Abstract
Background/Objectives: Bacteriophages infecting the genus Streptococcus play a crucial role in microbial ecology and have potential applications in biotechnology and medicine. Despite their importance, significant gaps remain in our understanding of their lysis modules. This study aims to address these deficiencies by analyzing the genomic diversity and lysis module organization in Streptococcus phages. Methods: A search was conducted in the NCBI RefSeq database to identify phage genomes infecting Streptococcus. A representative panel was selected based on taxonomic diversity. Lysis modules were annotated and visualized, functional domains in endolysins were identified, and holins were characterized. Results: A total of 205 phage genomes were retrieved from the NCBI RefSeq database, of which 185 complete genomes were analyzed. A subset of 34 phages was selected for in-depth analysis, ensuring the representation of taxonomic diversity. The lysis modules were annotated and visualized, revealing five distinct organizations. Among the 256 identified endolysins, 25 distinct architectural organizations were observed, with amidase activity being the most prevalent. Holins were classified into 9 of the 74 families listed in the Transporter Classification Database, exhibiting one to three transmembrane domains. Conclusions: This study provides insights into the structural diversity of lysis modules in Streptococcus phages, paving the way for future research and potential biotechnological applications.
1. Introduction
Antimicrobial resistance (AMR) poses a major global health threat, with antibiotic-resistant bacterial infections estimated to have caused 4.95 million deaths worldwide in 2019 [1]. In response to this growing crisis, phage therapy has re-emerged as a promising alternative or complement to traditional antibiotics.
Bacteriophages, or phages, are viruses that infect bacteria with high specificity, often targeting a single species or even a specific strain [2]. They can be broadly classified as lytic or temperate: lytic phages rapidly lyse and kill their bacterial hosts, whereas temperate phages may integrate into the bacterial genome as prophages. Numerous clinical cases have demonstrated the therapeutic potential of lytic phages, particularly against multi-drug-resistant bacterial infections [3].
One of the most promising components of phage therapy is the exploitation of the lysis module, a set of genes that orchestrate bacterial cell wall degradation at the end of the phage replication cycle. In phages infecting Gram-positive bacteria, such as Streptococcus, the lysis module typically comprises an endolysin, which enzymatically cleaves the peptidoglycan, and a holin, which forms pores in the cytoplasmic membrane to facilitate endolysin access. In contrast, phages infecting Gram-negative hosts often include an additional component, the spanin complex (Rz and Rz1 gene products), responsible for disrupting the outer membrane after peptidoglycan degradation [4]. Variants of this system also exist: some phages use pinholins, which create small lesions in the membrane to activate SAR (Signal–Arrest–Release) endolysins, a type of endolysin tethered to the membrane and released upon membrane depolarization [5]. These mechanistic variations underscore the functional diversity and evolutionary specialization of phage lysis strategies.
Endolysins, in particular, have drawn attention for their high specificity, being able to target selected bacterial genera, species, or even strains [6]. Their therapeutic potential has been notably demonstrated against pathogens such as Streptococcus spp. [7,8,9]. Bacteria from this genus are involved in a broad spectrum of diseases, ranging from mild infections to life-threatening conditions such as pneumonia and meningitis [10,11]. The increasing prevalence of antibiotic-resistant Streptococcus strains highlights the urgent need for alternative treatment options [12,13,14]. Notably, endolysins have also shown efficacy in eliminating intracellular Streptococcus, further expanding their clinical relevance [8].
Beyond their clinical importance, some Streptococcus spp., such as Streptococcus thermophilus, hold significant industrial value due to their use in fermented dairy products like yogurt and cheese, underlining their biotechnological and economic importance [15].
This study aims to conduct a comparative in silico analysis of the lysis modules encoded by phages infecting Streptococcus spp. By examining the genetic organization, domain architecture, and functional diversity of holins as well as endolysins, we seek to provide insights into their functional variability and potential for therapeutic application.
2. Materials and Methods
2.1. Retrieval of Phages Infecting the Streptococcus Genus
A search was performed in the NCBI RefSeq database [16] on 7 March 2025, using the keywords “Streptococcus phage” and “Streptococcus bacteriophage.” Only complete and annotated genomes were retained. Duplicates and evidently incomplete sequences were manually verified and excluded. When available, the bacterial host species was recorded.
2.2. Selection of a Representative Panel
Taxonomic classification (class, family, subfamily, genus, and species) was retrieved from the International Committee on Taxonomy of Viruses (ICTV) database [17] on 22 April 2025. For genera represented by multiple phages, a representative genome was selected based on citation frequency in the literature or the diversity/complexity of the lysis module. Phages unassigned at the genus level were systematically included. The phylogenetic diversity of the final panel was assessed using genome-based distance analyses with VICTOR [18] and VIRIDIC [19]. The resulting tree was visualized using iTOL v6 [20].
2.3. Detection and Annotation of Lysis Module
Phage genomes from the selected panel were annotated using Pharokka v1.7.5 [21] and visualized with LoVis4u version 0.1.2 [22] to identify genes associated with the lysis module. Annotations were cross-checked against RefSeq data. In ambiguous cases, unannotated proteins were analyzed using BLASTp [23] to determine potential homology to known holins or endolysins.
2.4. Detection of Functional Domains in Endolysins
Endolysin sequences were analyzed with phiBIOTICS [24] and SMART [25] to detect enzymatically active domains (EADs) and cell wall-binding domains (CBDs). Additional searches were conducted using HMMER, the Conserved Domain Database (CDD) [26], and MOTIF Search (https://www.genome.jp/tools/motif/, accessed on 28 May 2025), incorporating domain models from Pfam, COG, TIGRFAM, and SMART. An E-value cutoff of <0.01 was applied.
2.5. Characterization of Holins
Identified holins were submitted to BLASTp against the Transporter Classification Database [27] for functional classification. Transmembrane domain (TMD) number and topology were predicted using DeepTMHMM v1.0.42 [28] and confirmed via Protter version 1.0 [29].
3. Results
3.1. Diversity of Phages Infecting the Streptococcus Genus
A total of 205 Streptococcus-associated phages were initially retrieved from RefSeq. After excluding incomplete entries, 185 complete genomes were retained; their NCBI accession numbers are listed in Appendix A.1. A subset of 34 phages was selected for in-depth analysis, representing the observed taxonomic diversity (Figure 1). This selection included phages belonging to different genera when such classification was available, or otherwise phages that remained unclassified at the genus level according to the ICTV database. These 34 phages collectively infect 12 distinct Streptococcus species, although the host species remained unidentified for 1 of them. Notably, 20 phages in this subset were unassigned at the genus level. The four major groups of S. thermophilus-infecting phages were included: Pac (Brussowvirus), Cos (Moineauvirus), 5093 (Vansinderenvirus), and 9871 (Piorkowskivirus) (Appendix A.2).
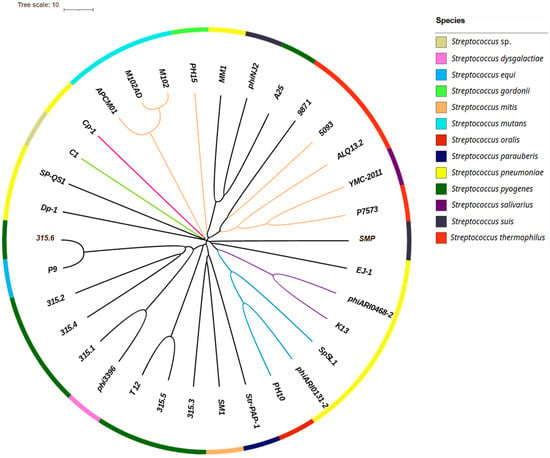
Figure 1.
Phylogenetic tree of the 34 phages selected for their taxonomic diversity. The tree was constructed using pairwise nucleotide alignments between phage genomes, based on a distance matrix computed by VIRIDIC and visualized with iTOL. Branches are colored according to the phages’ taxonomic classification. At the family level: Aliceevansviridae (orange), Madridviridae (fuchsia), and Rountreeviridae (green); at the subfamily level: Ferrettivirinae (purple) and Mcshanvirinae (blue). Branches shown in black indicate phages for which neither the family nor the subfamily is known. Bacterial hosts are represented by colored squares (as indicated in the color key), forming an outer ring.
3.2. Organization of Lysis Modules
Gene annotations revealed the presence of holin and endolysin genes, enabling the delineation of lysis modules in the 34 selected phages (Appendix A.3). Five distinct module organizations (A to E) were identified (Figure 2). These modules contain up to two ORFs each for holins and endolysins.
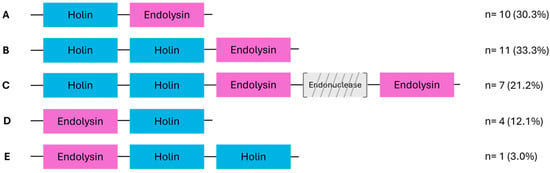
Figure 2.
Schematic representation of the different lysis module organizations identified among the analyzed phages (n = 33). Among the 34 phages examined, 33 harbored a recognizable lysis module. Five distinct organizations, labeled A to E, were defined based on the number and arrangement of lysis-related genes, holins (blue) and endolysins (pink). The number of phages exhibiting each organization is indicated on the right. In organizations A to C, holin genes precede endolysins, whereas in D and E, endolysins come first. Organization C includes an endonuclease gene (gray, diagonally striped) that is variably present between holin and endolysin. Each module is represented in the same transcriptional direction (left to right).
In 85% of phages, holin-encoding ORFs preceded endolysin-encoding ones, while this order was reversed in the remaining 15%. Lysis genes were generally contiguous, though some modules featured intercalated genes. For instance, phage ALQ13.2 (Brussowvirus) harbored an endonuclease and a hypothetical protein between two endolysins. Phage SP-QS1 uniquely lacked a clearly defined lysis module, with holin and endolysin genes separated by 18 unrelated genes, mostly encoding structural or hypothetical proteins.
3.3. Endolysin Architecture
Among the 185 phages, 114 encoded a single endolysin, while 71 encoded two. Endolysin genes ranged from 228 to 1386 bp, yielding a total of 256 distinct endolysins. Within the 34-phage subset, 26 had a single endolysin (Figure 3A).
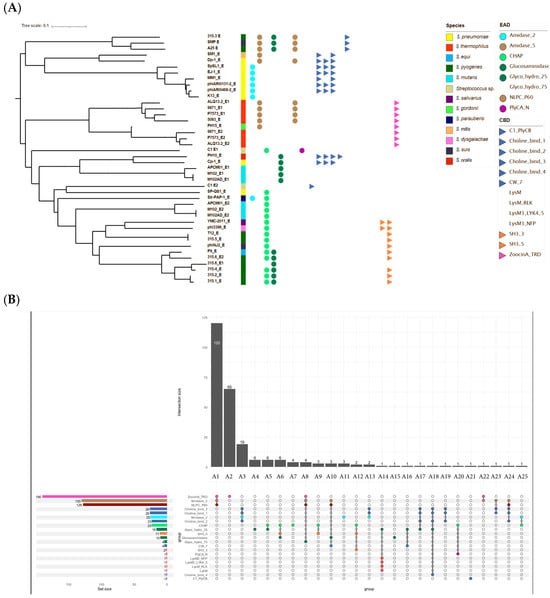
Figure 3.
Diversity and domain architecture of phage endolysins. (A) Phylogenetic tree of the 42 endolysins identified among the 34 selected phages. The tree was built using amino acid sequences aligned with ClustalW pairwise alignment, a slow algorithm, and the BLOSUM matrix). Endolysins are named using the abbreviated name of the originating phage, with the suffix _E when a single endolysin is present, or _E1/_E2 in the case of phages encoding two. For each endolysin, EADs and CBDs, when identified, are represented by colored circles and triangles, respectively (domain color codes are indicated in the figure). Colored squares indicate the bacterial species targeted by the phages encoding these endolysins. (B) Upset plot of the 256 analyzed endolysins. The plot displays the 25 distinct combinations of enzymatic and binding domains, labeled A1 to A25, based on the presence and arrangement of known EADs and CBDs. Each vertical bar represents the number of endolysins sharing a specific combination, while the connected dots below each bar indicate the motifs involved in that combination. On the left, a horizontal bar chart shows the overall frequency of each individual domain motif across all endolysins. A color code is used to distinguish motifs, with similar motifs sharing the same color.
These 256 proteins exhibited 25 architectural types, defined by EAD and CBD composition and arrangement (Figure 3B, Appendix A.4). The most common catalytic activity was amidase (Amidase_5 domain), and the most frequent CBD was ZoocinA_TRD. Seven architectures (A4, A5, A7, A11, A15, A20, and A25) lacked identifiable CBDs; three (A2, A19, and A21) lacked catalytic domains. Architectures were numbered by decreasing frequency (A1 = most common). Some were unique (A14–A25), and five (A5, A8, A14, A16, and A19) were absent from the representative subset.
Architectures lacking catalytic domains were usually co-encoded with catalytically active endolysins. For example, A2 (ZoocinA_TRD only) consistently co-occurred with A1 (Amidase_5 & NLPC_P60 & ZoocinA_TRD). Similarly, A20 (Chap & PlyCA) and A21 (PlyCB) were both encoded by phage C1, reflecting the PlyC endolysin structure.
An exception was phage IPP55, where a lone endolysin belonged to architecture A19. Further analysis using CDD and BLASTP revealed two conserved domains: a PGRP superfamily motif and a pneumo_PspA domain. The protein shared > 99% identity with S. pneumoniae LytA amidase (WP_073176617.1).
Notably, all three S. mutans phages encoding two endolysins lacked any identifiable CBDs.
Generally, catalytic domains were located at the N-terminus, with CBDs at the C-terminus. An exception was architecture A10, where CW7 motifs interrupted the Amidase_5/NLPC_P60 and Glucosaminidase domains. In some endolysins combining Amidase_5 and NLPC_P60, overlapping genes were observed, with the latter starting ~39 nt downstream of the former.
3.4. Characterization of Holins
A total of 340 holins were identified among the 185 phages analyzed. Within the subset of 34 phages, 53 holins were detected, distributed across 9 families (1.E.10, 1.E.11, 1.E.16, 1.E.18, 1.E.19, 1.E.21, 1.E.24, 1.E.26, and 1.E.65) out of the 74 currently listed in the Transporter Classification Database (TCDB). Fourteen holins remained unclassified. Phages encoding two holins always featured proteins from different families, consistent with divergent evolutionary origins (Figure 4).
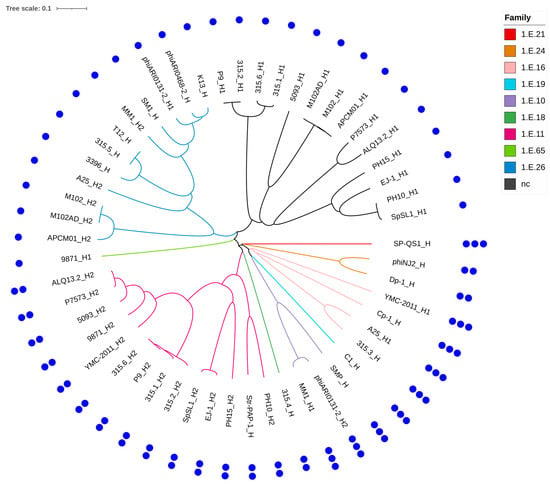
Figure 4.
Phylogenetic tree of the 53 holins identified among the 34 selected phages. The tree was built using amino acid sequences aligned with ClustalW (pairwise alignment, slow algorithm, BLOSUM matrix). Each holin is named using the abbreviated phage name, followed by the suffix _H when a single holin is present, or _H1/_H2 when two are encoded. Nine holin families were identified and are color-coded on the tree branches. Black branches indicate holins with no assigned family. The number of predicted transmembrane domains is shown by blue dots forming the outer ring of the tree.
The number of predicted TMDs was also assessed for all 53 holins and ranged from one to three, depending on the family. The number of TMDs was conserved within each family. Holins from families 1.E.26 and 1.E.65, as well as the unclassified holins, each contained a single TMD. Holins from families 1.E.11, 1.E.18, and 1.E.24 harbored two TMDs, whereas those from the remaining four families exhibited three TMDs.
Interestingly, among the 19 phages encoding two holins, only those infecting S. mutans featured holins with a single TMD in both cases.
4. Discussion
This study highlights significant deficiencies in the current taxonomic annotation of Streptococcus phages, with many genomes, particularly within the selected subset, remaining unclassified at the genus level. The lysis modules exhibited considerable diversity, including instances of intercalated ORFs such as endonucleases. Endolysin genomic sequences were generally short, even though they sometimes encode two structurally distinct proteins. This diversity appears to be facilitated by gene overlapping. Amidase activity was the most prevalent enzymatic function observed. While most endolysin gene architectures conformed to the typical N-terminal EAD and C-terminal CBD layout, some deviated from this pattern, including cases with a central CBD or a separate CBD-encoding ORF. Some CBDs also remained uncharacterized. Holins were predominantly found in tandem across the studied phages, although this trend was less pronounced in the subset. The diversity of holins was relatively limited, with those in the subset classified into only 9 of the 74 families currently listed in TCDB, and exhibiting between one and three transmembrane domains depending on the family.
Although all 185 phages were assigned to the class Caudoviricetes, the taxonomic information for several phages, particularly those in the subset, remains incomplete. This is hardly surprising given the trends reported by Turner et al. [30]. Viral taxonomy evolves rapidly, and the ICTV acknowledges that taxonomic assignment may remain partial for certain viruses, particularly when based solely on genomic data without cultivation, or in the context of metagenomic studies [31]. Other factors also complicate viral taxonomy, such as deficiencies in the curation of deposited viral genomes, especially for prophages. For example, phage phiNIH 1.1 (NC_003157.5) shares near-complete genomic identity with phage 315.4 (NC_004587.1), and, according to VIRIDIC analysis (data not shown), should arguably be classified within the same species. The only distinguishing feature is the presence of a gene (NC_003157.5:149-1255) encoding a peptide-methionine (R)-S-oxide reductase (MsrB), located upstream of the integrase gene in phiNIH 1.1 and likely acquired from the S. pyogenes host genome. These challenges underscore the need for improved viral genome curation, and some have proposed the use of artificial intelligence to assist with this task [32].
The findings from this study on the lysis modules of phages infecting Streptococcus species led to the proposal of a classification based on the position and number of ORFs encoding lysins and holins. This classification could be extended to phages infecting Gram-negative bacteria by incorporating the number and position of ORFs encoding spanins [5]. Although lysis-related genes are often contiguous, interspersed ORFs are not uncommon. Among these, endonucleases have been reported, under the designation lysin intergenic locus [33].
Regarding endolysin genomic sequences, the analysis revealed a notable rate of gene overlapping, allowing a single ORF to encode two distinct enzymatic functions. The most common association involved amidase activity (Amidase_5 domain) and endopeptidase activity (NLPC_P60 domain). This genomic arrangement supports the hypothesis of viral genome compression via overprinting, a mechanism widely used by phages to optimize coding efficiency [34,35].
Amidase activity emerged as the dominant enzymatic function in the endolysins of phages infecting the genus Streptococcus, in contrast with endolysins from Staphylococcus phages, where Cysteine, Histidine-dependent Amidohydrolases/Peptidases (CHAP) domains often predominate and amidase domains may play an auxiliary role, possibly contributing to peptidoglycan binding rather than catalysis [36]. This distinction may reflect functional adaptations to differences in host peptidoglycan structure and susceptibility.
The organization of endolysins observed here aligns with the classical architecture described for phages infecting Gram-positive bacteria, typically featuring an N-terminal EAD and a C-terminal CBD [37,38]. However, previously reported exceptions were also identified in this study, such as the well-characterized endolysin PlyC, whose CBD (PlyCB) oligomerizes into an octamer [33], or the presence of a central CBD flanked by EADs at both termini, as described for phage LambdaSa2 lysin and PlySK1249 lysin [39,40]. In some endolysins, particularly from phages infecting S. mutans, only EADs could be identified, which is consistent with a previous review [41]. The 25 architectural organizations described here therefore represent only a subset of the 89 known architectures [42,43].
The frequent presence of tandem holins supports previous observations by Escobedo et al. [44], who reported that the presence of a second holin may improve lytic activity. Nevertheless, further studies are needed to clarify the functional significance of membrane domain organization in holin regulation and activity, as current classification systems do not fully reflect the mechanistic diversity of these proteins [45,46].
5. Conclusions
This study revealed extensive diversity in the lysis modules of Streptococcus phages, with gene overlap and interspersed ORFs contributing to their structural complexity. These findings pave the way for future investigations, including the functional characterization of unannotated CBDs and the refinement of lysin engineering strategies for therapeutic purposes.
Author Contributions
M.S.-J.: writing—original draft, data curation, formal analysis. O.C.: writing—original draft, methodology, data curation, formal analysis. C.L.M.: writing—review and editing, methodology. J.S.: writing—original draft, writing—review and editing, supervision, project administration, conceptualization, methodology, formal analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CBD | Cell-wall Binding Domain |
| CDD | Conserved Domain Database |
| CHAP | Cysteine, Histidine-dependent Amidohydrolases/Peptidases |
| EAD | Enzymatically Active Domain |
| ICTV | International Committee on Taxonomy of Viruses |
| ORF | Open Reading Frame |
| SAR | Signal–Arrest–Release |
| TCDB | Transporter Classification Database |
| TMD | Transmembrane domain |
Appendix A
Appendix A.1

Table A1.
List of the 185 phages included in the study.
Table A1.
List of the 185 phages included in the study.
| Phage Name | Bacterial Target | NCBI RefSeq | kbp | Class | Family | Subfamily | Genus | Species |
|---|---|---|---|---|---|---|---|---|
| Streptococcus phage 20617 | Streptococcus thermophilus | NC_023503.1 | 48 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus 20617 | |
| Streptococcus phage ALQ13.2 | Streptococcus thermophilus | NC_013598.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus ALQ132 | |
| Streptococcus phage 2972 | Streptococcus thermophilus | NC_007019.1 | 34 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus bv2972 | |
| Streptococcus phage 858 | Streptococcus thermophilus | NC_010353.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus bv858 | |
| Streptococcus phage 01205 | Streptococcus thermophilus | NC_004303.1 | 43 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus bvO1205 | |
| Streptococcus phage CHPC1042 | Streptococcus thermophilus | NC_071061.1 | 42 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1042 | |
| Streptococcus phage CHPC1109 | Streptococcus thermophilus | NC_071062.1 | 33 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1109 | |
| Streptococcus phage CHPC1152 | Streptococcus thermophilus | NC_071063.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1152 | |
| Streptococcus phage CHPC1230 | Streptococcus thermophilus | NC_071064.1 | 39 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1230 | |
| Streptococcus phage CHPC1246 | Streptococcus thermophilus | NC_071065.1 | 36 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1246 | |
| Streptococcus phage CHPC1247 | Streptococcus thermophilus | NC_071066.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1247 | |
| Streptococcus phage CHPC1248 | Streptococcus thermophilus | NC_071067.1 | 38 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC1248 | |
| Streptococcus phage CHPC640 | Streptococcus thermophilus | NC_071068.1 | 40 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC640 | |
| Streptococcus phage CHPC869 | Streptococcus thermophilus | NC_071069.1 | 37 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC869 | |
| Streptococcus phage CHPC931 | Streptococcus thermophilus | NC_071070.1 | 36 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC931 | |
| Streptococcus phage CHPC951 | Streptococcus thermophilus | NC_071071.1 | 36 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC951 | |
| Streptococcus phage CHPC952 | Streptococcus thermophilus | NC_071072.1 | 36 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus CHPC952 | |
| Streptococcus phage P4761 | Streptococcus thermophilus | NC_071073.1 | 38 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P4761 | |
| Streptococcus phage P7571 | Streptococcus thermophilus | NC_071074.1 | 34 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P7571 | |
| Streptococcus phage P7951 | Streptococcus thermophilus | NC_071075.1 | 34 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P7951 | |
| Streptococcus phage P7952 | Streptococcus thermophilus | NC_071076.1 | 37 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P7952 | |
| Streptococcus phage P7953 | Streptococcus thermophilus | NC_071077.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P7953 | |
| Streptococcus phage P7954 | Streptococcus thermophilus | NC_071078.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P7954 | |
| Streptococcus phage P7955 | Streptococcus thermophilus | NC_071079.1 | 36 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P7955 | |
| Streptococcus phage P9853 | Streptococcus thermophilus | NC_071080.1 | 34 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus P9853 | |
| Streptococcus phage Sfi11 | Streptococcus thermophilus | NC_002214.1 | 39 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus Sfi11 | |
| Streptococcus phage SW1151 | Streptococcus thermophilus | NC_071081.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus SW1151 | |
| Streptococcus phage SW13 | Streptococcus thermophilus | NC_071082.1 | 36 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus SW13 | |
| Streptococcus phage SW14 | Streptococcus thermophilus | NC_071083.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus SW14 | |
| Streptococcus phage SW15 | Streptococcus thermophilus | NC_071084.1 | 34 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus SW15 | |
| Streptococcus phage SW18 | Streptococcus thermophilus | NC_071085.1 | 34 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus SW18 | |
| Streptococcus phage TP-778L | Streptococcus thermophilus | NC_022776.1 | 41 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus TP778L | |
| Streptococcus phage TP-J34 | Streptococcus thermophilus | NC_020197.1 | 45 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus TPJ34 | |
| MAG: Streptococcus phage VS-2018a | Streptococcus thermophilus | NC_071086.1 | 39 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus VS2018a | |
| Streptococcus phage Abc2 | Streptococcus thermophilus | NC_013645.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus Abc2 | |
| Streptococcus phage B0 | Streptococcus thermophilus | NC_070655.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus B0 | |
| Streptococcus phage CHPC1005 | Streptococcus thermophilus | NC_070656.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1005 | |
| Streptococcus phage CHPC1027 | Streptococcus thermophilus | NC_070657.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1027 | |
| Streptococcus phage CHPC1029 | Streptococcus thermophilus | NC_070658.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1029 | |
| Streptococcus phage CHPC1033 | Streptococcus thermophilus | NC_070659.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1033 | |
| Streptococcus phage CHPC1036 | Streptococcus thermophilus | NC_070660.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1036 | |
| Streptococcus phage CHPC1041 | Streptococcus thermophilus | NC_070661.1 | 38 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1041 | |
| Streptococcus phage CHPC1045 | Streptococcus thermophilus | NC_070662.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1045 | |
| Streptococcus phage CHPC1046 | Streptococcus thermophilus | NC_070663.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1046 | |
| Streptococcus phage CHPC1062 | Streptococcus thermophilus | NC_070664.1 | 40 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1062 | |
| Streptococcus phage CHPC1067 | Streptococcus thermophilus | NC_070665.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1067 | |
| Streptococcus phage CHPC1091 | Streptococcus thermophilus | NC_070666.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1091 | |
| Streptococcus phage CHPC1148 | Streptococcus thermophilus | NC_070667.1 | 39 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1148 | |
| Streptococcus phage CHPC1156 | Streptococcus thermophilus | NC_070668.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC1156 | |
| Streptococcus phage CHPC572 | Streptococcus thermophilus | NC_070669.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC572 | |
| Streptococcus phage CHPC595 | Streptococcus thermophilus | NC_070670.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC595 | |
| Streptococcus phage CHPC642 | Streptococcus thermophilus | NC_070671.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC642 | |
| Streptococcus phage CHPC663 | Streptococcus thermophilus | NC_070672.1 | 38 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC663 | |
| Streptococcus phage CHPC873 | Streptococcus thermophilus | NC_070673.1 | 38 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC873 | |
| Streptococcus phage CHPC875 | Streptococcus thermophilus | NC_070674.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC875 | |
| Streptococcus phage CHPC877 | Streptococcus thermophilus | NC_070675.1 | 39 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC877 | |
| Streptococcus phage CHPC879 | Streptococcus thermophilus | NC_070676.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC879 | |
| Streptococcus phage CHPC919 | Streptococcus thermophilus | NC_070677.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC919 | |
| Streptococcus phage CHPC925 | Streptococcus thermophilus | NC_070678.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC925 | |
| Streptococcus phage CHPC927 | Streptococcus thermophilus | NC_070679.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC927 | |
| Streptococcus phage CHPC928 | Streptococcus thermophilus | NC_070680.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC928 | |
| Streptococcus phage CHPC930 | Streptococcus thermophilus | NC_070681.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC930 | |
| Streptococcus phage CHPC950 | Streptococcus thermophilus | NC_070682.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC950 | |
| Streptococcus phage CHPC979 | Streptococcus thermophilus | NC_070683.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus CHPC979 | |
| Streptococcus phage D1024 | Streptococcus thermophilus | NC_070684.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus D1024 | |
| Streptococcus phage D1811 | Streptococcus thermophilus | NC_070685.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus D1811 | |
| Streptococcus phage D4276 | Streptococcus thermophilus | NC_070686.1 | 39 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus D4276 | |
| Streptococcus phage DT1 | Streptococcus thermophilus | NC_002072.2 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus DT1 | |
| Streptococcus phage L5A1 | Streptococcus thermophilus | NC_070687.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus L5A1 | |
| Streptococcus phage M19 | Streptococcus thermophilus | NC_070688.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus M19 | |
| Streptococcus phage MM25 | Streptococcus thermophilus | NC_070689.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus MM25 | |
| Streptococcus phage 128 | Streptococcus thermophilus | NC_070651.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus mv128 | |
| Streptococcus phage 53 | Streptococcus thermophilus | NC_070652.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus mv53 | |
| Streptococcus phage 7201 | Streptococcus thermophilus | NC_002185.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus mv7201 | |
| Streptococcus phage 73 | Streptococcus thermophilus | NC_070653.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus mv73 | |
| Streptococcus phage 7A5 | Streptococcus thermophilus | NC_070654.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus mv7A5 | |
| Streptococcus phage P0091 | Streptococcus thermophilus | NC_070690.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P0091 | |
| Streptococcus phage P3681 | Streptococcus thermophilus | NC_070691.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P3681 | |
| Streptococcus phage P3684 | Streptococcus thermophilus | NC_070692.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P3684 | |
| Streptococcus phage P5641 | Streptococcus thermophilus | NC_070693.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P5641 | |
| Streptococcus phage P5651 | Streptococcus thermophilus | NC_070694.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P5651 | |
| Streptococcus phage P7132 | Streptococcus thermophilus | NC_070695.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7132 | |
| Streptococcus phage P7133 | Streptococcus thermophilus | NC_070696.1 | 33 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7133 | |
| Streptococcus phage P7134 | Streptococcus thermophilus | NC_070697.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7134 | |
| Streptococcus phage P7151 | Streptococcus thermophilus | NC_070698.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7151 | |
| Streptococcus phage P7152 | Streptococcus thermophilus | NC_070699.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7152 | |
| Streptococcus phage P7154 | Streptococcus thermophilus | NC_070700.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7154 | |
| Streptococcus phage P7573 | Streptococcus thermophilus | NC_070701.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7573 | |
| Streptococcus phage P7574 | Streptococcus thermophilus | NC_070702.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7574 | |
| Streptococcus phage P7601 | Streptococcus thermophilus | NC_070703.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7601 | |
| Streptococcus phage P7602 | Streptococcus thermophilus | NC_070704.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7602 | |
| Streptococcus phage P7631 | Streptococcus thermophilus | NC_070705.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7631 | |
| Streptococcus phage P7632 | Streptococcus thermophilus | NC_070706.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7632 | |
| Streptococcus phage P7633 | Streptococcus thermophilus | NC_070707.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7633 | |
| Streptococcus phage P9851 | Streptococcus thermophilus | NC_070708.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P9851 | |
| Streptococcus phage P9854 | Streptococcus thermophilus | NC_070709.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P9854 | |
| Streptococcus phage P9901 | Streptococcus thermophilus | NC_070710.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P9901 | |
| Streptococcus phage P9902 | Streptococcus thermophilus | NC_070711.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P9902 | |
| Streptococcus phage P9903 | Streptococcus thermophilus | NC_070712.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P9903 | |
| Streptococcus phage Sfi19 | Streptococcus thermophilus | NC_000871.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus Sfi19 | |
| Streptococcus phage Sfi21 | Streptococcus thermophilus | NC_000872.1 | 40 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus Sfi21 | |
| Streptococcus phage STP1 | Streptococcus thermophilus | NC_070713.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus STP1 | |
| Streptococcus phage SW1 | Streptococcus thermophilus | NC_070714.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW1 | |
| Streptococcus phage SW11 | Streptococcus thermophilus | NC_070715.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW11 | |
| Streptococcus phage SW12 | Streptococcus thermophilus | NC_070716.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW12 | |
| Streptococcus phage SW2 | Streptococcus thermophilus | NC_070717.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW2 | |
| Streptococcus phage SW3 | Streptococcus thermophilus | NC_070718.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW3 | |
| Streptococcus phage SW5 | Streptococcus thermophilus | NC_070719.1 | 36 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW5 | |
| Streptococcus phage SW6 | Streptococcus thermophilus | NC_070720.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW6 | |
| Streptococcus phage SW7 | Streptococcus thermophilus | NC_070721.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW7 | |
| Streptococcus phage SW8 | Streptococcus thermophilus | NC_070722.1 | 33 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SW8 | |
| Streptococcus phage SWK3 | Streptococcus thermophilus | NC_070723.1 | 35 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SWK3 | |
| Streptococcus phage SWK6 | Streptococcus thermophilus | NC_070724.1 | 34 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus SWK6 | |
| Streptococcus phage vB_SthS_VA214 | Streptococcus thermophilus | NC_070725.1 | 38 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus VA214 | |
| Streptococcus phage vB_SthS_VA460 | Streptococcus thermophilus | NC_070726.1 | 41 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus VA460 | |
| Streptococcus phage 5093 | Streptococcus thermophilus | NC_012753.1 | 37 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus 5093 | |
| Streptococcus phage CHPC1198 | Streptococcus thermophilus | NC_071053.1 | 33 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus CHPC1198 | |
| Streptococcus phage CHPC1151 | Streptococcus thermophilus | NC_071057.1 | 33 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus CHPC1232 | |
| Streptococcus phage CHPC1282 | Streptococcus thermophilus | NC_071058.1 | 34 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus CHPC1282 | |
| Streptococcus phage P0092 | Streptococcus thermophilus | NC_071056.1 | 34 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus P0092 | |
| Streptococcus phage P0093 | Streptococcus thermophilus | NC_071054.1 | 34 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus P0093 | |
| Streptococcus phage P0095 | Streptococcus thermophilus | NC_071059.1 | 35 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus P0095 | |
| Streptococcus phage SW19 | Streptococcus thermophilus | NC_071060.1 | 35 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus SW19 | |
| Streptococcus phage SW24 | Streptococcus thermophilus | NC_071055.1 | 32 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus SW24 | |
| Streptococcus phage SW27 | Streptococcus thermophilus | NC_071052.1 | 33 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus SW27 | |
| Streptococcus phage SW4 | Streptococcus thermophilus | NC_071051.1 | 34 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus SW4 | |
| Streptococcus phage Cp-1 | Streptococcus pneumoniae | NC_001825.1 | 19 | Caudoviricetes | Madridviridae | Cepunavirus | Cepunavirus Cp1 | |
| Streptococcus phage Cp-7 | Streptococcus pneumoniae | NC_042114.1 | 19 | Caudoviricetes | Madridviridae | Cepunavirus | Cepunavirus Cp7 | |
| Streptococcus phage C1 | Group C Streptococci | NC_004814.1 | 16 | Caudoviricetes | Rountreeviridae | Fischettivirus | Fischettivirus C1 | |
| Streptococcus phage phiARI0004 | Streptococcus pneumoniae | NC_031920.1 | 41 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus ARI0004 | |
| Streptococcus phage phiARI0031 | Streptococcus pneumoniae | NC_031910.1 | 41 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus ARI0031 | |
| Streptococcus phage phiARI0462 | Streptococcus pneumoniae | NC_031942.1 | 40 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus ARI0062 | |
| Streptococcus phage phiARI0468-1 | Streptococcus pneumoniae | NC_031929.1 | 41 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus ARI04681 | |
| Streptococcus phage DCC1738 | Streptococcus pneumoniae | NC_024361.1 | 38 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus DCC1738 | |
| Streptococcus phage IC1 | Streptococcus pneumoniae | NC_024370.1 | 39 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus IC1 | |
| Streptococcus phage K13 | Streptococcus pneumoniae | NC_024357.1 | 39 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus K13 | |
| Streptococcus phage phiARI0131-1 | Streptococcus pneumoniae | NC_031901.1 | 42 | Caudoviricetes | Ferrettivirinae | Spinunavirus | Spinunavirus ARI01311 | |
| Streptococcus phage phiARI0460-1 | Streptococcus pneumoniae | NC_031913.1 | 41 | Caudoviricetes | Ferrettivirinae | Spinunavirus | Spinunavirus ARI04601 | |
| Streptococcus phage phiARI0468-2 | Streptococcus pneumoniae | NC_031923.1 | 42 | Caudoviricetes | Ferrettivirinae | Spinunavirus | Spinunavirus ARI04682 | |
| Streptococcus phage phiARI0923 | Streptococcus pneumoniae | NC_030946.1 | 33 | Caudoviricetes | Mcshanvirinae | Adrianbuildvirus | Adrianbuildvirus ARI0923 | |
| Streptococcus phage SpSL1 | Streptococcus pneumoniae | NC_027396.1 | 33 | Caudoviricetes | Mcshanvirinae | Adrianbuildvirus | Adrianbuildvirus SpSL1 | |
| Streptococcus phage phiARI0131-2 | Streptococcus pneumoniae | NC_031941.1 | 34 | Caudoviricetes | Mcshanvirinae | Medawarvirus | Medawarvirus ARI01312 | |
| Streptococcus phage PH10 | Streptococcus oralis | NC_012756.1 | 31 | Caudoviricetes | Mcshanvirinae | Phadecavirus | Phadecavirus PH10 | |
| Streptococcus phage phiBHN167 | Streptococcus pneumoniae | NC_022791.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus BHN167 | ||
| Streptococcus phage IPP14 | Streptococcus pneumoniae | NC_070925.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP14 | ||
| Streptococcus phage IPP39 | Streptococcus pneumoniae | NC_070926.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP39 | ||
| Streptococcus phage IPP48 | Streptococcus pneumoniae | NC_070928.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP48 | ||
| Streptococcus phage IPP52 | Streptococcus pneumoniae | NC_070929.1 | 36 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP52 | ||
| Streptococcus phage IPP54 | Streptococcus pneumoniae | NC_070923.1 | 38 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP54 | ||
| Streptococcus phage IPP55 | Streptococcus pneumoniae | NC_070924.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP55 | ||
| Streptococcus phage IPP65 | Streptococcus pneumoniae | NC_070922.1 | 39 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP65 | ||
| Streptococcus phage IPP66 | Streptococcus pneumoniae | NC_070930.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus IPP66 | ||
| Streptococcus pneumoniae bacteriophage MM1 | Streptococcus pneumoniae | NC_003050.2 | 40 | Caudoviricetes | Paclarkvirus | Paclarkvirus MM1 | ||
| Streptococcus phage SpGS-1 | Streptococcus pneumoniae | NC_070927.1 | 37 | Caudoviricetes | Paclarkvirus | Paclarkvirus SpGS-1 | ||
| Streptococcus phage CHPC577 | Streptococcus thermophilus | NC_070901.1 | 35 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus CHPC577 | ||
| Streptococcus phage CHPC926 | Streptococcus thermophilus | NC_070900.1 | 30 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus CHPC926 | ||
| Streptococcus virus 9871 | Streptococcus thermophilus | NC_031069.1 | 32 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus pv9871 | ||
| Streptococcus virus 9872 | Streptococcus thermophilus | NC_031094.1 | 33 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus pv9872 | ||
| Streptococcus virus 9874 | Streptococcus thermophilus | NC_031023.1 | 32 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus pv9874 | ||
| Streptococcus phage SW16 | Streptococcus thermophilus | NC_070899.1 | 32 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus SW16 | ||
| Streptococcus phage SW22 | Streptococcus thermophilus | NC_070898.1 | 31 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus SW22 | ||
| Streptococcus phage SP-QS1 | Streptococcus pneumoniae | NC_021868.1 | 58 | Caudoviricetes | Saphexavirus | Saphexavirus SPQS1 | ||
| Streptococcus phage A25 | Streptococcus pyogenes | NC_028697.1 | 33 | Caudoviricetes | Stonewallvirus | Stonewallvirus A25 | ||
| Streptococcus phage APCM01 | Streptococcus mutans | NC_029030.1 | 31 | Caudoviricetes | ||||
| Streptococcus phage Dp-1 | Streptococcus pneumoniae | NC_015274.1 | 56 | Caudoviricetes | ||||
| Streptococcus phage M102 | Streptococcus mutans | NC_012884.1 | 31 | Caudoviricetes | ||||
| Streptococcus phage M102AD | Streptococcus mutans | NC_028984.1 | 30 | Caudoviricetes | ||||
| Streptococcus phage P9 | Streptococcus equi | NC_009819.1 | 40 | Caudoviricetes | ||||
| Streptococcus phage PH15 | Streptococcus gordonii | NC_010945.1 | 39 | Caudoviricetes | ||||
| Streptococcus phage phi3396 | Streptococcus dysgalactiae | NC_009018.1 | 38 | Caudoviricetes | ||||
| Streptococcus phage phiNIH1.1 | Streptococcus pyogenes | NC_003157.5 | 43 | Caudoviricetes | ||||
| Streptococcus phage phiNJ2 | Streptococcus suis | NC_019418.1 | 37 | Caudoviricetes | ||||
| Streptococcus phage SM1 | Streptococcus mitis | NC_004996.1 | 34 | Caudoviricetes | ||||
| Streptococcus phage SMP | Streptococcus suis | NC_008721.2 | 36 | Caudoviricetes | ||||
| Streptococcus phage Str-PAP-1 | Streptococcus parauberis | NC_028666.1 | 36 | Caudoviricetes | ||||
| Streptococcus phage T12 | Streptococcus pyogenes | NC_028700.1 | 37 | Caudoviricetes | ||||
| Streptococcus phage YMC-2011 | Streptococcus salivarius | NC_018285.1 | 40 | Caudoviricetes | ||||
| Streptococcus pyogenes phage 315.3 | Streptococcus pyogenes | NC_004586.1 | 34 | Caudoviricetes | ||||
| Streptococcus virus 9873 | Streptococcus thermophilus | NC_047763.1 | 32 | Caudoviricetes | ||||
| Streptococcus prophage 315.1 | Streptococcus pyogenes | NC_004584.1 | 39 | Caudoviricetes | ||||
| Streptococcus prophage 315.2 | Streptococcus pyogenes | NC_004585.1 | 41 | Caudoviricetes | ||||
| Streptococcus prophage 315.4 | Streptococcus pyogenes | NC_004587.1 | 41 | Caudoviricetes | ||||
| Streptococcus prophage 315.5 | Streptococcus pyogenes | NC_004588.1 | 38 | Caudoviricetes | ||||
| Streptococcus prophage 315.6 | Streptococcus pyogenes | NC_004589.1 | 40 | Caudoviricetes | ||||
| Streptococcus prophage EJ-1 | Streptococcus pneumoniae | NC_005294.1 | 42 | Caudoviricetes | ||||
Taxonomic information according to ICTV is provided when available.
Appendix A.2

Table A2.
List of the 34 selected phages.
Table A2.
List of the 34 selected phages.
| Phage | Short Name for Figure Readability | Bacterial Target | NCBI RefSeq Accession | kbp | Class | Family | Subfamily | Genus | Species | |
|---|---|---|---|---|---|---|---|---|---|---|
| Streptococcus phage ALQ13.2 | ALQ13.2 | Streptococcus thermophilus | NC_013598.1 | 35 | Caudoviricetes | Aliceevansviridae | Brussowvirus | Brussowvirus ALQ132 | Pac group | |
| Streptococcus phage P7573 | P7573 | Streptococcus thermophilus | NC_070701.1 | 37 | Caudoviricetes | Aliceevansviridae | Moineauvirus | Moineauvirus P7573 | Cos group | |
| Streptococcus phage 5093 | 5093 | Streptococcus thermophilus | NC_012753.1 | 37 | Caudoviricetes | Aliceevansviridae | Vansinderenvirus | Vansinderenvirus 5093 | 5093 group | |
| Streptococcus phage Cp-1 | Cp-1 | Streptococcus pneumoniae | NC_001825.1 | 19 | Caudoviricetes | Madridviridae | Cepunavirus | Cepunavirus Cp1 | ||
| Streptococcus phage C1 | C1 | Group C Streptococci | NC_004814.1 | 16 | Caudoviricetes | Rountreeviridae | Fischettivirus | Fischettivirus C1 | ||
| Streptococcus phage K13 | K13 | Streptococcus pneumoniae | NC_024357.1 | 39 | Caudoviricetes | Ferrettivirinae | Hinxtonvirus | Hinxtonvirus K13 | ||
| Streptococcus phage phiARI0468-2 | phiARI0468-2 | Streptococcus pneumoniae | NC_031923.1 | 42 | Caudoviricetes | Ferrettivirinae | Spinunavirus | Spinunavirus ARI04682 | ||
| Streptococcus phage SpSL1 | SpSL1 | Streptococcus pneumoniae | NC_027396.1 | 33 | Caudoviricetes | Mcshanvirinae | Adrianbuildvirus | Adrianbuildvirus SpSL1 | ||
| Streptococcus phage phiARI0131-2 | phiARI0131-2 | Streptococcus pneumoniae | NC_031941.1 | 34 | Caudoviricetes | Mcshanvirinae | Medawarvirus | Medawarvirus ARI01312 | ||
| Streptococcus phage PH10 | PH10 | Streptococcus oralis | NC_012756.1 | 31 | Caudoviricetes | Mcshanvirinae | Phadecavirus | Phadecavirus PH10 | ||
| Streptococcus pneumoniae bacteriophage MM1 | MM1 | Streptococcus pneumoniae | NC_003050.2 | 40 | Caudoviricetes | Paclarkvirus | Paclarkvirus MM1 | |||
| Streptococcus virus 9871 | 9871 | Streptococcus thermophilus | NC_031069.1 | 32 | Caudoviricetes | Piorkowskivirus | Piorkowskivirus pv9871 | 9871 group | ||
| Streptococcus phage SP-QS1 | SP-QS1 | Streptococcus pneumoniae | NC_021868.1 | 58 | Caudoviricetes | Saphexavirus | Saphexavirus SPQS1 | |||
| Streptococcus phage A25 | A25 | Streptococcus pyogenes | NC_028697.1 | 33 | Caudoviricetes | Stonewallvirus | Stonewallvirus A25 | |||
| Streptococcus phage APCM01 | APCM01 | Streptococcus mutans | NC_029030.1 | 31 | Caudoviricetes | Aliceevansviridae | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage M102 | M102 | Streptococcus mutans | NC_012884.1 | 31 | Caudoviricetes | Aliceevansviridae | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage M102AD | M102AD | Streptococcus mutans | NC_028984.1 | 30 | Caudoviricetes | Aliceevansviridae | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage PH15 | PH15 | Streptococcus gordonii | NC_010945.1 | 39 | Caudoviricetes | Aliceevansviridae | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage YMC-2011 | YMC-2011 | Streptococcus salivarius | NC_018285.1 | 40 | Caudoviricetes | Aliceevansviridae | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage Dp-1 | Dp-1 | Streptococcus pneumoniae | NC_015274.1 | 56 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus prophage EJ-1 | EJ-1 | Streptococcus pneumoniae | NC_005294.1 | 42 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage P9 | P9 | Streptococcus equi | NC_009819.1 | 40 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage phi3396 | phi3396 | Streptococcus dysgalactiae | NC_009018.1 | 38 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage phiNJ2 | phiNJ2 | Streptococcus suis | NC_019418.1 | 37 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage SM1 | SM1 | Streptococcus mitis | NC_004996.1 | 34 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage SMP | SMP | Streptococcus suis | NC_008721.2 | 36 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage Str-PAP-1 | Str-PAP-1 | Streptococcus parauberis | NC_028666.1 | 36 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus phage T12 | T12 | Streptococcus pyogenes | NC_028700.1 | 37 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus pyogenes phage 315.3 | 315.3 | Streptococcus pyogenes | NC_004586.1 | 34 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus prophage 315.1 | 315.1 | Streptococcus pyogenes | NC_004584.1 | 39 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus prophage 315.2 | 315.2 | Streptococcus pyogenes | NC_004585.1 | 41 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus prophage 315.4 | 315.4 | Streptococcus pyogenes | NC_004587.1 | 41 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus prophage 315.5 | 315.5 | Streptococcus pyogenes | NC_004588.1 | 38 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
| Streptococcus prophage 315.6 | 315.6 | Streptococcus pyogenes | NC_004589.1 | 40 | Caudoviricetes | No ICTV assignation | No ICTV assignation | No ICTV assignation | No ICTV assignation | |
The abbreviated names used in graphical representations are indicated.
Appendix A.3

Figure A1.
Nucleotide alignment of the 34 selected phages. Alignment was performed using the Lovis4u softwarev0.0.12. ORF functions are shown using a color-coded scheme.
Appendix A.4

Table A3.
Summary of the architectures identified among the 256 endolysins.
Table A3.
Summary of the architectures identified among the 256 endolysins.
| Architecture | Count | EAD | CBD | Phages List |
|---|---|---|---|---|
| A1 | 120 | Amidase_5 & NLPC_P60 | ZoocinA_TRD | CHPC1151_E1, CHPC572_E1, CHPC1027_E1, SW27_E1, P7134_E1, P0093_E1, P0092_E1, CHPC1282_E1, SW14_E1, CHPC1033_E1, SW12_E1, SW24_E, STP1_E1, SW6_E1, P7955_E1, P7573_E1, P7571_E1, P0091_E, L5A1_E, SW4_E1, P9854_E1, CHPC927_E1, CHPC926_E1, CHPC925_E1, SW1151_E1, CHPC950_E1, CHPC1248_E1, CHPC1247_E1, CHPC1246_E1, CHPC1230_E1, CHPC1046_E1, SW11_E1, CHPC877_E, 128_E, P7133_E, MM25_E, 5093_E, CHPC979_E, CHPC928_E, P7602_E, P7633_E, P7601_E, P7632_E, P7631_E, SW7_E, P3684_E, CHPC875_E, CHPC1091_E, CHPC930_E, CHPC1067_E, 7A5_E, SW3_E, B0_E, P3681_E, SW15_E, P7574_E, SW19_E1, CHPC952_E, CHPC931_E1, CHPC869_E1, SW8_E1, P5651_E1, CHPC1029_E1, CHPC1042_E1, P9851_E1, P9853_E1, CHPC640_E1, P7152_E1, SW18_E1, 9874_E1, CHPC1041_E1, D4276_E1, P0095_E1, CHPC1005_E1, CHPC879_E, vB_SthS_VA460_E2, MAG-VS-2018a_E, CHPC663_E1, 20617_E, 858_E1, 2972_E1, P7151_E1, CHPC1156_E1, vB_SthS_VA214_E2, CHPC642_E, P7132_E, SWK3_E, CHPC1036_E, CHPC1045_E, SWK6_E, P7154_E, CHPC1148_E, 7201_E, CHPC1198_E1, CHPC577_E1, 9871_E1, SW22_E1, SW16_E, 9873_E1, M19_E, 53_E, CHPC1152_E, SW5_E, P9901_E, SW2_E, CHPC951_E, Abc2_E, 73_E, D1811_E1, D1024_E1, ALQ13.2_E1, 9872_E1, DT1_E1, TP-778L_E, TP-J34_E, Sfi19_E, CHPC919_E, Sfi21_E, Sfi11_E, 1205_E |
| A2 | 65 | ZoocinA_TRD | vB_SthS_VA460_E1, 858_E2, CHPC1246_E2, CHPC1230_E2, 9874_E2, CHPC663_E2, P7571_E2, CHPC1033_E2, CHPC1027_E2, CHPC931_E2, CHPC869_E2, SW4_E2, CHPC1282_E2, CHPC1198_E2, SW27_E2, SW11_E2, P9854_E2, P9851_E2, P7134_E2, P0093_E2, P0092_E2, CHPC1248_E2, CHPC1247_E2, CHPC1151_E2, 2972_E2, P9853_E2, CHPC926_E2, 9871_E2, SW22_E2, SW1151_E2, CHPC1042_E2, CHPC1029_E2, vB_SthS_VA214_E1, P4761_E2, 9873_E2, 9872_E2, SW19_E2, D1811_E2, D1024_E2, CHPC595_E2, DT1_E2, CHPC1156_E2, CHPC927_E2, P0095_E2, CHPC925_E2, CHPC640_E2, CHPC1041_E2, SW14_E2, D4276_E2, SW8_E2, CHPC950_E2, CHPC1046_E2, SW12_E2, ALQ13.2_E2, P7151_E2, CHPC577_E2, STP1_E2, CHPC572_E2, P7955_E2, P7573_E2, SW6_E2, SW18_E2, P7152_E2, P5651_E2, CHPC1005_E2 | |
| A3 | 19 | Amidase_2 | Choline_bind_1 & Choline_bind_2 & Choline_bind_3 | SpSL1_E, EJ-1_E, phiARI0462_E, phiARI0460-1_E, phiARI0131-1_E, phiARI0131-2_E, phiARI0923_E, phiARI0004_E, phiARI0468-2_E, phiARI0468-1_E, phiARI0031_E, IPP48_E, phiBHN167_E, IPP14_E, IPP52_E, IC1_E, IPP39_E, IPP54_E, SpGS-1_E |
| A4 | 6 | Glyco_hydro_25 | APCM01_E1, M102_E1, M102AD_E1, P7951_E, P5641_E, CHPC1062_E | |
| A5 | 6 | CHAP & Glyco_hydro_25 | P7954_E, SW13_E, P7953_E, P7952_E, SW1_E, CHPC873_E | |
| A6 | 6 | Glucosaminidase & CHAP | SH3_5 | P9_E, 315.6_E2, 315.2_E, 315.1_E, phiNIH1.1_E, 315.4_E |
| A7 | 4 | CHAP | SP-QS1_E, APCM01_E2, M102_E2, M102AD_E2 | |
| A8 | 4 | Amidase_5 & NLPC_P60 & Glyco_hydro_75 | ZoocinA_TRD | P4761_E1, CHPC595_E1, P9903_E, P9902_E |
| A9 | 3 | CHAP | SH3_5 | T12_E, 315.5_E, phiNJ2_E |
| A10 | 3 | Amidase_5 & NLPC_P60 & Glucosaminidase | CW7 | 315.3_E, SMP_E, A25_E |
| A11 | 3 | Amidase_2 | K13_E, DCC1738_E, IPP66_E | |
| A12 | 2 | CHAP | SH3_3 & SH3_5 | YMC-2011_E, phi3396_E |
| A13 | 2 | Amidase_2 | Choline_bind_1 & Choline_bind_3 | MM1_E, IPP65_E |
| A14 | 1 | Glyco_hydro_25 | LysM & LysM_RLK & LysM3_LYK4_5 & LysM3_NFP | CHPC1109_E |
| A15 | 1 | Glucosaminidase | 315.6_E1 | |
| A16 | 1 | Glyco_hydro_25 | CW7 | Cp-7_E |
| A17 | 1 | Glyco_hydro_25 | Choline_bind_1 & Choline_bind_2 & Choline_bind_3 | Cp-1_E |
| A18 | 1 | Glyco_hydro_25 | Choline_bind_1 & Choline_bind_2 & Choline_bind_3 & Choline_bind_4 | PH10_E |
| A19 | 1 | Choline_bind_1 & Choline_bind_2 & Choline_bind_3 | IPP55_E | |
| A20 | 1 | PlyCA_N & CHAP | C1 E1 | |
| A21 | 1 | C1_PlyCB | C1 E2 | |
| A22 | 1 | Amidase_5 | ZoocinA_TRD | PH15_E |
| A23 | 1 | Amidase_5 | Choline_bind_1 & Choline_bind_3 | SM1_E |
| A24 | 1 | Amidase_5 & NLPC_P60 | Choline_bind_1 & Choline_bind_2 & Choline_bind_3 | Dp-1_E |
| A25 | 1 | CHAP & Amidase_2 | Str-PAP-1_E |
Each identified endolysin is associated with its domain architecture. Endolysins from phages included in the 34-phage subset are highlighted in bold.
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Cahill, J.; Young, R. Phage Lysis: Multiple Genes for Multiple Barriers. Adv. Virus Res. 2019, 103, 33–70. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Zhu, X.; Dion, M.B.; Shi, R.; Moineau, S. Phage endolysins are adapted to specific hosts and are evolutionarily dynamic. PLoS Biol. 2022, 20, e3001740. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Barros, M.; Vennemann, T.; Gallagher, D.T.; Yin, Y.; Linden, S.B.; Heselpoth, R.D.; Spencer, D.J.; Donovan, D.M.; Moult, J.; et al. A bacteriophage endolysin that eliminates intracellular streptococci. eLife 2016, 5, e13152. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.D.; Andre, C.; Bispo, P.J.M. Targeted Killing of Ocular Streptococcus pneumoniae by the Phage Endolysin MSlys. Ophthalmol. Sci. 2022, 2, 100193. [Google Scholar] [CrossRef] [PubMed]
- Sims Sanyahumbi, A.; Colquhoun, S.; Wyber, R.; Carapetis, J.R. Global Disease Burden of Group A Streptococcus. In Streptococcus pyogenes: Basic Biology to Clinical Manifestations; Ferretti, J.J., Stevens, D.L., Fischetti, V.A., Eds.; University of Oklahoma Health Sciences Center: Oklahoma City, OK, USA, 2016. [Google Scholar]
- Weiser, J.N.; Ferreira, D.M.; Paton, J.C. Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 2018, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Lupo, A.; Madec, J.Y. Antimicrobial Resistance in Streptococcus spp. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Mohanty, S.; Feemster, K.; Yu, K.C.; Watts, J.A.; Gupta, V. Trends in Streptococcus pneumoniae Antimicrobial Resistance in US Children: A Multicenter Evaluation. Open Forum Infect. Dis. 2023, 10, ofad098. [Google Scholar] [CrossRef] [PubMed]
- Russomando, G.; Farina, N.; Amour, S.; Grau, L.; Guillen, R.; Abente, S.; Aldama, M.; Hahn, I.; Castro, H.; Messaoudi, M.; et al. Streptococcus pneumoniae carriage, antimicrobial resistance, and serotype distribution in children and adults from Paraguay in the post-vaccinal era. Front. Public Health 2025, 13, 1584857. [Google Scholar] [CrossRef] [PubMed]
- Hanemaaijer, L.; Kelleher, P.; Neve, H.; Franz, C.; de Waal, P.P.; van Peij, N.; van Sinderen, D.; Mahony, J. Biodiversity of Phages Infecting the Dairy Bacterium Streptococcus thermophilus. Microorganisms 2021, 9, 1822. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, T.; Kodali, V.K.; Pujar, S.; Brover, V.; Robbertse, B.; Farrell, C.M.; Oh, D.H.; Astashyn, A.; Ermolaeva, O.; Haddad, D.; et al. NCBI RefSeq: Reference sequence standards through 25 years of curation and annotation. Nucleic Acids Res. 2025, 53, D243–D257. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Goker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Bouras, G.; Nepal, R.; Houtak, G.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Pharokka: A fast scalable bacteriophage annotation tool. Bioinformatics 2023, 39, btac776. [Google Scholar] [CrossRef] [PubMed]
- Egorov, A.A.; Atkinson, G.C. LoVis4u: A locus visualization tool for comparative genomics and coverage profiles. NAR Genom. Bioinform. 2025, 7, lqaf009. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Hojckova, K.; Stano, M.; Klucar, L. phiBIOTICS: Catalogue of therapeutic enzybiotics, relevant research studies and practical applications. BMC Microbiol. 2013, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Muller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adams, M.J.; Benko, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Inglis, L.K.; Edwards, R.A. How Metagenomics Has Transformed Our Understanding of Bacteriophages in Microbiome Research. Microorganisms 2022, 10, 1671. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Schuch, R.; Chahales, P.; Zhu, S.; Fischetti, V.A. PlyC: A multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. USA 2006, 103, 10765–10770. [Google Scholar] [CrossRef] [PubMed]
- Chirico, N.; Vianelli, A.; Belshaw, R. Why genes overlap in viruses. Proc. Biol. Sci. 2010, 277, 3809–3817. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology 2020, 546, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Son, B.; Kong, M.; Ryu, S. The Auxiliary Role of the Amidase Domain in Cell Wall Binding and Exolytic Activity of Staphylococcal Phage Endolysins. Viruses 2018, 10, 284. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J. Bacteriophage endolysins--current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Daraspe, J.; Giddey, M.; Moreillon, P.; Resch, G. In vitro characterization of PlySK1249, a novel phage lysin, and assessment of its antibacterial activity in a mouse model of Streptococcus agalactiae bacteremia. Antimicrob. Agents Chemother. 2013, 57, 6276–6283. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, D.G.; Dong, S.; Kirk, M.C.; Cartee, R.T.; Baker, J.R. LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl. Environ. Microbiol. 2007, 73, 7150–7154. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nobrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Sao-Jose, C.; Azeredo, J. Phage-Derived Peptidoglycan Degrading Enzymes: Challenges and Future Prospects for In Vivo Therapy. Viruses 2018, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Escobedo, S.; Wegmann, U.; Perez de Pipaon, M.; Campelo, A.B.; Stentz, R.; Rodriguez, A.; Martinez, B. Resident TP712 Prophage of Lactococcus lactis Strain MG1363 Provides Extra Holin Functions to the P335 Phage CAP for Effective Host Lysis. Appl. Environ. Microbiol. 2021, 87, e0109221. [Google Scholar] [CrossRef] [PubMed]
- Desvaux, M. Contribution of holins to protein trafficking: Secretion, leakage or lysis? Trends Microbiol. 2012, 20, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.L.; Saier, M.H., Jr. Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim. Biophys. Acta 2013, 1828, 2654–2671. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).