.C.; Richter, J.; Stadler, M.B.; Bibel, M.; Schubeler, D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 2008, 30, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Filion, G.J.; van Bemmel, J.G.; Braunschweig, U.; Talhout, W.; Kind, J.; Ward, L.D.; Brugman, W.; de Castro, I.J.; Kerkhoven, R.M.; Bussemaker, H.J.; et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 2010, 143, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kheradpour, P.; Mikkelsen, T.S.; Shoresh, N.; Ward, L.D.; Epstein, C.B.; Zhang, X.; Wang, L.; Issner, R.; Coyne, M.; et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011, 473, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sequeira-Mendes, J.; Araguez, I.; Peiro, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 2014, 26, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Roudier, F.; Ahmed, I.; Berard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Francis, N.J.; Saurin, A.J.; Shao, Z.; Kingston, R.E. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 2001, 8, 545–556. [Google Scholar] [CrossRef]
- Grau, D.J.; Chapman, B.A.; Garlick, J.D.; Borowsky, M.; Francis, N.J.; Kingston, R.E. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011, 25, 2210–2221. [Google Scholar] [CrossRef] [PubMed]
- Eskeland, R.; Leeb, M.; Grimes, G.R.; Kress, C.; Boyle, S.; Sproul, D.; Gilbert, N.; Fan, Y.; Skoultchi, A.I.; Wutz, A.; et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 2010, 38, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Wildhaber, T.; Siretskiy, A.; Gruissem, W.; Hennig, L. Distinct modes of DNA accessibility in plant chromatin. Nat. Commun. 2012. [Google Scholar] [CrossRef] [PubMed]
- Bell, O.; Schwaiger, M.; Oakeley, E.J.; Lienert, F.; Beisel, C.; Stadler, M.B.; Schubeler, D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat. Struct. Mol. Biol. 2010, 17, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Chang, E.Y.; Shankar, S.; Kuchenbecker, K.M.; Simon, M.D.; Madhani, H.D.; Narlikar, G.J.; Al-Sady, B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 2011, 41, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Liao, M.; Naber, N.; Pate, E.; Larson, A.; Wu, S.; Marina, D.B.; Garcia, J.F.; Madhani, H.D.; Cooke, R.; et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 2013, 496, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Teif, V.B.; Kepper, N.; Yserentant, K.; Wedemann, G.; Rippe, K. Affinity, stoichiometry and cooperativity of heterochromatin protein 1 (HP1) binding to nucleosomal arrays. J. Phys. Condens. Matter 2015. [Google Scholar] [CrossRef] [PubMed]
- Blein, J.P.; Coutos-Thevenot, P.; Marion, D.; Ponchet, M. From elicitins to lipid-transfer proteins: A new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci. 2002, 7, 293–296. [Google Scholar] [CrossRef]
- Cheutin, T.; Cavalli, G. Polycomb silencing: From linear chromatin domains to 3D chromosome folding. Curr. Opin. Genet. Dev. 2014, 25, 30–37. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; de Laat, W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012, 26, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Hovel, I.; Louwers, M.; Stam, M. 3C technologies in plants. Methods 2012, 58, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Arpon, J.; Sakai, K.; Andrey, P.; Gaudin, V. Nuclear architecture and chromatin dynamics in interphase nuclei of Arabidopsis thaliana. Cytogenet. Genome Res. 2014, 143, 28–50. [Google Scholar] [CrossRef] [PubMed]
- Lanzuolo, C.; Roure, V.; Dekker, J.; Bantignies, F.; Orlando, V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 2007, 9, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.K.; McGarvey, K.M.; Licchesi, J.D.; Ohm, J.E.; Herman, J.G.; Schubeler, D.; Baylin, S.B. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008, 6, 2911–2927. [Google Scholar] [CrossRef] [PubMed]
- Williamson, I.; Berlivet, S.; Eskeland, R.; Boyle, S.; Illingworth, R.S.; Paquette, D.; Dostie, J.; Bickmore, W.A. Spatial genome organization: Contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014, 28, 2778–2791. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Schuettengruber, B.; Sexton, T.; Cavalli, G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl. Acad. Sci. USA 2011, 108, 2294–2299. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Ohno, K.; Gui, H.; Pirrotta, V. Insulators target active genes to transcription factories and polycomb-repressed genes to Polycomb bodies. PLoS Genet. 2013, 9, e1003436. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Rosa, S.; Dean, C. Nuclear organization changes and the epigenetic silencing of FLC during vernalization. J. Mol. Biol. 2014, 427, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Louwers, M.; Bader, R.; Haring, M.; van Driel, R.; de Laat, W.; Stam, M. Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 2009, 21, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Crevillen, P.; Sonmez, C.; Wu, Z.; Dean, C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013, 32, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.; de Lucia, F.; Mylne, J.S.; Zhu, D.; Ohmido, N.; Pendle, A.; Kato, N.; Shaw, P.; Dean, C. Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev. 2013, 27, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Tolhuis, B.; Blom, M.; Kerkhoven, R.M.; Pagie, L.; Teunissen, H.; Nieuwland, M.; Simonis, M.; de Laat, W.; van Lohuizen, M.; van Steensel, B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011, 7, e1001343. [Google Scholar] [CrossRef] [PubMed]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006, 38, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.; Holbro, N.; Reichert, H. Polycomb group genes are required for neural stem cell survival in postembryonic neurogenesis of Drosophila. Development 2007, 134, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Ciabrelli, F.; Cavalli, G. Chromatin-driven behavior of topologically associating domains. J. Mol. Biol. 2014, 427, 608–625. [Google Scholar] [CrossRef] [PubMed]
- Bantignies, F.; Roure, V.; Comet, I.; Leblanc, B.; Schuettengruber, B.; Bonnet, J.; Tixier, V.; Mas, A.; Cavalli, G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011, 144, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Li, L.; Qin, Z.S.; Corces, V.G. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 2012, 48, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Witt, H.; Katsumura, K.; Ye, Z.; Wang, Q.; Bresnick, E.H.; Farnham, P.J.; Jin, V.X. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res. 2012, 40, 7690–7704. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, D.; Leleu, M.; Splinter, E.; Rougemont, J.; de Laat, W.; Duboule, D. The dynamic architecture of Hox gene clusters. Science 2011, 334, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cokus, S.J.; Schubert, V.; Zhai, J.; Pellegrini, M.; Jacobsen, S.E. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 2014, 55, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, C.; Roqueiro, D.; Grimm, D.; Schwab, R.; Becker, C.; Lanz, C.; Weigel, D. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015, 25, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Grob, S.; Schmid, M.W.; Grossniklaus, U. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 2014, 55, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Moissiard, G.; Cokus, S.J.; Cary, J.; Feng, S.; Billi, A.C.; Stroud, H.; Husmann, D.; Zhan, Y.; Lajoie, B.R.; McCord, R.P.; et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 2012, 336, 1448–1451. [Google Scholar] [CrossRef] [PubMed]
- Grimaud, C.; Bantignies, F.; Pal-Bhadra, M.; Ghana, P.; Bhadra, U.; Cavalli, G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 2006, 124, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Vincenz, C.; Kerppola, T.K. Changes in the distributions and dynamics of Polycomb repressive complexes during embryonic stem cell differentiation. Mol. Cell. Biol. 2008, 28, 2884–2895. [Google Scholar] [CrossRef] [PubMed]
- Saurin, A.J.; Shiels, C.; Williamson, J.; Satijn, D.P.; Otte, A.P.; Sheer, D.; Freemont, P.S. The human Polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 1998, 142, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Vandenbunder, B.; Fourre, N.; Leray, A.; Mueller, F.; Volkel, P.; Angrand, P.O.; Heliot, L. PRC1 components exhibit different binding kinetics in Polycomb bodies. Biol. Cell 2014, 106, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.; Mateos-Langerak, J.; Thomas, A.; Cheutin, T.; Cavalli, G. Identification of regulators of the three-dimensional Polycomb organization by a microscopy-based genome-wide RNAi screen. Mol. Cell 2014, 54, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Muller, J. O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev. Cell 2014, 31, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Tessadori, F.; Germann, S.; Snijder, B.; Fransz, P.; Gaudin, V. The Arabidopsis LHP1 protein is a component of euchromatin. Planta 2005, 222, 910–925. [Google Scholar] [CrossRef] [PubMed]
- Parry, G. The plant nuclear envelope and regulation of gene expression. J. Exp. Bot. 2015, 66, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Goto, C.; Hara-Nishimura, I. Recent advances in understanding plant nuclear envelope proteins involved in nuclear morphology. J. Exp. Bot. 2015, 66, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Groves, N.R.; Meier, I. Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 2015, 6, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000, 18, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Germann, S.; Gaudin, V. Mapping in vivo protein-DNA interactions in plants by DamID, a DNA adenine methylation-based method. Methods Mol. Biol. 2011, 754, 307–321. [Google Scholar] [PubMed]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.; Solovei, I.; Brugman, W.; Graf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Egelhofer, T.A.; Strome, S.; Lieb, J.D. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Filion, G.J.; van Steensel, B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat. Genet. 2010. [Google Scholar] [CrossRef] [PubMed]
- Shevelyov, Y.Y.; Lavrov, S.A.; Mikhaylova, L.M.; Nurminsky, I.D.; Kulathinal, R.J.; Egorova, K.S.; Rozovsky, Y.M.; Nurminsky, D.I. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sc.i USA 2009, 106, 3282–3287. [Google Scholar] [CrossRef] [PubMed]
- Towbin, B.D.; Meister, P.; Pike, B.L.; Gasser, S.M. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Andrulis, E.D.; Neiman, A.M.; Zappulla, D.C.; Sternglanz, R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 1998, 394, 592–595. [Google Scholar] [PubMed]
- Dialynas, G.; Speese, S.; Budnik, V.; Geyer, P.K.; Wallrath, L.L. The role of Drosophila Lamin C in muscle function and gene expression. Development 2010, 137, 3067–3077. [Google Scholar] [CrossRef] [PubMed]
- Finlan, L.E.; Sproul, D.; Thomson, I.; Boyle, S.; Kerr, E.; Perry, P.; Ylstra, B.; Chubb, J.R.; Bickmore, W.A. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008, 4, e1000039. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.L.; Zullo, J.M.; Bertolino, E.; Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008, 452, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Casolari, J.M.; Brown, C.R.; Drubin, D.A.; Rando, O.J.; Silver, P.A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005, 19, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Horn, P.J.; Peterson, C.L. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007, 21, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Arib, G.; Laemmli, C.; Nishikawa, J.; Durussel, T.; Laemmli, U.K. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Mol. Cell 2006, 21, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Brickner, J.H.; Walter, P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004, 2, e342. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kumar, R.M.; Biggs, V.J.; Lee, H.; Chen, Y.; Kagey, M.H.; Young, R.A.; Abate-Shen, C. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev. Cell 2011, 21, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Fedorova, E.; Sadoni, N.; Dahlsveen, I.K.; Koch, J.; Kremmer, E.; Eick, D.; Paro, R.; Zink, D. The nuclear organization of Polycomb/Trithorax group response elements in larval tissues of Drosophila melanogaster. Chromosome Res. 2008, 16, 649–673. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.M.; Qiu, Y.; van Buskirk, E.K.; Yang, E.J.; Chen, M. Light-regulated gene repositioning in Arabidopsis. Nat. Commun. 2014. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Park, C.M. HOS1-mediated activation of FLC via chromatin remodeling under cold stress. Plant Signal. Behav. 2013. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Prete, S.; Mikulski, P.; Schubert, D.; Gaudin, V. One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation. Genes 2015, 6, 520-542. https://doi.org/10.3390/genes6030520
Del Prete S, Mikulski P, Schubert D, Gaudin V. One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation. Genes. 2015; 6(3):520-542. https://doi.org/10.3390/genes6030520
Chicago/Turabian StyleDel Prete, Stefania, Pawel Mikulski, Daniel Schubert, and Valérie Gaudin. 2015. "One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation" Genes 6, no. 3: 520-542. https://doi.org/10.3390/genes6030520
APA StyleDel Prete, S., Mikulski, P., Schubert, D., & Gaudin, V. (2015). One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation. Genes, 6(3), 520-542. https://doi.org/10.3390/genes6030520




