Abstract
Objective: Growth traits are important economic characteristics in livestock. Genetic polymorphism has great influences on the improvement of goat growth traits. As an important member of the myogenic regulatory factor (MRFs) family, MyoG gene polymorphisms can alter the growth characteristics in goats. In this study, we aimed to investigate the regulation mechanism of the MyoG gene promoter region from the perspective of single nucleotide polymorphisms (SNPs) and transcription factors. Methods: Genomic DNA sequencing was carried out to detect SNPs in the −1000 bp upstream to 300 bp downstream of the MyoG gene promoter region in 224 Guizhou White goats (Capra hircus), and the genetic parameters of novel SNPs were calculated. The association between SNPs and growth traits, comprising body weight, body length, body height, chest circumference and cannon circumference, were analyzed using one-way ANOVA by IBM SPSS 23.0 software according to the general linear model. Transcription factor binding sites in the promoter region of the MyoG gene before and after mutation were predicted using bioinformatics software programs. Results: Four SNPs, including g.–709C>T, g.–461G>T, g.–377G>T and g.–249G>A, were identified in the 1 246 bp promoter region of the MyoG gene in Guizhou White goats. Based on χ2 test, the g.–709C>T and g.–461G>T loci were consistent with Hardy–Weinberg equilibrium, while two other SNPs were deviated from Hardy–Weinberg equilibrium in Guizhou White goats. Association analysis revealed that the body weight of those with the CT genotype at the g.–709C>T locus was greater than of those with the CC and TT genotypes in Guizhou White goats (p < 0.05). At the g.–461G>T locus, the body weight of individuals with the GG genotype was significantly higher than that of those with GT genotype (p < 0.01). The body length of individuals with the GG genotype formed by the g.–249G>A locus was significantly higher than that of those with the GA genotype (p < 0.01). Online software programs found that four SNPs within the promoter region of the MyoG gene changed some transcription factor binding sites. Conclusions: Mutations of the MyoG gene promoter region may have a significant regulatory effect on the growth traits of Guizhou White goats. The small sample size may be one of the limitations for this study; nevertheless, these findings could provide a theoretical basis for further exploring the relationship between the four SNPs studied and the growth traits in Guizhou White goats, as well as the promoter function of the MyoG gene.
1. Introduction
Myogenic regulatory factors (MRFs) are known as vital members of the myogenesis regulatory family, which is involved in the proliferation of muscle precursor cells and in the formation of muscle fiber, as well as in postnatal muscular functions [1,2]. Previous studies have shown that the myogenin (MyoG) gene is the only member in the family of MRFs that can be expressed in animal skeletal muscle cell lines. It can activate muscle gene transcription, improve cell differentiation and is a key factor in regulating skeletal muscle growth and development. It plays a central regulatory role in the process of muscle cell generation [3]. At the same time, the MyoG gene interacts with other transcription regulatory factors in the MRFs family, such as MyoD, Myf5 and MRF4, to jointly regulate animal meat quality traits and meat yield [4]. Thus, genetic variation in the MyoG gene may affect the growth, carcass and meat traits in livestock. In previous studies, some MRFs were confirmed to be related to meat quality traits in swine [5,6], beef cattle [7] and sheep [8,9]. In sheep, variants in MyoD1 were reported as being associated with body traits such as thoracic girth and loin width [10]. Additionally, positive correlations between MyoG expression and body weight in Hu sheep were explored [11]. At present, candidate gene analysis has been achieved using relevant molecular methods [12], and the candidate gene method has been proven to be effective in identifying gene polymorphisms. If the genetic variation in quantitative traits in organisms is caused by causal mutations in candidate genes, it can be determined from this perspective that the analysis of gene physiology, biochemical function and metabolic pathways is helpful in selecting potential candidate genes for evaluation.
Guizhou White goats, with advantages such as strong adaptability, stable genetic performance, fresh flesh and having a delicious taste, make for an important germplasm resource of animal husbandry in the Guizhou province, China [13]. While being smaller in size, growing more slowly, and having lower meat production rates are contributing factors for the low economic efficiency of Guizhou White goats [14,15]. The growth characteristics of goats have high economic value, and increasing lamb yield in practical production largely depends on identifying and exploring how genetic variations in key genes that control growth rate regulate gene expression at the molecular level.
In recent years, the Genome-wide association study (GWAS) has gradually been applied to conduct research on goat body weight, body size and meat quality traits, identifying a large number of important genes and their associated SNPs for quantitative and qualitative traits, including Zhongwei goat [16], Karachai goat [17], Youzhou Dark goat [18] and Sub-Saharan Africa indigenous goat [19], enriching the scope of goat genomics research. However, there are relatively few SNPs screened in the promoter region, especially key factors related to gene transcription regulation mechanisms. Promoters are important regions for gene regulation, as they contain many sites and functional domains that bind to transcription regulatory factors. Therefore, the polymorphic sites in these regions are likely to affect the mRNA expression level of the gene to some extent, further affecting the protein function and causing changes in animal phenotypic traits.
So far, research on the relationship between the promoter region of the MyoG gene and growth traits is limited. The main objective of this study is to further investigate the association between novel SNPs within the promoter region of the MyoG gene and body size in goats. The MyoG gene in goats is located on chromosome 16, and there are several quantitative trait loci (QTLs) related to carcass weight [20]. Up to now, there have been no reports on the association between genetic variations in the promoter region of the MyoG gene in goats and growth traits. Thus, this study aims to explore the SNPs in the promoter region of the MyoG gene in Guizhou White goats and their effects on body measurement traits and transcription factor binding sites.
2. Materials and Methods
2.1. Statement of Ethics
The collection of goats samples involved in this study has been approved by the Ethics Committee of Tongren University, China (Permit number for conducting animal experiments: TRXY 2023-088; Approval date: 8 March 2023), and everything possible was done to minimize the pain of the goat as much as possible.
2.2. Animal Sources, Data Collection and DNA Extraction
Male Guizhou White (GZW) goats (n = 224 of about three years of age) were selected at random from Huazhen Animal Husbandry Co., Ltd. in Tongren City, Guizhou Province, China. According to farm records, all of the goats were GZW goats that were genetically distinct from each other and were subjected to the same environmental conditions and nutrition. According to the same standards, peripheral blood samples (5 mL) were collected from each goat through the jugular vein using a vacuum tube and frozen at −20 °C for DNA extraction. The growth traits of all selected goat individuals have been recorded by the breeding farm. Body size indicators include the following: body weight (BW), body length (BL), body height (BH), chest circumference (ChC) and cannon circumference (CaC). The blood genomic DNA of Guizhou White goats was extracted with the blood genomic DNA extraction kit (TransGen Biotech, Beijing, China), and its concentration and purity were detected by ultraviolet spectrophotometer and 0.7% agarose gel electrophoresis diluted to 80 ng/μL and stored at −20 °C.
2.3. Primer Design and PCR Amplification
Based on the flanking regions of the MyoG gene in C. h. (Accession number FJ607135) and Bos taurus (Accession number HM452338) as reference sequences, a pair of specific primers (Forward: 5′-CTTCCCTCCTCACCCACATT-3′, Reverse: 5′-TCCACAGACACCGACTTCCT-3′) were designed using Oligo 7.0 and Primer Premier 5.0 software to amplify the promoter region of the MyoG gene in Guizhou White goats. The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). PCR amplification was performed in 40 μL reaction mixture, which was made up of 20 μL 2 × SanTaq Fast Mix (Sangon Biotech Co., Ltd., Shanghai, China), 1.0 μL of 80 ng/μL template DNA, 1.0 μL of 10 pmol/μL each primer and 17 μL sterilized ultrapure water. The PCR thermal cycle was programmed as follows: pre-denaturation at 94 °C for 5 min, denaturation at 94 °C for 30 s, annealing at 58.8 °C for 30 s, extension at 72 °C for 75 s and 40 cycles and a final extension at 72 °C for 10 min, stored at 4 °C. PCR products were detected by 1.2% agarose gel electrophoresis and then sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequencing. SNPs in the MyoG gene were detected by bidirectional direct sequencing.
2.4. SNPs Screening and Genotype Determination
The obtained MyoG gene sequences of 224 Guizhou White goats were aligned by the ClusterW Multiple alignment program in BioEdit 7.0 software and detected SNPs sites. The genotype of SNPs loci was determined using the sequencing peak mapping software Chromos 5.0, with a single peak representing a homozygous genotype and a nested peak representing a heterozygous genotype.
2.5. Genotype, Allele Frequency and Association Analysis Between SNPs Loci and Growth Traits
The genotype, allele frequency and Hardy–Weinberg equilibrium (HWE) of SNPs loci were analyzed based on the online software SHEsis (http://analysis.bio-x.cn, accessed on 8 June 2024). Polymorphism information content (PIC) was calculated through online software (http://www.msrcall.com/Gdicall.aspx, accessed on 18 June 2024). The mean and standard error of the different growth traits of 224 individual Guizhou White goats with different SNPs loci of the MyoG gene were statistically analyzed by means of IBM SPSS 23.0 software. At the same time, according to the general linear model, which is used to make a significant test on the effects of various genotypes on growth traits, one-way ANOVA was used to conduct a homogeneity of variance test and a multiple comparison test and to further analyze the correlation between SNPs locus and growth traits of Guizhou White goats. A general linear model is yij = μ + αi + εij, where yij is the individual growth trait (body weight, etc.) record, μ is population mean for each growth trait, αi is the genotype effect and εij is the random error. The data is displayed as mean ± standard error (SE), with p < 0.05 indicating statistically significant differences and p < 0.01 indicating extremely significant differences.
2.6. Identification of Transcription Factor Binding Sites Before and After SNPs Mutation
The possible transcription factor binding sites in the promoter region of the MyoG gene in Guizhou White goats were predicted using AliBaba 2.1 software (https://gene-regulation.com/pub/programs/alibaba2/index.html, accessed on 6 July 2024). Predicting changes in transcription factors before and after SNPs mutation in the promoter region of the MyoG gene by means of a transcription factor binding database profile (JASPAR2024: https://ngdc.cncb.ac.cn/databasecommons/database/id/9171, accessed on 10 August 2024).
3. Results
3.1. Detection of MyoG Gene PCR Products
The designed primer was used to amplify the MyoG gene of Guizhou White goats, and the PCR products were separated by 1.2% agarose gel electrophoresis. As shown in Figure 1, the size of the amplified fragment is consistent with the target fragment. The amplified band is clear and dense, which could be used for subsequent direct sequencing analysis.
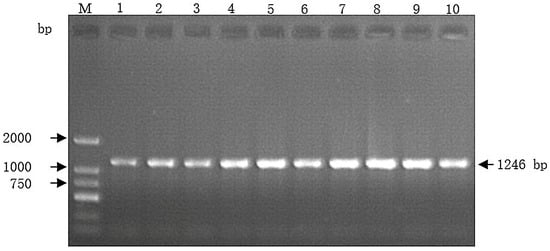
Figure 1.
Agarose electrophoretic map of the MyoG gene specific fragment in Guizhou White goats. Lane M represents DL 2000 Marker. Lane 1 to 10 denotes target segment. The arrow indicates the 1246 bp of the MyoG gene fragment.
3.2. SNP Identification in the Promoter Region of the MyoG Gene
Based on the sequence structure of the bovine MyoG gene in the GenBank database, a total of 1246 bp nucleotide sequences of the MyoG gene in Guizhou White goats were obtained, including 1004 bp 5′UTR and 242 bp exon 1. The Clustal W Multiple alignment program in BioEdit 7.0 software was used to compare the sequencing results of 224 Guizhou White goats, screen for SNPs and determine the genotype of each locus based on the sequencing peak maps. The naming principle of SNPs is that the A base in the start codon ATG is marked as “+1”. At the same time, the obtained 1246 bp sequence was aligned with the goat genome reference sequence in the NCBI database, using BLAST (version 2.17.0) to determine the specific location of the identified SNPs in the goat chromosome. The MyoG gene is located on chromosome 16 of Guizhou White goats. Four SNPs were screened in the promoter region of the MyoG gene in Guizhou White goats, including g.–709C>T, g.–461G>T, g.–377G>T and g.–249G>A, which are located at 193,987 bp, 193,739 bp, 193,655 bp and 193,528 bp of the goat reference genome sequence NC_030823.1, and the variant IDs are rs666139870, rs636117637, rs652431992 and rs639389894, respectively. Two genotypes were formed at the g.–461G>T and g.–249G>A loci in Guizhou White goats, while three genotypes were found at the g.–709C>T and g.–377G>T loci. The sequencing results of the four SNPs are shown in Figure 2 and Supplementary Materials, Figure S1.
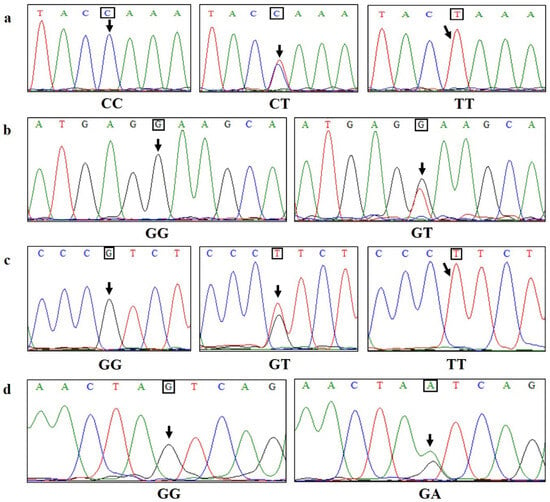
Figure 2.
The sequencing chromatogram of four SNPs for the MyoG gene in Guizhou White goats; SNPs are indicated by arrows. (a–d) represent sequencing results of each genotype at g.–709C>T, g.–461G>T, g.–377G>T and g.–249G>A, respectively.
3.3. Polymorphic Parameter, Genotype and Allele Frequencies for Different SNPs
The genotype and allele frequencies of MyoG gene SNPs in Guizhou White goats are shown in Table 1. Three genotypes were found at the g.–709C>T locus, namely, CC, CG and GG; GG, GT and TT were identified at the g.–377G>T locus. Two genotypes were found at the g.–461G>T locus, namely, GG and GT; GG and GA were screened at the g.–249G>A locus. The χ2-test showed that Guizhou White goats conform to the Hardy–Weinberg equilibrium at the g.–709C>T and g.–461G>T loci (p > 0.05), while the g.–377G>T and g.–249G>A loci significantly departed from the Hardy–Weinberg equilibrium (p < 0.01). At the g.–709C>T, g.–461G>T, g.–377G>T and g.–249G>A loci in the promoter region of the MyoG gene, the frequencies of the C, G, G and G alleles are higher than those of alleles T, T, T and A. As shown in Table 1, the population genetic parameters of four SNPs of the promoter region of the MyoG gene in Guizhou White goats were calculated. The PIC, Ne, Ho and He of the g.–709C>T locus were 0.36, 1.92, 0.52 and 0.48 and of the g.–377G>T locus were 0.29, 1.53, 0.65 and 0.35, respectively, indicating that Guizhou White goats have moderately polymorphisms (0.25 < PIC < 0.5) at the g.–709C>T and g.–377G>T loci of the MyoG gene promoter region.

Table 1.
Population genetic analysis of four SNPs of the MyoG gene promoter region in Guizhou White goats.
3.4. Association of MyoG Gene Promoter Region Polymorphism and Growth Traits in Guizhou White Goats
In order to investigate the association between the above mentioned SNPs and the growth traits of Guizhou White goats, a general linear model was used for association analysis by means of IBM SPSS 23.0 software. As shown in Table 2, the individuals with the CT genotype formed by the g.–709C>T locus in the promoter region of the MyoG gene in Guizhou White goats had significantly higher body weight than those with the CC and TT genotypes (p < 0.05), and the chest circumference of individuals with CT genotype was significantly higher than that of individuals with the CC and TT genotypes (p < 0.05). The body length, body height and circumference of individuals with the CT genotype tended to be higher than that of those with the CC and TT genotypes, but these differences are not significant (p > 0.05). The body weight of individuals with the GG genotype caused by the g.–461G>T locus was extremely significantly higher than those with the GT genotype (p < 0.01), and no significant differences were observed in body length, height, chest circumference or cannon circumference among individuals with different genotypes. (p > 0.05). No significant differences were observed in body weight, body length, body height, chest circumference or cannon circumference among genotypes at the g.−377G>T locus (p > 0.05). The body length of individuals with the GG genotype caused by the g.–249G>A locus was significantly higher than those with the GA genotype (p < 0.01), and no significant differences were observed in body weight, body height, chest circumference and cannon circumference among individuals with different genotypes (p > 0.05).

Table 2.
The correlation between four SNPs in the promoter region of the MyoG gene and the growth traits of Guizhou White goats.
3.5. SNPs in the Promoter Region of the MyoG Gene Cause Alterations in Transcription Factors
The alterations of transcription factors in the MyoG gene promoter region, calculated by the online software JASPAR2024, are shown in Table 3. The g.–709C>T locus may result in the vanishing of both the C/EBPalp and serum response factor (SRF) transcription factor binding sites and in the increase of a new TATA binding protein (TBP) site. The g.–461G>T locus may result in the vanishing of the PU.1 transcription factor binding sites and in the increase of activator protein 1 (AP-1). The g.–377G>T locus may lead to the vanishing of the specific protein 1 (SP1) transcription factor binding sites and to the increase of octamer binding transcription factor 1 (OCT-1) binding sites. The g.–249G>A mutation site may result in the vanishing of the ELA1 transcription factor binding site and in the increase of ALX1 binding sites. Thus, these SNPs, especially at newly identified loci, may form new transcription factor binding sites while disrupting primitive existing binding sites and disturbing transcriptional regulation and expression of the MyoG gene.

Table 3.
Alterations of transcription factors before and after SNPs mutation of the MyoG gene in Guizhou White goats.
4. Discussion
4.1. Correlation Between MyoG Gene SNPs and Growth Traits of Caprinae
The Guizhou White goat is an excellent local breed in the Guizhou province, with advantages such as strong adaptability, tolerance to rough feeding, tender meat and delicious meat taste. However, this breed has disadvantages such as small size, low slaughter rate, low net meat rate and slow growth rate, which limit the large-scale breeding and industrial development of local goat breeds. Screening candidate genes and related molecular markers for goat growth traits at the molecular level and conducting marker-assisted selection breeding will be one of the effective means to improve goat meat production capacity and meat quality. It is also a good way to improve local goat breeds with Guizhou characteristics or cultivate new meat breeds [21]. There are few SNPs in the MyoG gene of goats, all of which are significantly correlated with meat quality and growth traits, indicating that the MyoG gene is a key candidate gene for mutton development [22]. Screening of genetic variant molecular markers of the MyoG gene associated with growth traits is of significant theoretical importance and has implications for the development of molecular breeding or meat production traits. At present, research on MyoG gene SNPs is mainly focused on pigs [23,24,25], cattle [26,27] and sheep [28,29], mainly involving the association between SNPs and meat quality and carcass traits. Previous studies have analyzed sheep MyoG, including three exons and two introns, and identified some polymorphisms. Han et al. [30] reported that there was one single nucleotide missense mutation of A109C at exon 1 of a Tibetan sheep MyoG gene. Statistical analysis indicated that in the Henan Oula Tibetan sheep population, individuals with the CC genotype have higher mean values for body weight, body height and body length than those with the AA genotype. Li et al. [9] analyzed the polymorphism in intron 2 of MyoG gene in Chinese sheep local breeds using the PCR-RFLP method. The dominant genotype was AB, and the Eco 72 I locus could serve as a molecular marker affecting sheep growth traits. The same SNPs of the MyoG gene intron 2 have also been reported in Boer goat populations, which indicates that the MyoG gene has a certain influence on the growth traits of goats [31]. Thus, investigating the SNPs in the promoter region of the MyoG gene and their associations with growth traits in Guizhou White goats represents a significant approach for discovering new molecular genetic markers used in goat breeding practices.
Previous studies have found that PIC value is an important parameter for marker site polymorphism in linkage analysis [32,33]. In this study, four SNPs in the promoter region of the MyoG gene were identified, and the relationship between SNPs and growth traits in Guizhou White goats was studied. These SNPs appear to be new and have never appeared in previous databases. In addition, their impact on the MyoG gene expression or their association with goat growth traits have not been explored. For the g.–709C>T and g.–377G>T loci, the PIC value has been obtained and were identified as moderate polymorphic in the Guizhou White goat population (0.25 < PIC < 0.5). This indicates that breeders can utilize significant genetic diversity throughout the entire evolutionary history of animals to select superior varieties. For the g.–461G>T and g.–249G>A loci, the PIC value exhibits low level polymorphism. Simultaneously, the goat population in this study was in Hardy–Weinberg equilibrium at the g.–709C>T and g.–461G>T loci. This equilibrium may be due to the stable genetic status of the Guizhou White goat population after long-term breeding, which suggests that selective breeding measures may be taken in the future to improve production traits such as growth rate, meat quality and carcass traits. Nevertheless, the g.–377G>T and g.–249G>A loci exhibited a significant deviation from the Hardy–Weinberg equilibrium, which may be due to an insufficient sample size that does not adequately represent the phenotypic distribution of these loci within these broader populations. Therefore, it is imperative that the Guizhou White goat sample size be expanded in future studies to validate this discovery.
The CT genotypes of the g.–709C>T locus appear to have a relatively higher impact on body weight and chest circumference in Guizhou White goats (p < 0.01). The genetic variation at this locus is directly associated with the weight and chest circumference of Guizhou White goats, which may involve dominant effects or heterozygous advantages. Similarly, in Nile tilapia, heterozygosity at certain SNP loci of the MYF5 gene is significantly associated with growth traits [34]. Therefore, heterozygous state may enhance growth potential through gene interactions. The significant effect of heterozygous genotypes suggests that these SNPs may affect growth traits through dominant or heterozygous dominance, providing a key target for genetic improvement. This result suggests that screening heterozygous genotypes in molecular marker-assisted breeding of goats has important theoretical significance and practical value, which is helpful for breeding excellent meat breeds of Guizhou White goats with rapid growth. The GG genotypes of the g.–249G>A locus appear to have a relatively higher impact on body length in Guizhou White goats (p < 0.01). The GG genotypes of the g.–461G>T locus appear to have a relatively higher impact on body weight in Guizhou White goats (p < 0.01).
Nevertheless, it must be clarified that body weight, chest circumference and body length are quantitative traits determined by many genes, including MYOD1, MSTN, IGFs [35,36,37] and so on. Therefore, the impact of single genes and mutations may be minimal. In addition, the application of MyoG gene polymorphism in Guizhou White goat breeding may be hindered by bias in genotype and allele distribution, even lacking some genotypes in goat populations (such as the TT genotype of the g.–461G/T locus). Therefore, their value in breeding and selection must still be validated on sufficient goat populations, preferably on different goat breeds. In addition, the next step of this study in the future should be to expand the sample size of goats and further analyze the association between SNPs haplotypes in the MyoG gene promoter region and goat growth traits.
The SNPs identified in the untranslated region and exons of the MyoG gene may have a significant impact on the expression of this gene. In Jinghai yellow chickens, individuals with the BB genotype of the MyoG gene, which was formed by g.36T>C mutation in exon 3, had higher bodyweight at different weeks [38]. In zebrafish embryos, the expression of the MyoG gene is regulated by lots of regulatory elements, especially multiple transcription factors in the promoter region [39]. Current research has only analyzed the correlation between SNPs and growth traits of Guizhou White goats. The correlation analysis between SNPs and the expression level of this gene will be the focus of our subsequent research. However, as far as we know, this study is the first to investigate the association between SNPs in the promoter region of the MyoG gene and growth traits in goats.
In the screening of SNPs in the exons and introns of the MyoG gene in goats, individuals with the AA genotype formed by the g.1823C>T mutation in intron 2 had significantly lower birth weight than individuals with the AB and BB genotypes. Selecting individuals with the B allele is expected to improve growth traits related to goat body mass [31]. In terms of other members of the myogenic regulatory factor family, myogenic differentiation factor 1 is also a gene locus significantly associated with growth traits. The Copy Number Variation loci containing the MyoD1 gene exon in Shanbei white cashmere goats is significantly correlated with body weight, body height, hip height, chest circumference and hip circumference (p < 0.05), indicating that this locus is a marker-assisted selection breeding locus for goats [35]. In addition, insulin-like growth factor 1 (IGF-1) is an important member of the animal growth axis, mainly regulating fetal growth and development, and is a key factor for linear growth in animals after birth. Several QTLs related to growth and body shape traits were identified on goat chromosome 5, including the IGF-1 gene [40]. Many researchers have directly sequenced different goat breeds and found a large number of SNPs in the 5′untranslated region of the goat IGF-1 gene, some of which are significantly correlated with growth traits such as birth weight, chest width and chest depth [41,42]. However, whether the SNPs of the MyoG gene discovered in this study jointly affect the growth traits of Guizhou White goats with the above mentioned mutation sites still needs further verification through subsequent gene interaction and related analyses.
In future research, we will expand to genome-wide association analysis (GWAS) to systematically identify more SNPs loci related to the growth traits of Guizhou White goats. In addition, it is necessary to combine functional validation experiments, such as gene editing or expression analysis, to clarify the biological mechanisms of candidate genes. Strengthening the integration of genetic resource protection and molecular breeding techniques will help achieve precise breeding for Guizhou White goats.
4.2. Effects of Transcription Factor Binding Sites Alteration in the Promoter Region on Transcriptional Regulation of MyoG Gene
The promoter region is a specific DNA sequence located upstream of a gene, which is a key and important region for RNA polymerase and transcription factors to bind and initiate gene transcription. SNPs in the promoter region may regulate gene expression by altering the binding mode of some transcription factors, which is one of the important mechanisms for understanding individual genetic differences and susceptibility to complex diseases [43,44]. SNPs in the promoter region may indeed alter transcription factor binding sites by directly disrupting or creating transcription factor binding sites and indirectly affecting binding affinity or conformation, thereby affecting gene expression [45,46].
In this study, the transcription factor prediction of the MyoG gene promoter region showed that the mutation of g.–709C>T resulted in the disappearance of the initial C/EBPalp and SRF transcription factor binding sites and in the formation of a new binding site for the TBP transcription factor. TATA binding protein (TBP) is essential for RNA polymerase III transcription. Previous studies have found that TBP plays a central role in transcriptional initiation, which was first discovered to recognize and bind to the TATA box in the gene promoter region. In addition, TBP is a key factor necessary for initiating the three major eukaryotic RNA polymerases [47,48]. Regarding the newly added TBP transcription factor binding sites, this study only used bioinformatics software for predictive analysis. Whether there is a positive over-effect still needs to be verified in the future. Similarly, the sample size should also be increased for analysis in the future.
The mutation of g.–461G>T locus resulted in the vanishing of the initial PU.1 transcription factor binding site and in the increase of a new binding site for the AP-1 transcription factor. Activation protein-1 (AP-1) is an inducible transcription factor which is composed of multiple protein complexes, including members of the fos and jun gene families. Many cells and viral genes contain AP-1 binding sites in their promoters; therefore, AP-1 has been investigated to determine whether it plays a role in regulating these genes and inducing transcription [49]. Further in-depth research is needed to determine whether the increased AP-1 transcription factor caused by the g.461G>T mutation in the promoter region of the MyoG gene in Guizhou White goats will affect the transcription and functional expression of this gene.
The mutation of g.–377G>T locus, led to the vanishing of the initial SP1 transcription factor binding site; meanwhile, a new binding site for the OCT-1 transcription factor was raised. OCT-1 was first found as a member of the POU transcription factor family and is widely expressed in most cells and tissues. OCT-1 plays an extremely important role in development and differentiation processes [50]. The specific role of the increased OCT-1 transcription factor in the promoter region of the MyoG gene in Guizhou White goats also requires further verification and exploration. The mutation of g.–249G>A locus resulted in the disappearance of the initial ELA1 transcription factor binding site and a new binding site for the ALX1 transcription factor was formed. Alx1 is a transcription factor containing homologous domains and is a highly conserved regulatory factor for bone formation in echinoderms. In sea urchins, Alx1 plays a central role in the differentiation of embryonic primary mesenchymal cells (PMCs) and actively regulates the transcription of most biomineralization genes expressed in these cells [51]. The function of the Alx1 transcription factor in the development of muscle and bone tissue in vertebrates, especially goats, also needs to be further explored.
These four SNPs in the promoter region of the MyoG gene have altered the initial transcription factor interaction, which may, therefore, be leading to alterations in promoter activity and gene function. Alterations of the transcription factor are likely to decrease or increase the expression level of the MyoG gene, which indirectly affects the growth traits of Guizhou White goats. Thus, it is speculated that the alterations of transcription factors may affect the meat production capacity of Guizhou White goats, but the specific mechanism still needs to be explored further in-depth.
5. Conclusions
In the current study, the MyoG gene promoter region sequences of 224 Guizhou White goats were explored. Four SNPs were screened in the region –1000 bp upstream to 300 bp downstream of the goat MyoG gene transcription start site. The mutation within the promoter region is being reported for the first time in goats. Association analysis suggests that g.–709C>T and g.–461G>T loci in the promoter region are molecular marker sites that affect the weight index of Guizhou White goats. The g.–249G>A site is a molecular marker site that affects the body length index of Guizhou White goats. These SNPs might lead to alterations in their initial transcription factor binding sites, thus potentially changing gene expression. However, the relationship between the SNPs identified in this research and the expression level of MyoG gene in goats should be further studied in the future. Similarly, we will use a dual luciferase reporter gene vector to detect the promoter activity and determine the promoter active region of Guizhou White goats and verify the transcription factors of the mutation sites through EMSA or Chip methods. In short, these findings provide a theoretical basis for further exploring the regulation mechanisms of MyoG expression and its relationship with goat breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes17010014/s1, Figure S1: Sanger sequencing chromatograms of different genotype for 4 mutation sites (a–d) in Guizhou White goat MyoG gene promoter region. Red rectangles indicate the positions of the mutation sites.
Author Contributions
X.S. and H.L. analyzed the data and drafted the manuscript. J.M. and Y.Z. prepared the blood samples for sequencing. Z.W. and H.L. performed the software and formal analysis. Q.A. initiated this study, designed the experiments and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Guizhou Science and Technology Project (Qian Kehe Foundation No. ZK [2022] common 559), Doctoral Research Initiation Foundation of Tongren University (trxyDH2001), Guizhou Thousand Level Innovative Talent Project (2024-[2022]-048), Doctoral Talents project of Science and Technology Bureau of Tongren, Guizhou Province ([2020]126). Science and Technology Program of Guizhou Province (support projects MS2025-093), Doctoral Talent Program of Tongren (Tongren Scientific Research 2023-4).
Institutional Review Board Statement
All experimental procedures and sample collection in this study involving goats were approved by the Ethics Committee of Tongren University, China (Permit number for conducting animal experiments: TRXY 2023-088; Approval date: 8 March 2023), and all efforts were made to minimize the suffering of the goats.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kopantseva, E.E.; Belyavsky, A.V. Key regulators of skeletal myogenesis. Mol. Biol. 2016, 50, 169–192. [Google Scholar] [CrossRef]
- Te Pas, M.F.W.; Soumillion, A.; Harders, F.L.; Verburg, F.J.; van den Bosch, T.J.; Galesloot, P.; Meuwissen, T.H. Influences of myogenin genotypes on birth weight, growth rate, carcass weight, backfat thickness, and lean weight of pigs. J. Anim. Sci. 1999, 77, 2352–2356. [Google Scholar] [CrossRef]
- Zhan, S.; Zhai, H.; Tang, M.; Xue, Y.; Li, D.; Wang, L.; Zhong, T.; Dai, D.; Cao, J.; Guo, J.; et al. Profiling and functional analysis of mRNAs during skeletal muscle differentiation in goats. Animals 2022, 12, 1048. [Google Scholar] [CrossRef]
- Wyszyńska-Koko, J.; Pierzchała, M.; Flisikowski, K.; Kamyczek, M.; Różycki, M.; Kurył, J. Polymorphisms in coding and regulatory regions of the porcine MYF6 and MYOG genes and expression of the MYF6 gene in m. longissimus dorsi versus productive traits in pigs. J. Appl. Genet. 2006, 47, 131–138. [Google Scholar] [CrossRef]
- Li, M.; Liu, Q.; Xie, S.; Fu, C.; Li, J.; Tian, C.; Li, X.; Li, C. LncRNA TCONS_00323213 promotes myogenic differentiation by interacting with PKNOX2 to upregulate MyoG in porcine satellite cells. Int. J. Mol. Sci. 2023, 24, 6773. [Google Scholar] [CrossRef]
- Verner, J.; Humpolíček, P.; Knoll, A. Impact of MYOD family genes on pork traits in Large White and Landrace pigs. J. Anim. Breed. Genet. 2007, 124, 81–85. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.A.; Kim, N.K.; Cho, Y.M.; Yoon, D.; Kim, K.S.; Jeon, J.T.; Lee, J.H. Identification of SNPs in MYOD gene family and their associations with carcass traits in cattle. Livest. Sci. 2009, 126, 292–297. [Google Scholar] [CrossRef]
- Sousa, L.P.B., Jr.; Meira, A.N.; Azevedo, H.C.; Muniz, E.N.; Coutinho, L.L.; Mourão, G.B.; Leão, A.G.; Pedrosa, V.B.; Pinto, L.F.B. Variants in myostatin and MyoD family genes are associated with meat quality traits in Santa Inês sheep. Anim. Biotechnol. 2022, 33, 201–213. [Google Scholar] [CrossRef]
- Li, J.Y.; Wang, X.L.; Zhang, Y.F.; Bai, J.Y.; Li, G.; Lei, Y.; Dong, Z.H.; Chen, Y.; Fan, H.D.; Wang, L.W.; et al. Polymorphism of MyoG gene intron II and its association with growth traits in sheep. China Anim. Husb. Vet. Med. 2022, 49, 1364–1373. [Google Scholar] [CrossRef]
- Trukhachev, V.; Skripkin, V.; Telegina, E.; Yatsyk, O.; Golovanova, N.; Krivoruchko, A. Associations between newly discovered polymorphisms of the MyoD1 gene and body parameters in Stavropol breed rams. Bulg. J. Vet. Med. 2018, 21, 28–39. [Google Scholar] [CrossRef]
- Sun, W.; Wang, P.; Ding, J.T.; Ma, Y.H.; Guan, W.J.; Chu, M.X.; Li, B.C.; Wu, W.Z.; Chen, L. Development changes of gene expression of Myostatin and Myogenin genes and their association analysis with carcass traits in Hu sheep. Sci. Agric. Sin. 2010, 43, 5129–5136. [Google Scholar] [CrossRef]
- Mukherjee, A.; Gali, J.; Kar, I.; Datta, S.; Roy, M.; Acharya, A.P.; Patra, A.K. Candidate genes and proteins regulating bull semen quality: A review. Trop. Anim. Health Prod. 2023, 55, 212. [Google Scholar] [CrossRef]
- Xu, H.Q. Local Livestock and Poultry Genetic Resources in Guizhou, 1st ed.; China Agriculture Press: Beijing, China, 2019. [Google Scholar]
- Ruan, Y.; Dai, L.; Huang, J.; Xiao, M.; Xu, J.; An, D.; Chen, J.; Chen, X. A novel nonsynonymous SNP in the OLR1 gene associated with litter size in Guizhou white goats. Theriogenology 2023, 200, 1–10. [Google Scholar] [CrossRef]
- Shi, S.Y.; Li, L.J.; Zhang, Z.J.; Wang, E.Y.; Wang, J.; Xu, J.W.; Liu, H.B.; Wen, Y.F.; He, H.; Lei, C.Z.; et al. Copy number variation of MYLK4 gene and its growth traits of Capra hircus (goat). Anim. Biotechnol. 2020, 31, 532–537. [Google Scholar] [CrossRef]
- Han, M.; Wang, X.; Du, H.; Cao, Y.; Zhao, Z.; Niu, S.; Bao, X.; Rong, Y.; Ao, X.; Guo, F.; et al. Genome-wide association study identifies candidate genes affecting body conformation traits of Zhongwei goat. BMC Genom. 2025, 26, 37. [Google Scholar] [CrossRef]
- Easa, A.A.; Selionova, M.; Aibazov, M.; Mamontova, T.; Sermyagin, A.; Belous, A.; Abdelmanova, A.; Deniskova, T.; Zinovieva, N. Identification of genomic regions and candidate genes associated with body weight and body conformation traits in Karachai goats. Genes 2022, 13, 1773. [Google Scholar] [CrossRef]
- Sun, X.; Niu, Q.; Jiang, J.; Wang, G.; Zhou, P.; Li, J.; Chen, C.; Liu, L.; Xu, L.; Ren, H. Identifying candidate genes for litter size and three morphological traits in Youzhou Dark goats based on genome-wide SNP markers. Genes 2023, 14, 1183. [Google Scholar] [CrossRef]
- Ncube, K.T.; Nephawe, K.A.; Mpofu, T.J.; Monareng, N.J.; Mofokeng, M.M.; Mtileni, B. Genomic advancements in assessing growth performance, meat quality, and carcass characteristics of goats in Sub-Saharan Africa: A systematic review. Int. J. Mol. Sci. 2025, 26, 2323. [Google Scholar] [CrossRef]
- Wang, Z.; Lv, Q.; Li, W.; Huang, W.; Gong, G.; Yan, X.; Liu, B.; Chen, O.; Wang, N.; Zhang, Y.; et al. Chromosome-level genome assembly of the cashmere goat. Sci. Data 2024, 11, 1107. [Google Scholar] [CrossRef]
- An, Q.; Zeng, L.; Wang, W.; Yang, J.; Meng, J.; Zhao, Y.; Song, X. Identification of FASN gene polymorphisms, expression and their relationship with body size traits in Guizhou White goat (Capra hircus) with different genders. Genes 2024, 15, 656. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, F.; Zhang, Y.; Li, W.; Yin, Y.; Zhu, C.; Du, L.; Elsayed, A.K.; Li, B. Cloning and expression of MyoG gene from Hu sheep and identification of its myogenic specificity. Mol. Biol. Rep. 2014, 41, 1003–1013. [Google Scholar] [CrossRef]
- Xue, H.L.; Zhou, Z.X. Effects of the MyoG gene on the partial growth traits in pigs. Acta Genet. Sin. 2006, 33, 992–997. [Google Scholar] [CrossRef]
- Anton, I.; Zsolnai, A.; Komlósi, I.; Király, A.; Fésüs, L. Effect of MYOG genotypes on growth rate and production traits in Hungarian large white pigs. Acta Vet. Hung. 2006, 54, 393–397. [Google Scholar] [CrossRef]
- Soumillion, A.; Erkens, J.H.F.; Lenstra, J.A.; Rettenberger, G.; Te Pas, M.F.W. Genetic variation in the porcine myogenin gene locus. Mamm. Genome 1997, 8, 564–568. [Google Scholar] [CrossRef]
- Beever, J.E.; Fisher, S.R.; Lewin, H.A. Polymorphism identification in the ACADM, AT3, IL10, MYOG and TSHB genes of cattle. Anim. Genet. 1997, 28, 373–374. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, J.; Raza, S.H.A.; Song, Y.; Jiang, C.; Song, X.; Wu, H.; Alotaibi, M.A.; Albiheyri, R.; Al-Zahrani, M.; et al. Interaction of MyoD and MyoG with Myoz2 gene in bovine myoblast differentiation. Res. Vet. Sci. 2022, 152, 569–578. [Google Scholar] [CrossRef]
- Sousa, L.P.B., Jr.; Meira, A.N.; Azevedo, H.C.; Muniz, E.N.; Coutinho, L.L.; Mourão, G.B.; Pedrosa, V.B.; Pinto, L.F.B. Polymorphismsin MyoD1, MyoG, MyF5, MyF6, and MSTN genes in Santa Inês sheep. Pesqui. Agropecu. Bras. 2019, 54, e01132. [Google Scholar] [CrossRef]
- Wang, X.; Bai, J.Y.; Yang, Y.B.; Lei, X.Q.; Pang, Y.Z.; Li, H.W.; Wang, H.L.; Tang, Y.L.; Shao, J.H.; Yin, H.Y. Study on the polymorphism of MyoG exon 2 and MyoD 5′ Flanking region of Small tail han sheep. J. Henan Agric. Sci. 2017, 26, 138–141. [Google Scholar] [CrossRef]
- Han, Y.C.; Sun, Y.G.; Chen, Z. Single nucleotide polymorphism of MyoG gene in Tibetan sheep populations and its association with body measurement traits. J. Anim. Ecol. 2016, 37, 19–23. [Google Scholar] [CrossRef]
- Liu, Z.Z.; Gong, Y.F.; Fu, Z.X.; Zhang, C.S.; Zhang, W.X.; Song, Y.; Ma, Y.H. Genetic variation of goat MyoG gene intron II and its genetic effects on body weight. Acta Vet. Zootech. Sin. 2011, 42, 1015–1021. [Google Scholar]
- Singh, S.P.; Kumar, R.; Kumari, P.; Kumar, S.; Mitra, A. Characterization of 5′ upstream region and investigation of TTTTA deletion in 5′ UTR of myostatin (MSTN) gene in Indian goat breeds. Anim. Biotechnol. 2014, 25, 55–68. [Google Scholar] [CrossRef]
- Zhou, X.; Yan, Q.; Liu, L.; Chen, G.; Tang, S.; He, Z.; Tan, Z. Maternal undernutrition alters the skeletal muscle development and methylation of myogenic factors in goat offspring. Anim. Biosci. 2022, 35, 847–857. [Google Scholar] [CrossRef]
- Wei, L.; Xiao, W.; Chen, B.; Zou, Z.; Zhu, J.; Li, D.; Yu, J.; Yang, H. Single nucleotide polymorphisms in the MRFs gene family associated with growth in Nile tilapia. Mol. Biol. Rep. 2024, 51, 128. [Google Scholar] [CrossRef]
- Ren, H.; Wei, Z.; Li, X.; Wang, Q.; Chen, H.; Lan, X. Goat MyoD1: mRNA expression, InDel and CNV detection and their associations with growth traits. Gene 2023, 866, 147348. [Google Scholar] [CrossRef]
- Na, R.; Ni, W.; E, G.; Zeng, Y.; Han, Y.; Huang, Y. SNP screening of the MSTN gene and correlation analysis between genetic polymorphisms and growth traits in Dazu black goat. Anim. Biotechnol. 2021, 32, 558–565. [Google Scholar] [CrossRef]
- Pehlivan, E. Relationship between insulin-like growth factor-1 (IGF-1) concentrations and body trait measurements and climatic factors in prepubertal goat kids. Arch. Anim. Breed. 2019, 62, 241–248. [Google Scholar] [CrossRef]
- Zhang, G.; Tang, Y.; Zhang, T.; Wang, J.; Wang, Y. Expression profiles and association analysis with growth traits of the MyoG and Myf5 genes in the Jinghai yellow chicken. Mol. Biol. Rep. 2014, 41, 7331–7338. [Google Scholar] [CrossRef]
- Du, S.J.; Gao, J.; Anyangwe, V. Muscle-specific expression of myogenin in zebrafish embryos is controlled by multiple regulatory elements in the promoter. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 134, 123–134. [Google Scholar] [CrossRef]
- Trukhachev, V.; Belyaev, V.; Kvochko, A.; Kulichenko, A.; Kovalev, D.; Pisarenko, S.; Volynkina, A.; Selionova, M.; Aybazov, M.; Shumaenko, S.; et al. Genes expression profiles in the loin muscle of manych merino sheep with different live weight. Bulg. J. Vet. Med. 2016, 19, 19–29. [Google Scholar] [CrossRef]
- Naicy, T.; Venkatachalapathy, R.T.; Aravindakshan, T.V.; Kurian, E. Association of a Cac8I polymorphism in the IGF1 gene with growth traits in Indian goats. J. Genet. Eng. Biotechnol. 2017, 15, 7–11. [Google Scholar] [CrossRef]
- Song, T.; Tan, Y.; Cuomu, R.; Liu, Y.; Ba, G.; Suo, L.; Wu, Y.; Cao, X.; Zeng, X. Polymorphisms and mRNA expression levels of IGF-1, FGF5, and KAP 1.4 in Tibetan cashmere goats. Genes 2023, 14, 711. [Google Scholar] [CrossRef]
- Vega, W.H.O.; Quirino, C.R.; Bartholazzi, A., Jr.; Rua, M.A.S.; Serapião, R.V.; Oliveira, C.S. Variants in the CYP19A1 gene can affect in vitro embryo production traits in cattle. J. Assist. Reprod. Genet. 2018, 35, 2233–2241. [Google Scholar] [CrossRef]
- Cosenza, G.; Iannaccone, M.; Pico, B.A.; Gallo, D.; Capparelli, R.; Pauciullo, A. Molecular characterisation, genetic variability and detection of a functional polymorphism influencing the promoter activity of OXT gene in goat and sheep. J. Dairy Res. 2017, 84, 165–169. [Google Scholar] [CrossRef]
- Mishra, C.; Kumar, S.; Panigrahi, M.; Yathish, H.M.; Chaudhary, R.; Chauhan, A.; Kumar, A.; Sonawane, A.A. Single nucleotide polymorphisms in 5′ upstream region of bovine TLR4 gene affecting expression profile and transcription factor binding sites. Anim. Biotechnol. 2018, 29, 119–128. [Google Scholar] [CrossRef]
- Wang, P.; Li, W.; Liu, Z.; He, X.; Lan, R.; Liu, Y.; Chu, M. Analysis of the association of two SNPs in the promoter regions of the PPP2R5C and SLC39A5 genes with litter size in Yunshang black goats. Animals 2022, 12, 2801. [Google Scholar] [CrossRef]
- Kwan, J.Z.J.; Nguyen, T.F.; Uzozie, A.C.; Budzynski, M.A.; Cui, J.; Lee, J.M.C.; Van Petegem, F.; Lange, P.F.; Teves, S.S. RNA Polymerase II transcription independent of TBP in murine embryonic stem cells. eLife 2023, 12, e83810. [Google Scholar] [CrossRef]
- Cormack, B.P.; Struhl, K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 1992, 69, 685–696. [Google Scholar] [CrossRef]
- Foletta, V.C. Transcription factor AP-1, and the role of Fra-2. Immunol. Cell Biol. 1996, 74, 121–133. [Google Scholar] [CrossRef]
- Hwang, S.S.; Kim, L.K.; Lee, G.R.; Flavell, R.A. Role of OCT-1 and partner proteins in T cell differentiation. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 825–831. [Google Scholar] [CrossRef]
- Guerrero-Santoro, J.; Khor, J.M.; Açıkbaş, A.H.; Jaynes, J.B.; Ettensohn, C.A. Analysis of the DNA-binding properties of Alx1, an evolutionarily conserved regulator of skeletogenesis in echinoderms. J. Biol. Chem. 2021, 297, 100901. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.