Abstract
Posttranslational modification of proteins by means of attachment of a small globular protein ubiquitin (i.e., ubiquitylation) represents one of the most abundant and versatile mechanisms of protein regulation employed by eukaryotic cells. Ubiquitylation influences almost every cellular process and its key role in coordination of the DNA damage response is well established. In this review we focus, however, on the ways ubiquitylation controls the process of unperturbed DNA replication. We summarise the accumulated knowledge showing the leading role of ubiquitin driven protein degradation in setting up conditions favourable for replication origin licensing and S-phase entry. Importantly, we also present the emerging major role of ubiquitylation in coordination of the active DNA replication process: preventing re-replication, regulating the progression of DNA replication forks, chromatin re-establishment and disassembly of the replisome at the termination of replication forks.
1. Introdution
During ubiquitylation, a small (76 amino acid) polypeptide, ubiquitin, is attached covalently to the substrate protein. The ubiquitin sequence and its three-dimensional structure are highly conserved through all eukaryotes, which allows the whole system of ubiquitin attachment and signalling to be very well preserved throughout evolution. Ubiquitin contains a conserved C-terminal glycine that is attached by an isopeptide bond, usually to a lysine residue in the substrate protein. This reaction is catalysed by a cascade of three enzymes: a ubiquitin activating enzyme (E1) passes activated ubiquitin to a ubiquitin conjugating enzyme (E2), which can attach it to the substrate, usually with the help of a ubiquitin ligase (E3) [1]. Substrates can be modified with ubiquitin in many ways: they can be monoubiquitylated, multimonoubiquitylated or polyubiquitylated (Figure 1). Polyubiquitylation, i.e., attachment of a ubiquitin chain to the substrate, is possible as ubiquitin contains seven lysine residues within its sequence (K6, K11, K27, K29, K33, K48 and K63) that can be used for further ubiquitin attachment and formation of ubiquitin chains. Depending on which lysine within ubiquitin is modified to form the chain, homogenous polyubiquitin chains can exhibit seven different linkages (all ubiquitins linked through the same lysine position in each ubiquitin), as well as: linear chains linked through its N-terminal methionine, mixed heterogenous linkage chains and even branched structures (Figure 1) [1]. Attachment of ubiquitin or a ubiquitin chain to the substrate changes the overall three-dimensional structure of the substrate and affects the substrate’s activity, localization and fate. Each type of chain has its own unique three-dimensional structure and thus constitutes a different signal and a different outcome for the substrate. The best-studied forms of ubiquitylation are observed when a protein is modified with K48 and K11 linked chains that targets it for proteasomal degradation, and modification with K63 linked chains which plays a crucial role in DNA damage response (DDR) signalling [2]. Ubiquitin can also be removed from the substrate or edited through action of the de-ubiquitylating enzymes (DUBs). Ubiquitylation as a posttranslational modification is, therefore, very flexible and versatile. It is also one of the most abundant types of protein modification in the cell [1].
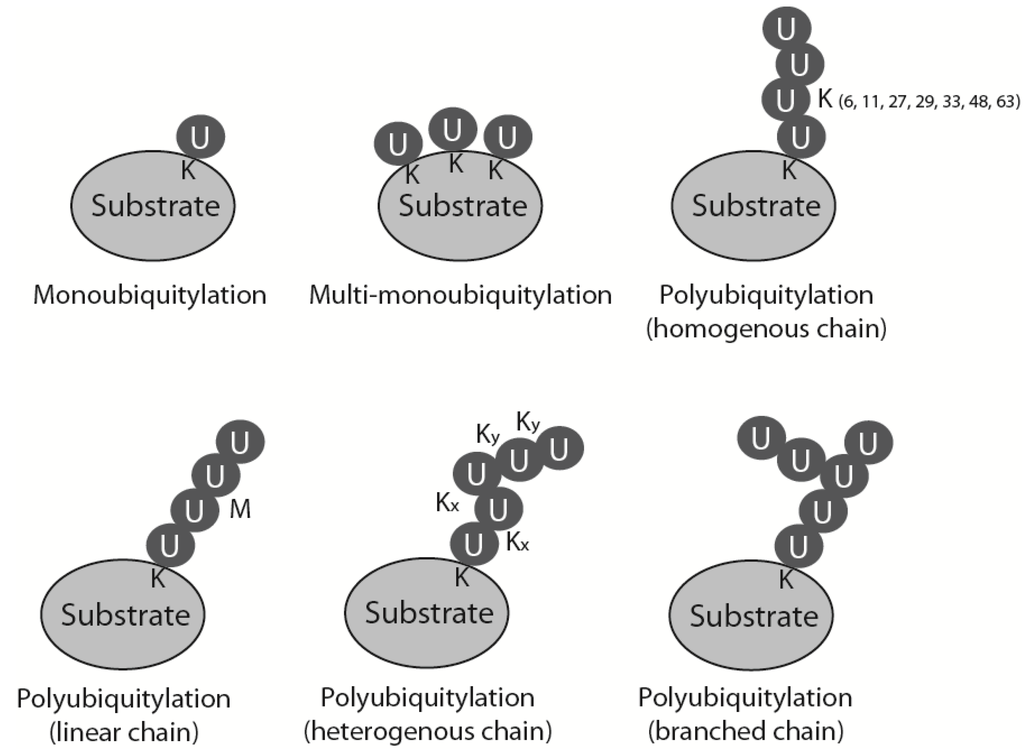
Figure 1.
Types of substrate ubiquitylation. Mono-, multimono- and polyubiquitylation is presented. Different types of polyubiquitin chains: linked through specific but the same lysine throughout the chain (homogenous chains), linked through N-terminal methionine (linear chains), linked through alternative lysines (heterogenous chains) and chains with one of the chain’s ubiquitin modified by two further ubiquitins linked through different lysines (branched chains).
The essential role for ubiquitylation in the regulation of the DDR and replicative stress processes has been studied for years [2], but its importance in regulation of unperturbed DNA replication has been established more recently. In this review, we will focus on describing the role of ubiquitylation specifically during unperturbed DNA replication. We will summarise the role for ubiquitin-driven degradation in origin licensing and entry into S-phase, discuss its importance in blocking re-replication during S-phase and chromatin re-establishment and finally focus on recent findings in the regulation of replisome factors during replication elongation and termination.
2. Setting the Scene for Origin Licensing
The licensing of replication origins occurs before the onset of S-phase: at the end of mitosis and through the G1-stage of the cell cycle. During licensing Origin Recognition Complex (ORC), Cdt1 and Cdc6 load multiple Mcm2-7 complexes onto origins forming pre-replicative complexes (pre-RCs) (reviewed in [3]). This loading reaction requires a low activity of Cyclin Dependent Kinases (CDK). Therefore, to allow licensing, the high CDK activity of mitotic CDKs needs to be abolished. The low CDK activity in G1-phase of cell cycle is achieved through (i) degradation of mitotic cyclins, (ii) degradation of CDK activator: tyrosine phosphatase Cdc25 and (iii) accumulation of CDK inhibitors (CKIs). Both mitotic cyclins and Cdc25 are degraded in a proteasome dependent manner upon their ubiquitylation by a master cell cycle regulator: the Anaphase Promoting Complex or Cyclosome (APC/C) [4]. APC/C is a multisubunit ubiquitin ligase that polyubiquitylates proteins with K48 and K11 linked ubiquitin chains, targeting them for proteasomal degradation [5]. APC/C utilises two substrate recognising adaptor proteins, Cdc20 and Cdh1 [6], which target proteins containing specific recognition motifs: D-boxes and KEN-boxes [7]. Cdc20 binds APC/C during mitosis in a CDK phosphorylation dependant manner [8] and is responsible for the initial degradation of mitotic cyclins (and other mitotic proteins) [9]. As the level of cyclins drops, the mitotic phosphatase Cdc14 has a chance to dephosphorylate and activate Cdh1, which can then compete with Cdc20 for binding to APC/C [10]. APC/C-Cdh1 has a vast repertoire of substrates, and importantly leads to ubiquitylation and degradation of Cdc20, further decline of mitotic cyclins, degradation of the Cdc25 phosphatase (CDK activator) and progression to G1-phase [11,12]. APC/C-Cdh1 also allows accumulation of CDK inhibitors: CKIs and the INKa family of proteins (p15, p16, p17 and p19) [13,14]. CKIs accumulate as APC/C promotes degradation of Skp2—a substrate receptor of ubiquitin ligase ubiquitylating CKIs for degradation [13,15] (Figure 2).
Apart from ensuring that the CDK activity is abolished to allow licensing of origins, APC/C-Cdh1 also triggers the accumulation and activity of licensing factors in G1-phase. In metazoans, APC/C-Cdh1 regulates the stability of the large subunit of the origin recognition complex, Orc1 [16], Cdc6 [17] and targets Cdt1 inhibitor geminin for degradation [18]. Altogether, APC/C activity creates favourable conditions for origin licensing by eliminating cyclins and, in metazoans, geminin (Figure 2).
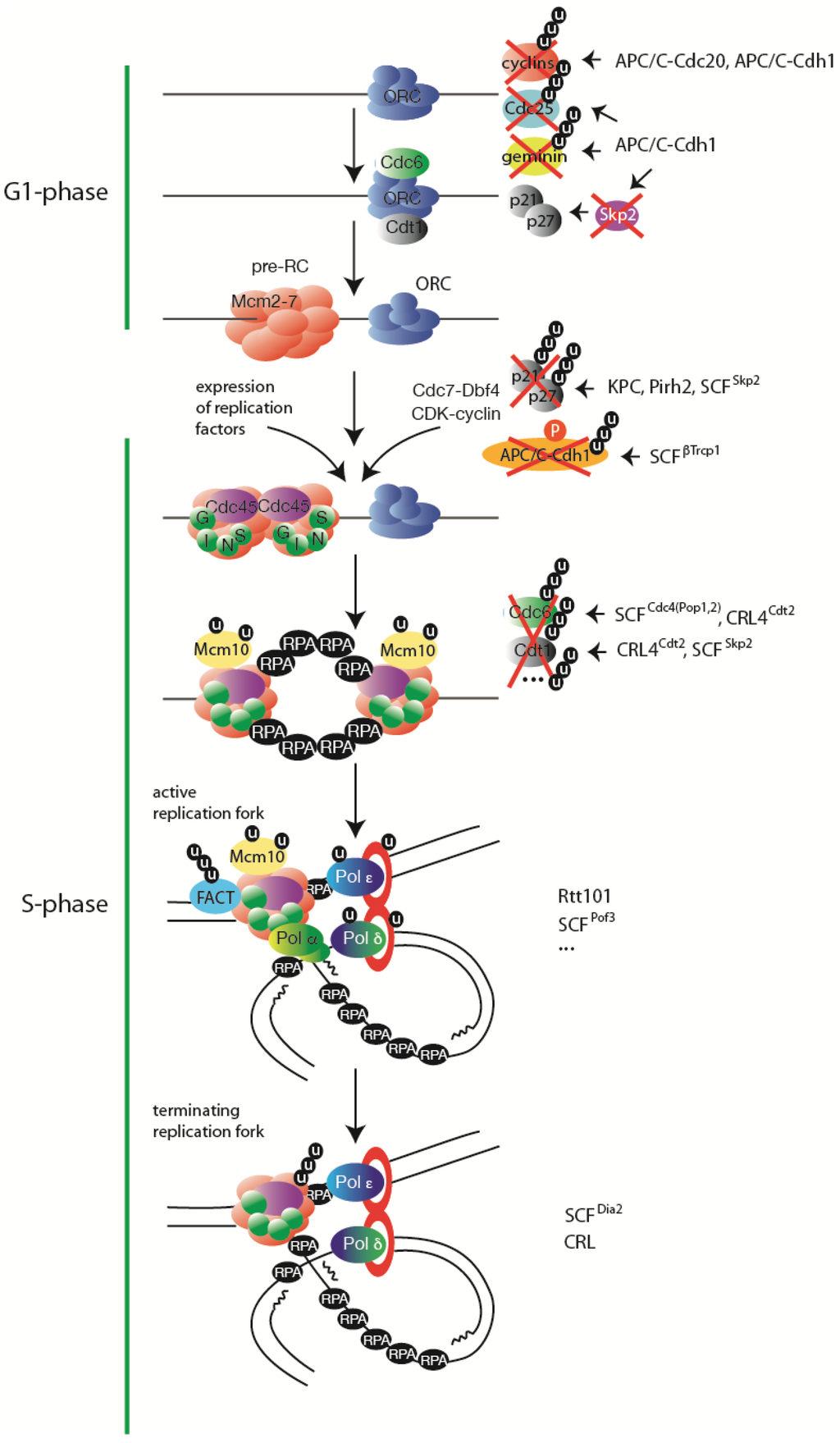
Figure 2.
Ubiquitylation regulates every stage of eukaryotic DNA replication. A simplified view of steps in DNA replication with indicated major substrates of ubiquitylation regulation. On the right hand side the respective ubiquitin ligases are listed, if known. Further explanations can be found in the text.
3. Initiation of DNA Replication
Initiation of DNA replication requires increased activity of the S-phase kinases (CDK and Cdc7) and expression of cell-cycle regulated initiation factors. To allow the rise in CDK level, CDK inhibitors have to be degraded and APC/C-Cdh1 activity diminished. Both are mainly driven through ubiquitylation directed proteasomal degradation and require activation of G1-CDKs at the mammalian G1 “restriction point” (“START point” in yeast), which is possible as G1 cyclins are not generally APC/C substrates. In mammalian cells, Cdk4 and Cdk6 are involved in early G1, while Cdk2 is believed to complete G1 and initiate S-phase. Positive growth stimuli in G1 induce expression of Cyclin D, which, together with Cdk4 and Cdk6, activates a programme of gene expression of S-phase factors. Key regulators of this programme are: the Retinoblastoma protein family (Rb), transcription factors E2F1-3 and Myc (reviewed in [19]). This transcriptional programme induces expression of Cyclin E-Cdk2, which is inactive through most of G1 due to high levels of CKIs (p27 and p21) [20]. In mammalian cells, p27 is ubiquitylated and degraded by three ubiquitin ligases: (i) KPC ligase targets p27 exported from the nucleus to the cytoplasm in G0 and G1-phases; (ii) Pirh2, whose expression increases from G1 to S-phase; and (iii) SCFSkp2, which from early S-phase, targets nuclear p27 (reviewed in [21]). SCF (Skp1-Cullin1-Fbox) belongs to the cullin-RING ligase (CRL) family of multisubunit ubiquitin ligases (Figure 3) [22]. The F-box substrate receptor Skp2 of the SCFSkp2 ubiquitin ligase specifically recognises Thr187-phosphorylated p27, which is modified by any Cyclin E-Cdk2 that escaped CKI inhibition [23]. SCFSkp2 also targets another CKI, p21, for proteasomal degradation (reviewed in [21]) (Figure 2).
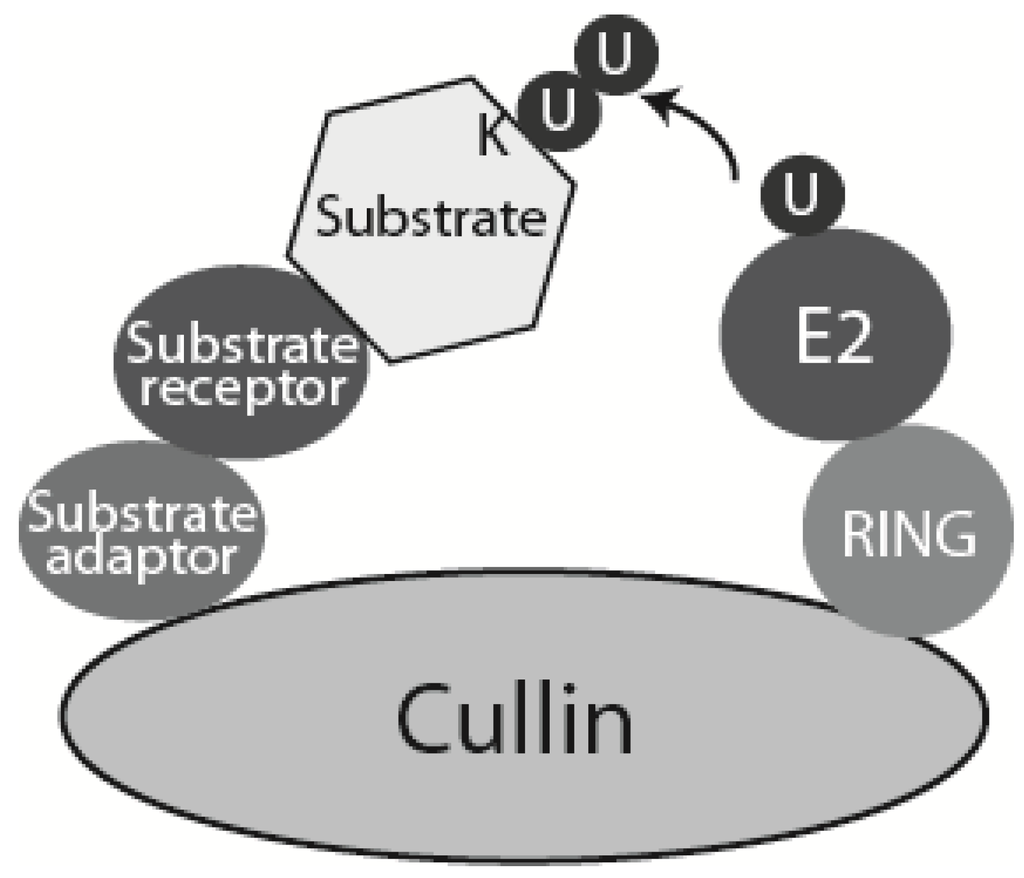
Figure 3.
Schematic model of CRLs. Cullin-RING ligases (CRLs) are multisubunit ubiquitin ligases built around a scaffold cullin subunit, which interacts at its C-terminus with RING domain ubiquitin ligase subunit Rbx1 or Rbx2, and at its N-terminus with substrate adaptor and substrate receptor. Higher eukaryotes express seven different cullins (Cul1, 2, 3, 4A, 4B, 5 and 7). Each of them interacts with a specific set of substrate adaptor and receptor proteins. In case of SCFs (Cullin 1 based CRL: Skp1-Cullin 1-F box) the substrate adaptor is Skp1 and substrate receptor is one of many F-box containing proteins [22]. CRL4 contains Cullin 4, Ddb1 as substrate adaptor and DCAF domain protein as substrate receptor.
Apart from degrading CKIs, APC/C-Cdh1 activity has to be blocked to allow S-phase entry. This happens in a number of ways: (i) Cdh1 is phosphorylated by Cdk2-Cyclin E, which blocks Cdh1 activity [24]; (ii) Cdh1 is phosphorylated by Polo-like kinase Plk1 and Cdk2-Cyclin A, which directs it for polyubiquitylation by another ubiquitin ligase, SCFβTrcp1 [25]; (iii) accumulation of the APC/C-Cdh1 inhibitor Emi1 [26]; and (iv) finally APC/C-Cdh1 can autoubiquitylate itself using the E2, UbcH10 [27]. As a result, the transition from G1 to S-phase brings the end of the reign of APC/C ubiquitin ligase and the beginning of rule by CRL ubiquitin ligases (Figure 2).
Once cells enter S-phase they use a wide number of SCF ligases (CRL1) to ubiquitylate and degrade factors essential for G1 to S-phase transition but are now dispensable or detrimental. SCFFBX4 promotes the degradation of Cyclin D [28], SCFFbw7 ubiquitylates Cyclin E [29], SCFSkp2 promotes the degradation of E2F [30], while c-Myc abundance is controlled by two SCF-type E3 ligases—SCFSkp2 and SCFFbw7 [21].
Another CRL ubiquitin ligase that plays a key role during S-phase is a Cullin 4 based ligase CRL4 (Figure 3) (reviewed in [31,32]). CRL4 acts in a PCNA-dependent ubiquitylation pathway. PCNA is a clamp loaded onto DNA to increase processivity of DNA polymerases at the replication forks. In addition to this role, PCNA creates an organising centre and a platform for multiple factors interacting with replication forks. Some of the PCNA interacting proteins (PIPs) are substrates of the CRL4Cdt2 ubiquitin ligase. Proteins interacting with PCNA most often contain a short PCNA binding motif (PIP box) [33], while CRL4Cdt2 substrates contain not only a PIP box but also four amino acids downstream of the PIP box, a “B + 4” basic residue (PIP + TD motif, or PIP-degron), essential for recruitment of CRL4Cdt2 to PCNA bound substrate [34]. PCNA itself also plays an active role in recruitment of CRL4Cdt2 to chromatin [35].
The first bona fide substrate of CRL4Cdt2 identified was Spd1—the fission yeast inhibitor of ribonucleotide reductase (RNR) enzyme that catalyzes the synthesis of dNTPs [36]. Degradation of Spd1 in S-phase is the key function of CRL4Cdt2 in S.pombe and a lack of this degradation activity leads to DNA mutagenesis, slow growth and inhibition of double strand break repair [37].
In Drospohila, the transcription factor E2f1 promotes transcription of genes in G1 whose expression is essential to enter S-phase. Once cells enter S-phase, E2f1 is degraded by CRL4Cdt2 [38]. Drosophila E2f1 is unique in the fact that it contains a PIP + TD motif, and thus alternative destruction pathways are utilised in other organisms [39].
The most recently discovered substrate of CRL4Cdt2 is a uracil DNA glycosylase (TDG) involved in the base excision repair pathway [40]. In human cells, this enzyme accumulates during G2-M and G1 phases, while its levels are compromised during S-phase in a proteasomal degradation dependent manner [41]. TDG has a conserved PIP + TD degron motif and inhibition of TDG degradation slows down S-phase progression and leads to an increase in γH2AX foci and DNA breaks [42]. Consistent with this, it has been also shown that TDG is destroyed during S-phase in early Xenopus development [43].
The most striking role of CRL4Cdt2 is, however, the degradation of origin licensing factors to prevent re-replication during S-phase, as described below [31,32].
4. Blocking Re-Replication
Upon initiation of DNA replication it becomes essential that no more origins of replication can be licensed to avoid re-licensing already replicated DNA and re-replication. Replication of the same DNA twice is likely to lead to chromosomal instability and cell death [44]. To avoid re-replication, cells degrade or inhibit the activity of origin licensing factors upon entering S-phase. In general, all organisms use multiple overlapping mechanisms to prevent DNA re-replication, but the choice of pathways differs significantly between species. Here we will focus on the ones dependent on ubiquitin-mediated proteolysis.
S. pombe and vertebrates degrade Cdt1 in a PCNA-dependent manner through ubiquitylation by CRL4Cdt2 [45]. Mammalian cells degrade Cdt1 also through an additional mechanism—utilising SCFSkp2 [46,47]. The interaction of Cdt1 and Skp2 requires phosphorylation of Cdt1 by S-phase CDKs, providing S-phase specificity of this degradation [47]. Both of the above pathways overlap in mammalian cells and thus mutation of one pathway motifs is not sufficient to inhibit Cdt1 degradation, but deletion of the N-terminal part of Cdt1 (which contains all regulatory elements for both pathways) results in stable Cdt1 [48,49,50]. Finally, any Cdt1 remaining through S-phase until G2 can be ubiquitylated by SCFFbx°31 and degraded [51] (Figure 2).
Budding yeast Cdc6 and its fission yeast homolog Cdc18 are degraded in S-phase, upon their phosphorylation by CDK, through ubiquitylation by SCFCdc4 and SCFP°p1,2, respectively [52,53,54]. In human cells, Cdc6 activity is inhibited mainly through CDK stimulated nuclear export in S-phase [55,56], but a recent report suggests a degradation of Cdc6 through CRL4Cdt2 pathway due to a PIP box within Cdc6 [57]. Finally, ubiquitylation and degradation of the CDK inhibitor p21, by CRL4Cdt2 and SCFSkp2, blocks re-replication as it allows full activity of S-phase CDKs, leading to CDK driven phosphorylation of Cdc6 and its nuclear export [56,58].
The Orc1-6 complex is regulated in different ways throughout evolution: mostly the levels of Orc1-6 remain stable throughout the cell cycle. It has been reported, however, that in transformed human cells the majority of Orc1 can be selectively degraded during S-phase in a proteasomal dependent manner, most likely through SCFSkp2 ubiquitin ligase activity [59,60]. The Orc1-6 interacting partner ORCA, which plays a role in origin licensing in human cells, has also been shown to be ubiquitylated by CRL4A(Ddb1) at the onset of S-phase, but the role of this ubiquitylation is not yet clear [61]. Finally, interaction of the Orc1-6 complex with chromatin and licensing ability of the cell depends on chromatin structure and epigenetic marks, i.e., Orc1 binds to methylated histones H3 and H4 [62]. The degradation of the enzyme responsible for these modifications: histone methyl-transferase Set8, in a PCNA- and CRL4Cdt2-dependent manner, also contributes to blocking re-replication, while non-degradable Set8 mutant causes uncontrolled re-replication [63,64].
Taking into account the key and widespread role of CRL4Cdt2 in blocking origin licensing, it is not surprising that its activity is highly controlled in order to restrict its activity to S-phase. This control is executed not only through the fact that PCNA needs to be chromatin bound to stimulate substrate recognition, but also through the modulation of Cdt2 turnover. In human cells, Cdt2 is ubiquitylated and degraded by two mechanisms: by SCFFbx°11 and by PCNA-independent autoubiquitylation by CRL4A [65]. To prevent Cdt2 from being ubiquitylated in S-phase, it is phosphorylated by Cyclin B-Cdk1, which inhibits its interaction with SCFFbx°11 [65,66,67].
5. Ubiquitylation of Replisome Components during the Elongation Stage of DNA Replication
The progression of replication forks can be affected by a number of different post-translational modifications to the replisome factors. Moreover, a whole raft of modifications is applied when forks encounter DNA damage (reviewed in [68]). We will focus here on the ubiquitylation events regulating replisome components during unperturbed DNA replication (Figure 2).
In all eukaryotic systems examined to date, PCNA undergoes monoubiquitylation of a highly conserved Lys164 (K164) by the RAD18-RAD6 ubiquitin ligase in response to replication block (reviewed in [69]). Monoubiquitylation of K164 on PCNA plays a key role in promoting the interaction of PCNA with Y-family translesion DNA synthesis (TLS) of DNA polymerases and lesion bypass. Sometimes, this monoubiquitylation can be extended to create a K63 linked ubiquitin chain. Such polyubiquitylation of PCNA promotes error-free repair of the lesion involving template switch DNA synthesis. Other lysines within PCNA can also be ubiquitylated upon DNA damage, but the role of such ubiquitylation is less well-defined [69]. However, in Xenopus laevis egg extract a proportion of PCNA can be ubiquitylated constitutively on K164 during unperturbed DNA replication. This ubiquitylation is important for efficient DNA replication and its inhibition leads to reduction of the level of lagging strand polymerase, Pol∂, associated with chromatin [70].
Another replisome factor ubiquitylated during normal S-phase is Mcm10. Mcm10 has been shown to play important roles during both the initiation and the elongation stages of DNA replication. Although there is some controversy in the field, the current consensus is that Mcm10 is loaded onto chromatin after origin licensing and is needed for the activation of the replicative helicase [71]. Mcm10 has been also found to interact with and maintain the stability of DNA polymerase α (priming polymerase) as well as its association with chromatin [72]. Sapna Das-Bradoo et al showed that Mcm10 is monoubiquitylated at two distinct lysine residues during G1-S phase, and this modification allowed the interaction of ubiquitylated Mcm10 with PCNA. This interaction event has been considered of essential importance during DNA replication since its impairment compromised cell growth [73]. The fact that ubiquitylated Mcm10 is unable to interact with Polα raises the possibility that this modification of Mcm10 could promote the release of Polα after the completion of the RNA-DNA primer synthesis and the recruitment of PCNA [71,73].
Another polymerase affected by ubiquitylation is the lagging strand polymerase, Pol∂. Pol∂ is formed of 4 subunits: the catalytic p125 and regulatory p50, p66 and p12. Both p12 and p66 have been shown to be ubiquitylated (predominantly monoubiquitylated) in human U2OS osteosarcoma cells in unperturbed S-phase [74]. Neither of these ubiquitylations leads to proteasomal degradation of the subunits - it is more likely that they can instead regulate protein-protein interactions [74].
A study performed in S. pombe showed also that Pol2 (a catalytic subunit of the leading strand polymerase, Polε) is continuously degraded by the proteasome during unperturbed S-phase in a manner dependent on the ubiquitin ligase SCFPof3 - an effect clearly visible especially in cells harbouring Swi1 deletion and thus experiencing replisome stability problems. However, the level of Pol2 in the cells is constant due to continuous Pol2 translation. The authors suggested that this degradation mechanism could serve to “refresh” Pol2 enzymes during S-phase: provide newly synthesised Pol2 enzyme to be reloaded onto the leading strand [75,76].
It has also been reported that S.cerevisiae Spt16, a subunit of the FACT complex (FAcilitates Chromatin Transcription), is a substrate of the E3 ubiquitin ligase complex containing Rtt101 [77]. FACT, a heterodimeric complex of Spt16 and SSRP1, has nucleosome remodeling activity, which makes it an important player during transcriptional regulation [78]. Apart from this, FACT has been found to be an integral component of the Replisome Progression Complex (RPC) in budding yeast [79] and an interaction partner of the MCM complex [80]. Ubiquitylation of Spt16 does not target it for proteasomal degradation but it has been proposed to play a role in the association of FACT with replication origins and MCM complexes. Moreover, it was suggested that it promotes the recruitment of MCM complexes to replication origins during G1 and early S-phase. Interestingly, the impairment of Spt16 ubiquitylation does not affect the role of FACT in transcription, which suggests that this mechanism specifically regulates FACT function during DNA replication [77].
6. Chromatin Re-Establishment during DNA Replication
While DNA is replicated, the chromatin structure becomes locally disassembled ahead of the replication forks and re-assembled behind the forks. This process involves eviction of nucleosomes in front of the fork by fork-associated histone chaperones: FACT and Asf1, followed by their re-assembly on newly formed sister chromatids. The parental histones are then joined by newly synthesised ones to provide enough nucleosomes to organise the doubled quantity of DNA [81]. As the new nucleosomes do not contain the histone modifications present on the parental nucleosomes these modifications have to be reconstituted behind the fork to provide the daughter cells with the same epigenome profile. The present model implies that histone-modifying enzymes can recognise the modifications on parental nucleosomes and replicate them on the neighboring, new nucleosomes. DNA replication also produces hemimethylated DNA, as the newly synthesised DNA strand contains unmodified cytosines. In this case, DNA methyltransferase 1 (Dnmt1) methylates the hemimethylated fragments of DNA behind the fork to produce a DNA strand identical to the parental one [82]. Importantly to this review, ubiquitylation plays a key role in many aspects of this chromatin reconstitution process.
Firstly, it is important that the expression and translation of histones is tightly regulated and coupled to the DNA replication process. It has been shown that the lack of proper regulation of histones levels during the cell cycle leads to defects in chromosome transmission fidelity and genomic instability [83,84]. The synthesis of histones is regulated by ubiquitin driven degradation of their transcription factors. In fission yeast, Ams2—a transcription factor responsible for core histone gene expression, is degraded in G1 by APC/C-Cdh1 [85] and after S-phase by SCFP°f3 [84]. This results in the synthesis of histones only in S-phase.
It was shown also that levels of “free” (non-chromatin bound) histones are regulated directly by ubiquitin driven proteasomal degradation. In budding yeast, non-chromatin bound histones were found to be degraded with a half-life of 30–40 min [86] in a manner dependent on the activity of the checkpoint kinase Rad53. The mechanism employs Ubc4/5 E2 enzymes and Tom1, Pep5, Snt2, Hel1 and Hel2 ubiquitin ligases [87,88]. Interestingly, the human homologue of Tom1: HUWE1, has also been reported to be able to ubiquitylate all four core histones in vitro [89].
The assembly of histones into nucleosomes on DNA is also regulated by ubiquitylation; specifically the deposition of newly synthesized histone H3 onto replicated DNA. Budding yeast Rtt101Mms1 E3 ubiquitin ligase preferentially ubiquitylates new histone H3 acetylated at lysine 56 (H3K56Ac-H4) in vitro and in vivo. This ubiquitylation of histone H3 at K121, 122 and 125 leads to release of H3-H4 from Asf1 to other histone chaperones followed by their incorporation into chromatin [90]. The human orthologue of Rtt101Mms1, Cul4ADDB1 appears to have an analogous role, since human cells depleted of Cul4ADDB1 activity show a similar defect in H3 deposition [90].
Another histone ubiquitylation event that regulates unperturbed DNA replication is the monoubiquitylation of histone H2B (H2Bub1) driven by the E3 ligase Bre1. A key role for H2Bub1 is to control the transcription of genes through regulation of Pol II elongation [91]. Transcription-independent functions of H2Bub1 include roles in meiosis, the UV-induced checkpoint, DNA double-strand breaks repair, apoptosis and Set1-directed methylation of the kinetochore associated protein Dam1 [92,93,94,95]. Moreover, H2Bub1 also coordinates activation of the intra-S checkpoint and chromatin assembly during replication under stress [96]. In budding yeast, H2Bub1 (on lysine 123) has been shown to occur on chromatin adjacent to replication origins localized at non-transcribed regions. This ubiquitylation event was found to be required for appropriate nucleosome assembly or nucleosome stability during DNA replication in times of replication stress (in the presence of hydroxyurea, HU). Under these conditions, impairment of H2B ubiquitylation also led to a defect in replication fork progression and replisome stability. Importantly however, during unperturbed DNA replication, htb-K123R cells displayed a delay in the completion of DNA replication implicating the important role of the H2Bub1 in normal replication processes [96].
Finally, a recent study suggested that Uhrf1 dependent polyubiquitylation of histone H3 on lysine 23 (H3K23nUb) serves as a platform for Dnmt1 to bind the hemimethylated DNA behind the replication fork in order to fully methylate it [97]. Uhrf1 is an ubiquitin ligase which can bind hemimethylated DNA through its SRA domain and has also been shown to interact with PCNA [98,99]. These two interactions bring Uhrf1 to the replicating fork where it ubiquitylates H3, which in turn is preferentially bound by Dnmt1. Interestingly, due to a negative feedback loop (once DNA is methylated, H3K23nUb is deubiquitylated), the polyubiquitylated form of H3 could be detected only upon inhibition of Dnmt1 activity [97].
7. Termination of Replication Forks
Recent studies in budding yeast and Xenopus laevis egg extract have shown that polyubiquitylation plays a key role in disassembly of the replisome machinery at the termination of DNA replication forks [100,101]. In both model systems, only one subunit of the replicative helicase—Mcm7—has been found polyubiquitylated with K48 linked chains upon replication fork termination (Figure 2). This ubiquitylation was then followed by dissolution of the replicative helicase (CMG complex), which is dependent on the activity of the protein segregase, Cdc48/p97/VCP. The segregase is a homohexameric ring-shaped ATPase, which can recognise proteins modified with K48 linked ubiquitin chains and remodel them in an ATP-dependent manner [102,103]. Such remodelling can lead to removal of the ubiquitylated protein from the endoplasmic reticulum membrane (ERAD—endoplasmic-reticulum-associated protein degradation) or chromatin [104,105]. The ubiquitylated protein can then be directed for proteasomal degradation or deubiquitylated by one of the DUBs interacting with the segregase [106]. The fate of ubiquitylated Mcm7 upon its recognition and remodelling by the segregase has not yet been established.
In budding yeast, Mcm7 is ubiquitylated by SCFDia2 ubiquitin ligase [100], while the ligase in higher eukaryotes is not yet known, although the ubiquitylation process in Xenopus egg extract is dependent on the CRLs [101]. SCFDia2 has been previously shown to interact with the replisome and travel with the replication fork with the RPCs [79,107]. Nevertheless, SCFDia2 recognises only the terminated helicase for ubiquitylation, and not the active form. In cells lacking Dia2 the replicative helicase complexes persist on chromatin until the subsequent cell cycle [100] and at the same time, Dia2Δ cells exhibit a high level of genomic instability and have problems with replicating through difficult-to-replicate DNA regions [108,109]. Dia2 was also shown to accumulate when the intra-S-phase checkpoint was activated, and to promote ubiquitylation of Mrc1 leading to its degradation [109,110]. Clearly, SCFDia2 is likely to have many functions at the replication forks.
8. Concluding Remarks
Recent years have brought a small explosion of examples of substrates and mechanisms through which ubiquitylation regulates the process of unperturbed DNA replication. With this knowledge a multitude of questions arises about the regulation, fine-tuning and coordination of all of these ubiquitin dependent mechanisms. Many of the enzymes involved also have additional functions upon replication stress, bringing another level of complexity to the emerging ubiquitin regulation network.
It is tempting to speculate about whether the emerging network of ubiquitylation enzymes, essential for faultless execution of DNA replication, could lead to the next generation of cancer therapy targets [111]. Many of the traditional therapies target DNA replication and DNA repair processes but they tend not to be cancer specific and thus exhibit a restricted therapeutic window due to side effects. Many of the enzymes involved in the ubiquitin regulatory network are deregulated in cancer, providing the opportunity to design and use drugs targeted against them [112]. One example of such a potential drug is MLN4924—a small molecule inhibitor of the Nedd8 activating enzyme, which blocks activity of the cullin-RING ubiquitin ligases CRLs. Treatment of variety of human tumour cell lines with MLN4924, results in uncontrolled DNA synthesis through re-replication, leading to DNA damage and induction of apoptosis [113,114]. Importantly, the potential of antitumor activity was shown in human colon and lung xenograft models at doses and schedules that were well tolerated and which led to multiple phase I trials of MLN4924 [113,115]. Future research will establish a better understanding of the ubiquitin regulation of DNA replication and the opportunity to use it to treat cancer.
Acknowledgements
This work has been supported by U.K. Medical Research Council Career.
Development Award MR/K007106/1 and Birmingham Fellows Fellowship to Agnieszka Gambus. Agnieszka Gambus would like to thank Dr. Daniel Tennant for support.
Conflict of Interests
The authors declare no conflict of interest.
References
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.D.; Walden, H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010, 11, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Riera, A.; Tognetti, S.; Speck, C. Helicase loading: How to build a MCM2-7 double-hexamer. Semin. Cell Dev. Biol. 2014, 30, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Wasch, R.; Engelbert, D. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene 2005, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, A.; Stengel, F.; Zhang, Z.; Enchev, R.I.; Kong, E.H.; Morris, E.P.; Robinson, C.V.; da Fonseca, P.C.; Barford, D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 2011, 470, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Visintin, R.; Prinz, S.; Amon, A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science 1997, 278, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, C.M.; Kirschner, M.W. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000, 14, 655–665. [Google Scholar] [PubMed]
- Rahal, R.; Amon, A. Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev. 2008, 22, 1534–1548. [Google Scholar] [CrossRef] [PubMed]
- Shirayama, M.; Toth, A.; Galova, M.; Nasmyth, K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 1999, 402, 203–207. [Google Scholar] [PubMed]
- Jaspersen, S.L.; Charles, J.F.; Morgan, D.O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 1999, 9, 227–236. [Google Scholar] [CrossRef]
- Robbins, J.A.; Cross, F.R. Regulated degradation of the APC coactivator Cdc20. Cell Div. 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.; Morgan, D.O. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007, 8, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Bashir, T.; Dorrello, N.V.; Amador, V.; Guardavaccaro, D.; Pagano, M. Control of the SCF (Skp2-Cks1) ubiquitin ligase by the APC/C (Cdh1) ubiquitin ligase. Nature 2004, 428, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ayad, N.G.; Wan, Y.; Zhang, G.J.; Kirschner, M.W.; Kaelin, W.G., Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 2004, 428, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Narbonne-Reveau, K.; Senger, S.; Pal, M.; Herr, A.; Richardson, H.E.; Asano, M.; Deak, P.; Lilly, M.A. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 2008, 135, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Diffley, J.F. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005, 122, 915–926. [Google Scholar] [CrossRef] [PubMed]
- McGarry, T.J.; Kirschner, M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Bartek, J.; Lukas, J. Pathways governing G1/S transition and their response to DNA damage. FEBS Letters 2001, 490, 117–122. [Google Scholar] [CrossRef]
- Abukhdeir, A.M.; Park, B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Exp. Rev. Mol. Med. 2008, 10. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Kotake, Y.; Kitagawa, M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci. 2009, 100, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, E.S.; Schulman, B.A.; Zheng, N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010, 20, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Sheaff, R.J.; Groudine, M.; Gordon, M.; Roberts, J.M.; Clurman, B.E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997, 11, 1464–1478. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.W.; Inuzuka, H.; Fukushima, H.; Wan, L.; Liu, P.; Gao, D.; Sun, Y.; Wei, W. Regulation of APC(Cdh1) E3 ligase activity by the Fbw7/cyclin E signaling axis contributes to the tumor suppressor function of Fbw7. Cell Res. 2013, 23, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.; Ogura, K.; Wan, L.; Lu, Y.; Li, V.; Gao, D.; Liu, P.; Lau, A.W.; Wu, T.; Kirschner, M.W.; et al. SCF-mediated Cdh1 degradation defines a negative feedback system that coordinates cell-cycle progression. Cell Rep. 2013, 4, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, A.G.; Loktev, A.V.; Hansen, D.V.; Verschuren, E.W.; Reimann, J.D.; Jackson, P.K. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell 2006, 124, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Rape, M.; Kirschner, M.W. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 2004, 432, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.I.; Barbash, O.; Kumar, K.G.; Weber, J.D.; Harper, J.W.; Klein-Szanto, A.J.; Rustgi, A.; Fuchs, S.Y.; Diehl, J.A. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell 2006, 24, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Koepp, D.M.; Schaefer, L.K.; Ye, X.; Keyomarsi, K.; Chu, C.; Harper, J.W.; Elledge, S.J. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 2001, 294, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Marti, A.; Wirbelauer, C.; Scheffner, M.; Krek, W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1999, 1, 14–19. [Google Scholar] [PubMed]
- Abbas, T.; Dutta, A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 2011, 10, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Havens, C.G.; Walter, J.C. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011, 25, 1568–1582. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Havens, C.G.; Walter, J.C. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 2009, 35, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Havens, C.G.; Shobnam, N.; Guarino, E.; Centore, R.C.; Zou, L.; Kearsey, S.E.; Walter, J.C. Direct role for proliferating cell nuclear antigen in substrate recognition by the E3 ubiquitin ligase CRL4Cdt2. J. Biol. Chem. 2012, 287, 11410–11421. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Powell, K.A.; Mundt, K.; Wu, L.; Carr, A.M.; Caspari, T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003, 17, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Poitelea, M.; Watson, A.; Yoshida, S.H.; Shimoda, C.; Holmberg, C.; Nielsen, O.; Carr, A.M. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J. 2005, 24, 3940–3951. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.T.; de la Cruz, A.F.; Tran, V.; Turbyfill, W.J., 3rd.; Reis, T.; Edgar, B.A.; Duronio, R.J. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell. 2008, 15, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Dimova, D.K.; Dyson, N.J. The E2F transcriptional network: Old acquaintances with new faces. Oncogene 2005, 24, 2810–2826. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, U.; Bentele, M.; Lettieri, T.; Steinacher, R.; Jiricny, J.; Schar, P. Thymine DNA glycosylase. Prog. Nucl. Acid. Res. Mol. Biol. 2001, 68, 235–253. [Google Scholar]
- Hardeland, U.; Kunz, C.; Focke, F.; Szadkowski, M.; Schar, P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 2007, 35, 3859–3867. [Google Scholar] [CrossRef] [PubMed]
- Shibata, E.; Dar, A.; Dutta, A. CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J. Biol. Chem. 2014, 289, 23056–23064. [Google Scholar] [CrossRef] [PubMed]
- Slenn, T.J.; Morris, B.; Havens, C.G.; Freeman, R.M., Jr.; Takahashi, T.S.; Walter, J.C. Thymine DNA glycosylase is a CRL4Cdt2 substrate. J. Biol. Chem. 2014, 289, 23043–23055. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005, 6, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.E.; Walter, J.C. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 2006, 8, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Li, X.; Yan, F.; Zhao, Q.; Wu, X. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 2004, 279, 17283–17288. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Tatsumi, Y.; Tsurumi, T.; Matsukage, A.; Kiyono, T.; Nishitani, H.; Fujita, M. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 2004, 279, 19691–19697. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Lygerou, Z.; Nishimoto, T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 2004, 279, 30807–30816. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Sugimoto, N.; Roukos, V.; Nakanishi, Y.; Saijo, M.; Obuse, C.; Tsurimoto, T.; Nakayama, K.I.; Nakayama, K.; Fujita, M.; et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006, 25, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Takeda, D.Y.; Parvin, J.D.; Dutta, A. Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J. Biol. Chem. 2005, 280, 23416–23423. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Jeffery, J.; Al-Ejeh, F.; Schulz, R.B.; Callen, D.F.; Kumar, R.; Khanna, K.K. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J. Biol. Chem. 2014, 289, 18514–18525. [Google Scholar] [CrossRef] [PubMed]
- Drury, L.S.; Perkins, G.; Diffley, J.F. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 2000, 10, 231–240. [Google Scholar] [CrossRef]
- Elsasser, S.; Chi, Y.; Yang, P.; Campbell, J.L. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol. Biol. Cell. 1999, 10, 3263–3277. [Google Scholar] [CrossRef] [PubMed]
- Kominami, K.; Toda, T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes Dev. 1997, 11, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Chen, J.; Thome, K.C.; Lawlis, S.J.; Hou, Z.H.; Hendricks, M.; Parvin, J.D.; Dutta, A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell Biol. 1998, 18, 2758–2767. [Google Scholar] [PubMed]
- Kim, J.; Kipreos, E.T. Control of the Cdc6 replication licensing factor in metazoa: The role of nuclear export and the CUL4 ubiquitin ligase. Cell Cycle. 2008, 7, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Clijsters, L.; Wolthuis, R. PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase. J. Cell Sci. 2014, 127, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Shiomi, Y.; Iida, H.; Michishita, M.; Takami, T.; Tsurimoto, T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 2008, 283, 29045–29052. [Google Scholar] [CrossRef] [PubMed]
- Mendez, J.; Zou-Yang, X.H.; Kim, S.Y.; Hidaka, M.; Tansey, W.P.; Stillman, B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 2002, 9, 481–491. [Google Scholar] [CrossRef]
- Ohta, S.; Tatsumi, Y.; Fujita, M.; Tsurimoto, T.; Obuse, C. The ORC1 cycle in human cells: II. Dynamic changes in the human ORC complex during the cell cycle. J. Biol. Chem. 2003, 278, 41535–41540. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Prasanth, S.G. Orc2 protects ORCA from ubiquitin-mediated degradation. Cell Cycle. 2012, 11, 3578–3589. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; Brustel, J.; Kirsh, O.; Lefevbre, C.; Callanan, M.; Sardet, C.; Julien, E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010, 12, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Shibata, E.; Park, J.; Jha, S.; Karnani, N.; Dutta, A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 2010, 40, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Centore, R.C.; Havens, C.G.; Manning, A.L.; Li, J.M.; Flynn, R.L.; Tse, A.; Jin, J.; Dyson, N.J.; Walter, J.C.; Zou, L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 2010, 40, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Mueller, A.C.; Shibata, E.; Keaton, M.; Rossi, M.; Dutta, A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol. Cell 2013, 49, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.; Wu, D.; Lee, N.; Shibata, E.; Dutta, A. 14-3-3 proteins play a role in the cell cycle by shielding cdt2 from ubiquitin-mediated degradation. Mol. Cell Biol. 2014, 34, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Duan, S.; Jeong, Y.T.; Horn, M.; Sarafs, A.; Florens, L.; Washburn, M.P.; Antebi, A.; Pagano, M. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol. Cell 2013, 49, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Zech, J.; Dalgaard, J.Z. Replisome components—Post-translational modifications and their effects. Semin. Cell Dev. Biol. 2014, 30, 144–153. [Google Scholar]
- Mailand, N.; Gibbs-Seymour, I.; Bekker-Jensen, S. Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 2013, 14, 269–82. [Google Scholar] [CrossRef] [PubMed]
- Leach, C.A.; Michael, W.M. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J. Cell Biol. 2005, 171, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Thu, Y.M.; Bielinsky, A.K. MCM10: One tool for all-Integrity, maintenance and damage control. Semin. Cell Dev. Biol. 2014, 30, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Ricke, R.M.; Bielinsky, A.K. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 2004, 16, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Das-Bradoo, S.; Ricke, R.M.; Bielinsky, A.K. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell Biol. 2006, 26, 4806–4817. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Warbrick, E. The p66 and p12 subunits of DNA polymerase delta are modified by ubiquitin and ubiquitin-like proteins. Biochem. Biophys. Res. Commun. 2006, 349, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Roseaulin, L.C.; Noguchi, C.; Martinez, E.; Ziegler, M.A.; Toda, T.; Noguchi, E. Coordinated degradation of replisome components ensures genome stability upon replication stress in the absence of the replication fork protection complex. PLoS Genet. 2013, 9, e1003213. [Google Scholar] [CrossRef] [PubMed]
- Roseaulin, L.C.; Noguchi, C.; Noguchi, E. Proteasome-dependent degradation of replisome components regulates faithful DNA replication. Cell Cycle 2013, 12, 2564–2569. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Li, Q.; McCullough, L.; Kettelkamp, C.; Formosa, T.; Zhang, Z. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 2010, 24, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Reinberg, D.; Sims, R.J., 3rd. de FACTo nucleosome dynamics. J. Biol. Chem. 2006, 281, 23297–23301. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Calzada, A.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Chien, C.T.; Hirose, S.; Lee, S.C. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006, 25, 3975–3985. [Google Scholar] [CrossRef] [PubMed]
- Jasencakova, Z.; Groth, A. Restoring chromatin after replication: How new and old histone marks come together. Semin. Cell Dev. Biol. 2010, 21, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Alabert, C.; Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012, 13, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Meeks-Wagner, D.; Hartwell, L.H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 1986, 44, 43–52. [Google Scholar] [CrossRef]
- Takayama, Y.; Mamnun, Y.M.; Trickey, M.; Dhut, S.; Masuda, F.; Yamano, H.; Toda, T.; Saitoh, S. Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell. 2010, 18, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Trickey, M.; Fujimitsu, K.; Yamano, H. Anaphase-promoting complex/cyclosome-mediated proteolysis of Ams2 in the G1 phase ensures the coupling of histone gene expression to DNA replication in fission yeast. J. Biol. Chem. 2013, 288, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Gunjan, A.; Verreault, A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 2003, 115, 537–549. [Google Scholar] [CrossRef]
- Singh, R.K.; Kabbaj, M.H.; Paik, J.; Gunjan, A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 2009, 11, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Gonzalez, M.; Kabbaj, M.H.; Gunjan, A. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS ONE 2012, 7, e36295. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Oughtred, R.; Wing, S.S. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell Biol. 2005, 25, 2819–2831. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, H.; Wang, Z.; Zhou, H.; Zhang, Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 2013, 155, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.B.; Kao, C.F.; Hillyer, C.; Pikaart, M.; Osley, M.A. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 2008, 31, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, M.; Lazzaro, F.; Plevani, P.; Muzi-Falconi, M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005, 280, 9879–9886. [Google Scholar] [CrossRef] [PubMed]
- Game, J.C.; Chernikova, S.B. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair 2009, 8, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Chernikova, S.B.; Dorth, J.A.; Razorenova, O.V.; Game, J.C.; Brown, J.M. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat. Res. 2010, 174, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Latham, J.A.; Chosed, R.J.; Wang, S.; Dent, S.Y. Chromatin signaling to kinetochores: Transregulation of Dam1 methylation by histone H2B ubiquitination. Cell 2011, 146, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, K.M.; Osley, M.A. A role for H2B ubiquitylation in DNA replication. Mol. Cell 2012, 48, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Yamaguchi, L.; Sharif, J.; Johmura, Y.; Kawamura, T.; Nakanishi, K.; Shimamura, S.; Arita, K.; Kodama, T.; Ishikawa, F.; et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 2013, 502, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Sharif, J.; Koseki, H. Recruitment of Dnmt1 roles of the SRA protein Np95 (Uhrf1) and other factors. Prog. Mol. Biol. Transl. Sci. 2011, 101, 289–310. [Google Scholar] [PubMed]
- Tauber, M.; Fischle, W. Conserved linker regions and their regulation determine multiple chromatin-binding modes of UHRF1. Nucleus 2015, 6, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Maric, M.; Maculins, T.; De Piccoli, G.; Labib, K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 2014, 346. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.P.; Bailey, R.; Campion, N.; Herron, S.; Gambus, A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 2014, 346, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Walsh, M.J.; Franke, W.W. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J. 1990, 9, 1757–1767. [Google Scholar] [PubMed]
- Rouiller, I.; DeLaBarre, B.; May, A.P.; Weis, W.I.; Brunger, A.T.; Milligan, R.A.; Wilson-Kubalek, E.M. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 2002, 9, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Jarosch, E.; Taxis, C.; Volkwein, C.; Bordallo, J.; Finley, D.; Wolf, D.H.; Sommer, T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell. Biol. 2002, 4, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Vaz, B.; Halder, S.; Ramadan, K. Role of p97/VCP (Cdc48) in genome stability. Front. Genet. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Rumpf, S.; Jentsch, S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 2006, 21, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, H.; Maculins, T.; Labib, K. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr. Biol. 2009, 19, 1943–1949. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.; Luke, B.; Kanellis, P.; Jorgensen, P.; Goh, T.; Penfold, S.; Breitkreutz, B.J.; Durocher, D.; Peter, M.; Tyers, M. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics 2006, 174, 1709–1727. [Google Scholar] [CrossRef] [PubMed]
- Koepp, D.M.; Kile, A.C.; Swaminathan, S.; Rodriguez-Rivera, V. The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell 2006, 17, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Mimura, S.; Komata, M.; Kishi, T.; Shirahige, K.; Kamura, T. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J. 2009, 28, 3693–3705. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Tcherpakov, M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 2010, 143, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr. Pharm. Des. 2013, 19, 3215–3225. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Milhollen, M.A.; Smith, P.G.; Narayanan, U.; Dutta, A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010, 70, 10310–10320. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; Smith, P.G.; Rolfe, M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin. Cancer Res. 2009, 15, 3912–3916. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).