Abstract
Background/Objectives: Sugarcane (Saccharum spp.) is a major global sugar crop, and improving sucrose accumulation is critical for industry and bioenergy. Due to its high Brix content, Erianthus fulvus (E. fulvus) is valuable for genetic improvement of sugarcane. The bZIP transcription factor family critically regulates plant sucrose metabolism, but its roles in sugarcane remain largely unexplored. Methods: Through bioinformatics methods, Efbzip gene family members were systematically identified within the genome of E. fulvus. Gene expression patterns in distinct plant tissues were examined by RNA-seq and quantitative real-time PCR (qRT-PCR). Furthermore, genes potentially involved in sucrose metabolism were screened using transient expression assays and subcellular localization studies conducted in tobacco. Results: Seventy-nine Efbzip genes were identified and classified into nine subgroups, showing uneven distribution across ten chromosomes. Among ten conserved motifs, Motif1 was most conserved. Subcellular localization and physicochemical analyses showed most Efbzip proteins were hydrophilic and nuclear-localized. Cis-regulatory element analysis suggested Efbzip proteins regulate sucrose metabolism through hormone and light-responsive pathways. Segmental duplication primarily drove Efbzip gene family expansion. qRT-PCR showed predominant expression in stems and leaves, with subgroup-specific patterns. Nuclear localization of Efbzip52 was confirmed. Transient overexpression of Efbzip52, Efbzip61, and Efbzip64 significantly increased sucrose content in tobacco leaves, with highly statistically significant (p < 0.0001). Conclusions: In this study, the Efbzip gene family of E. fulvus was systematically characterized for the first time. Key candidate genes potentially involved in sucrose metabolism were identified, providing potential targets for the genetic improvement of sugarcane.
1. Introduction
The bZIP transcription factor family has a wide distribution among eukaryotes [,,]. The domains of these proteins are essentially the basic region and the leucine zipper motif. The basic region of the transcription factor recognizes the specific target DNA, and the leucine zipper is involved in dimerization, thus having the ability to bind the target gene’s DNA efficiently [,]. bZIP transcription factors are divided into various subfamilies, such as the A, B, C, and D subfamilies, depending on the characteristics and evolutionary distances of the proteins []. Previous studies have identified the bZIP gene family in various species, such as rice and sugarcane [,,,,,,,,]. Functionally, bZIP transcription factors participate broadly in biological processes, including developmental regulation [], hormonal signaling pathways [], sucrose metabolism [], and plant responses to abiotic stresses (e.g., salt, drought, cold, heat) [,,,] as well as biotic stresses [,].
Critically, bZIP transcription factors are central to regulating sugar signaling responses and metabolic gene transcription. Sugar-dependent translational regulation inhibits transcription factors such as AtbZIP11/ATB2 []. Enhanced sugar accumulation, including sucrose, glucose, and fructose, results from the expression of SlbZIP1 and SlbZIP2 gene variants lacking upstream open reading frames (uORFs), specifically during tomato fruit development []. In apple, phosphorylated bZIP39 activates sorbitol metabolism by interacting with the promoter regions of the sorbitol dehydrogenase 1 (SDH1) and aldose-6-phosphate reductase (A6PR) genes []. The expression of MtATB2 also varies with the level of sucrose in the root nodules of legumes and corresponds with changes in root growth and senescence []. Overexpression of TBZ17 in tobacco leads to a significant increase in leaf sucrose content [], while PpybZIP43 promotes sucrose synthesis in pear by inducing PpySPS3 expression [].
Sugarcane is a worldwide sugar crop. Increasing the concentration of sugars in sugarcane is very important not only for the yield of the plant but also for the food and energy sectors. Contemporary research on sugar metabolism demonstrates that sugars serve not only as energy storage molecules but also as significant regulators of plant growth and stress response mechanisms at the cellular level [,,,]. Consequently, developing sugarcane varieties with high sugar content has become a fundamental breeding objective [,,]. E. fulvus, a wild type of sugarcane, belongs to the Saccharinae subfamily within the Poaceae family and is notably the only diploid species in the Saccharum Complex. This type of grass is found in tropical, subtropical, and temperate climates and grows in the valleys of mountains with elevations of 1300–2400 m above sea level []. The beneficial characteristics of E. fulvus found through former research studies include its high sugar (high Brix), resistance to drought, resistance to poor soil conditions, and cold resistance [,,,]. The search for genes linked to the high-sugar trait of E. fulvus could be a good approach for improving the sugar level of a sugarcane plant.
Due to the complexity of the sugarcane genome [], systematic identification of bZIP transcription factors in sugarcane remains unreported. Given the potential of E. fulvus for genetic improvement in sugarcane breeding programs and the significant role of bZIP transcription factors in sucrose metabolism, this study aimed to systematically characterize the Efbzip gene family. This research further explored candidate genes linked to sucrose metabolism, thereby addressing existing gaps in the understanding of bZIP transcription factors in E. fulvus.
2. Materials and Methods
2.1. Plant Materials
The ZM584 clonal line of E. fulvus (2n = 20, genome size 902 Mb) was obtained and preserved by the Sugarcane Research Institute of Yunnan Agricultural University. Plants were grown in pots under controlled greenhouse conditions at the same institute. This genotype reaches a Brix value of 18.6% at maturity, representing one of the high-sugar wild germplasms. In September 2025, mature roots, stems, and leaves were harvested, rapidly frozen in liquid nitrogen, and stored at −80 °C for RNA isolation and subsequent cDNA synthesis. These tissues were used for gene cloning and qRT-PCR analyses. Nicotiana benthamiana plants maintained in our laboratory were employed for subcellular localization assays and transient expression studies.
2.2. Identification of Gene Family and Physicochemical Analysis of Proteins
Genome data for E. fulvus (YN2009-3) were retrieved from the Sugarcane Genome Database []. Efbzip family members were identified using TBtools (V1.116) based on the hidden Markov model PF00170 obtained from InterPro []. Conserved domains of putative genes were verified through CDD search and Pfam. Physiochemical characteristics of the encoded proteins were evaluated using ExPasy (http://web.expasy.org/protparam/, accessed on 28 November 2025), and subcellular localization was inferred with the WoLF PSORT tool.
2.3. Phylogenetic Analysis of Efbzip Protein Family
The phylogenetic relationships among Efbzip proteins were assessed using Clustal X2, and the resulting tree was visualized with ChiPlot. The Neighbor-Joining method was applied with default settings, including a bootstrap analysis of 1000 replicates.
2.4. Promoter Cis-Acting Element Analysis of Efbzip Genes
The promoter regions, comprising 2000 bp upstream sequences from each Efbzip gene, were extracted from the E. fulvus genome utilizing TBtools (V1.116). The PlantCARE database was employed to predict cis-regulatory elements under default parameters, and the annotated results were subsequently visualized using TBtools.
2.5. Analysis of Gene Structure, Conserved Protein Motifs, and Domains in Efbzip Family Members
Gene structural analysis and conserved protein motifs were determined using TBtools (V1.116). The parameters for motif identification included a motif count of 10, minimum motif width of 6, and maximum motif width of 50, with other settings maintained at default. Conserved protein domains were identified using the CDD search tool, and all results related to gene structures, motifs, and domains were visualized through TBtools (V1.116).
2.6. Synteny Analysis of Efbzip Genes
Synteny analyses, both within E. fulvus for the Efbzip gene family and between E. fulvus and species such as Arabidopsis thaliana, Saccharum spontaneum, Erianthus rockii, and the sugarcane cultivar XTT22, were conducted utilizing the MCScanX plugin within the TBtools (V1.116) software (E values ≤ 1 × 10−10, with all other parameters set to their default values). Visualization of these synteny relationships was also accomplished through TBtools.
2.7. Expression Pattern Analysis of Efbzip Genes in Different Tissues
To verify expression patterns of representative Efbzip genes, those exhibiting elevated expression levels within each subfamily were initially selected based on transcriptome data previously reported []. Expression validation was conducted using quantitative real-time PCR (qRT-PCR), with experiments performed in triplicate. Statistical analysis was completed by one-way ANOVA, followed by Tukey’s multiple-comparison test, to assess significant differences among the groups.
2.8. RNA Extraction, cDNA Synthesis, Gene Cloning, Vector Construction, and qRT-PCR Assays
TRIzol reagent (Tiangen, Beijing, China) was employed to extract total RNA, and complementary DNA (cDNA) synthesis was performed from this RNA using the FastQuant RT Super Mix Kit (Tiangen, Beijing, China). Primers required for cloning and qRT-PCR were generated using Primer 5 software (Table S6), while those designed for homologous recombination were prepared using SnapGene 3.2.1 software (Table S3). Gene amplification was executed with PrimeSTAR Max DNA Polymerase (Takara, Beijing, China), whereas homologous recombination procedures utilized the In-Fusion Snap Assembly Master Mix (Takara, Beijing, China). qRT-PCR reactions were carried out on the ABI 7500 Real-Time PCR System using SuperReal PreMix Plus (SYBR Green) reagent (Tiangen, Beijing, China). The housekeeping gene, 25S rRNA, was employed as an internal control, and relative gene expression was calculated using the 2−ΔΔCT method.
2.9. Subcellular Localization Assay
The pCAMBIA1300-Efbzip52-eGFP subcellular localization vector was constructed using homologous recombination. The vector was sequentially transformed into Escherichia coli and Agrobacterium tumefaciens competent cells. It was subsequently injected into Nicotiana benthamiana leaves for transient expression. Fluorescence signals were detected 72 h after injection using an Olympus FV3000 laser scanning confocal microscope (Olympus, Tokyo, Japan). Each treatment was performed in triplicate.
2.10. Identification of Efbzip Family Members Regulating Sucrose Metabolism
Previous studies suggest that structurally similar genes often share analogous functions. To identify potential Efbzip family members involved in sucrose metabolism, a phylogenetic tree was constructed using known bzip genes (Ppybzip43, TBZ17, Atbzip11) regulating sucrose metabolism in other species as references. Four E. fulvus genes (Efbzip13, Efbzip52, Efbzip61, and Efbzip64) clustered closely with these reference genes. These genes were cloned, and four overexpression vectors (pCAMBIA3301-Ubi-Efbzip13, pCAMBIA3301-Ubi-Efbzip52, pCAMBIA3301-Ubi-Efbzip61, and pCAMBIA3301-Ubi-Efbzip64) were constructed by homologous recombination using KpnI and BamHI restriction sites. For transient expression analysis, vectors were introduced into Nicotiana benthamiana plants, initially kept under dark conditions for 24 h before subsequent light exposure. Leaf tissues were harvested at two time points, 24 h (control) and 72 h post-infiltration, for both qRT-PCR assessment and sucrose quantification. Sucrose concentrations were determined using the Plant Sucrose Content Assay Kit (BOXBIO, Beijing, China). Agrobacterium tumefaciens culture with an OD600 value between 0.4 and 0.5 was used for the transformation experiments. Healthy Nicotiana benthamiana plants aged 6 weeks (with 4–6 true leaves) were used, with three biological replicates performed for each treatment. Two-way ANOVA was performed, followed by Tukey’s multiple-comparisons test, to determine significant differences between groups.
2.11. Data Analysis
All statistical analyses and graphical representations were produced using Microsoft Excel 2010 and GraphPad Prism 8.0.1 software.
3. Results
3.1. Identification of Efbzip Gene Family Members and Their Physicochemical Properties
A genome-wide identification of bZIP gene family members in E. fulvus was performed using TBtools (V1.116), resulting in the identification of 79 genes. Based on chromosomal location, these genes were named Efbzip1-79. They were unevenly distributed across 10 chromosomes: Chr01 (8), Chr02 (14), Chr03 (12), Chr04 (11), Chr05 (4), Chr06 (2), Chr07 (7), Chr08 (6), Chr09 (8), and Chr10 (8) (Figure 1A). Physicochemical analysis revealed substantial variation among Efbzip proteins (Table S1): amino acid counts ranged from 144 to 669, and molecular weights ranged from 15,395.39 to 73,565.89 kDa, exhibiting consistent trends. Instability indices ranged from 40.33 to 82.75, indicating these proteins are unstable. Analysis of aliphatic indices showed values ranging between 39.49 and 93.99, while GRAVY scores varied from −1.042 to −0.122, indicating the hydrophilic nature of these proteins. The proteins exhibited a wide isoelectric point (pI) distribution, spanning from 4.48 to 10.96, with 38 proteins demonstrating a pI below 7, suggesting an absence of distinct acidic or alkaline predominance within this family. Predicted subcellular localization predominantly placed members within the nucleus, although exceptions were noted for Efbzip19 (endoplasmic reticulum), Efbzip48 (mitochondrion), Efbzip49 (chloroplast), Efbzip56 (peroxisome), Efbzip65 (endoplasmic reticulum), Efbzip78 (chloroplast), and Efbzip79 (cytoplasm). Experimental validation confirmed that Efbzip52 localized to the nucleus, consistent with the prediction (Figure 1C).
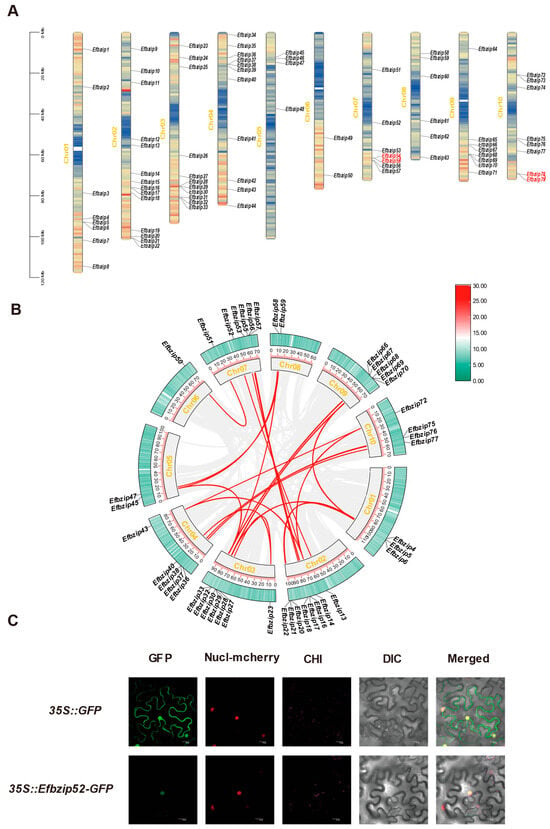
Figure 1.
Chromosomal localization, intraspecific synteny, and subcellular localization analysis of Efbzip genes. (A) The chromosomal localization map of the Efbzip gene family members. (B) Intraspecific synteny analysis. (C) Subcellular localization of Efbzip52. The subcellular localization maps from left to right are GFP green fluorescent protein (GFP), nucleus marker (Nucl mcherry), chloroplast fluorescence channel (CHI), brightfield (DIC), and merged image. Gray lines indicate collinear genes within E. fulvus, while red highlights Efbzip collinear genes.
3.2. Gene Family Evolution Analysis
To examine the phylogenetic relationships of the E. fulvus Efbzip gene family, the genes were classified into nine subfamilies using Arabidopsis thaliana as a reference (Figure 2). The number of members varied across subfamilies: A (18), B (2), C (5), D (19), E (5), F (4), G (6), I (11), and S (9). Members of the H subfamily in Arabidopsis did not cluster with E. fulvus bZIP genes and formed a separate branch. Within the Efbzip family, the A, D, and S subfamilies contained the most members, whereas the B subfamily had the fewest. These results indicate substantial variation in gene distribution among subfamilies.
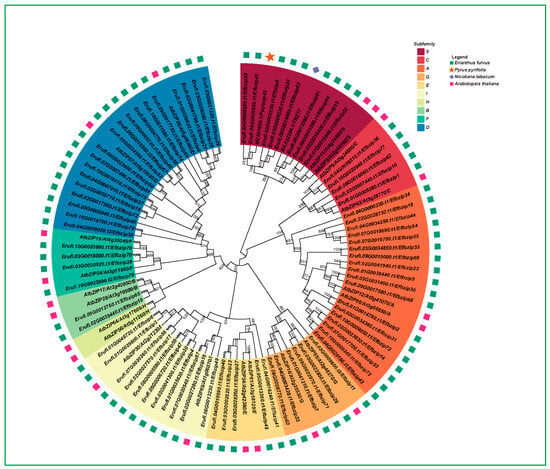
Figure 2.
Evolutionary tree of the bZIP gene family (the bootstrap value is 1000).
3.3. Analysis of Promoter Cis-Acting Elements in Efbzip Gene Family Members
Analysis of the 2000 bp promoter regions from all 79 Efbzip genes identified 26 major cis-acting elements (Figure 3). These elements included those responsive to light, MeJA, abscisic acid, salicylic acid, cold, and anaerobic induction. Light-responsive elements were the most abundant (785 occurrences), followed by MeJA-responsive (332) and abscisic acid-responsive (261) elements (Table S3). These findings suggest that the Efbzip gene family may regulate sucrose metabolism through light signaling and hormone-mediated pathways.
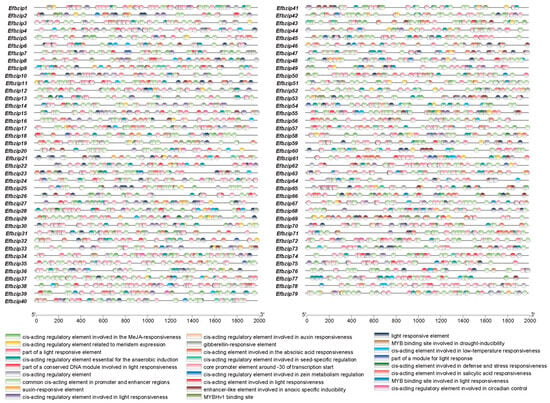
Figure 3.
The cis-acting element functions in the promoter of Efbzip gene family members.
3.4. Analysis of Gene Structure, Conserved Motifs, and Domains in Efbzip Family Members
Conserved motif prediction using TBtools (V1.116) identified 10 motifs (Motif 1–10) in Efbzip proteins (Figure 4A). All genes contained Motif 1. The A subfamily mainly contained Motif 7, Motif 9, and Motif 10; subfamilies E and I primarily contained Motif 4; and the D subfamily contained Motifs 2, 3, 5, 6, and 8. Domain analysis (Figure 4B) showed that: Subfamily A mainly contained the bZIP_plant_BZIP46 domain; Subfamilies S and C predominantly contained the bZIP_plant_GBF1 domain; Subfamily G primarily contained the MFMR and MFMR_assoc domains; Subfamily E mainly contained the bZIP_plant_RF2 domain; Subfamily B predominantly contained the bZIP_HY5-like domain; Subfamily D primarily contained the bZIP superfamily domain.
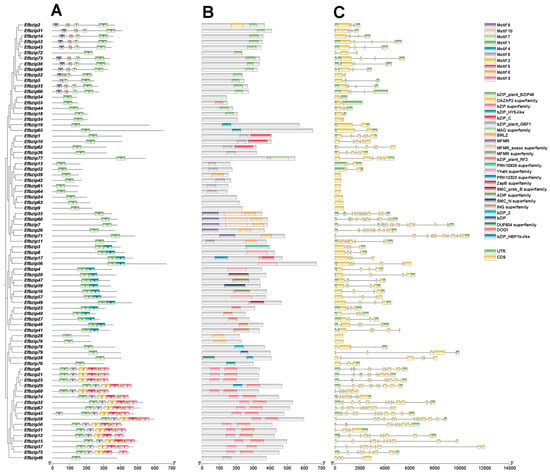
Figure 4.
Sequence characteristics of Efbzip genes. (A) Conserved motifs; (B) Domains; (C) Gene structure.
Gene structure analysis (Figure 4C) showed that members within the same subfamily had conserved gene structures. Seventeen genes lacked UTR sequences, whereas 78% of genes contained UTRs. These results indicate that gene structures are relatively conserved within subfamilies, while differences among subfamilies reflect functional diversification.
3.5. Synteny Analysis of the Efbzip Gene Family
Synteny analysis using TBtools (V1.116) identified 28 duplication events within the Efbzip family (Table S4), including 26 segmental duplications (Figure 1B) and 2 tandem duplications (Figure 1A). All chromosomes participated in segmental duplication events, while tandem duplications occurred only on Chr07 and Chr10. These results indicate that segmental duplication likely served as the primary driver of Efbzip gene family expansion in E. fulvus.
The divergence times of duplicated gene pairs ranged from 5.58 to 170 MYA, with 8 events within the past 60 MYA and 20 events occurring earlier. All Ka/Ks ratios were <1, suggesting that the duplicated genes experienced purifying selection.
To examine evolutionary relationships across species, interspecific synteny analysis was performed between E. fulvus (Ef) and Arabidopsis thaliana (At), S. spontaneum (Ss), Erianthus rockii (Er), and XTT22 (Figure 5). The numbers of collinear gene pairs (Table S5) were 5, 114, 262, and 597, respectively, indicating stronger synteny with species phylogenetically closer to E. fulvus.
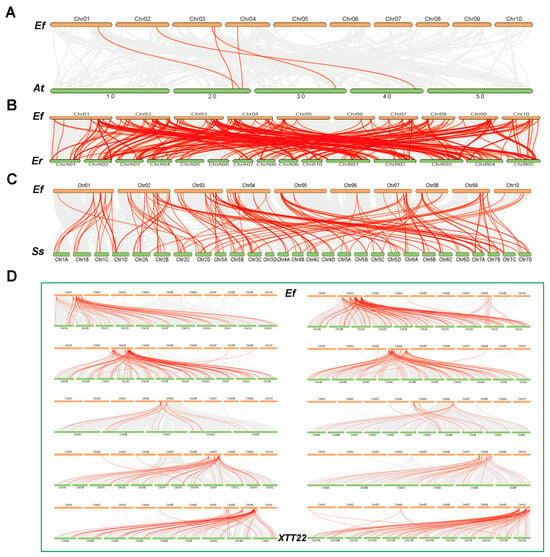
Figure 5.
Interspecies collinearity analysis. (A) Collinearity analysis between Ef and At; (B) Collinearity analysis between Ef and Er; (C) Collinearity analysis between Ef and Ss; (D) Collinearity analysis between Ef and Ss. The gray line represents the collinearity genes between E. fulvus and other species, while the red line highlights the collinearity genes between E. fulvus Efbzip and other species.
3.6. Expression Patterns of the Efbzip Gene Family in Different Tissues
To further validate the tissue-specific expression characteristics of the Efbzip gene family, 18 genes showing high transcript levels from each subfamily were identified based on available transcriptomic data [] (Figure 6A, Table S6). A combination of qRT-PCR assays and transcriptome analysis was utilized. Within subfamily A, Efbzip33 and Efbzip43 exhibited the greatest expression in leaves, notably higher than in stems and roots. For subfamily B, Efbzip19 and Efbzip65 were predominantly expressed in stems, significantly surpassing expression levels in leaves and roots. Subfamily C’s Efbzip36 was chiefly expressed in stems, substantially higher compared to leaves and roots. In subfamily D, elevated leaf expression was noted for Efbzip21 and Efbzip50, markedly greater than root expression levels. In subfamily E, Efbzip41 and Efbzip48 demonstrated peak expression in stems and leaves, respectively. Efbzip78 from subfamily F presented highest stem expression, significantly exceeding that observed in leaves and roots. Within, Efbzip60 had prominent expression in stems, markedly higher than in leaves and roots; Efbzip71 and Efbzip26 were predominantly expressed in leaves, significantly exceeding expression in stems and roots. In subfamily I, Efbzip47 was most expressed in leaves, considerably higher than in stems and roots. Subfamily S members Efbzip13, Efbzip52, Efbzip61, and Efbzip64 showed significantly elevated transcript levels in stems and roots compared to leaves. Overall, the Efbzip gene family in E. fulvus was mainly expressed in stems and leaves, with the fewest highly expressed genes found in roots. Members of the same subfamily generally exhibited similar expression patterns.
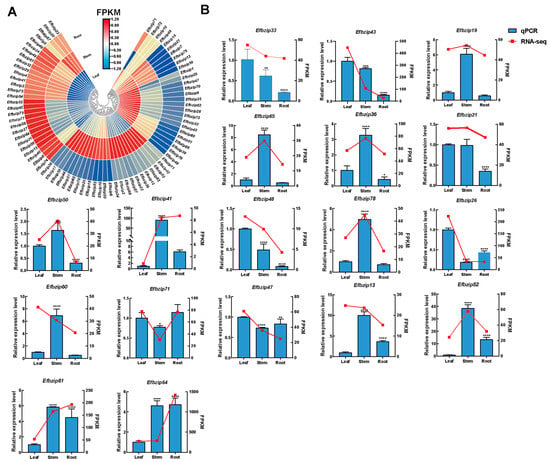
Figure 6.
Tissue-specific expression patterns. (A) Transcriptome data heatmap. (B) Expression patterns of Efbzip genes in leaves, stems, and roots. Statistical significance is indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Error bar: SD.
3.7. Identification of Genes Regulating Sucrose Metabolism Among Efbzip Family Members
Previous studies suggest structurally similar genes may share functional similarities. To identify potential regulators of sucrose metabolism within the Efbzip family, a phylogenetic tree was constructed (Figure 3), including known sucrose-related bZIP genes (Ppybzip43, TBZ17, Atbzip11) from other species and E. fulvus Efbzip members. Protein sequence alignment was also visualized (Figure 7A). Phylogenetic analysis revealed Efbzip13, Efbzip52, Efbzip61, and Efbzip64 clustered closely. These four genes underwent cloning, and overexpression vectors were subsequently generated using homologous recombination techniques. Nicotiana benthamiana plants were utilized for transient expression assays. Compared with the control (CK, Figure 7B), a marked increase in transcript abundance for all four genes was observed at 72 h post-transfection. Furthermore, the transient overexpression of Efbzip52, Efbzip61, and Efbzip64 notably elevated sucrose levels in tobacco leaves (Figure 7C), while Efbzip13 displayed no significant impact. These findings suggest that Efbzip52, Efbzip61, and Efbzip64 may play crucial roles in sucrose metabolism within sugarcane, marking them as potential genetic targets for developing high-sugar cultivars.
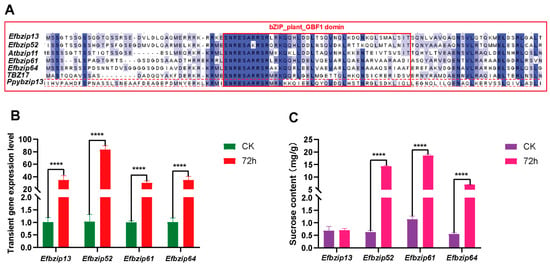
Figure 7.
Analysis of transient expression impacts of Efbzip genes on sucrose metabolism. (A) Protein sequence alignment. (B) Quantification of gene expression. (C) Determination of sucrose content. Statistical significance was set as follows: **** p < 0.0001. Error bar: SD.
4. Discussion
bZIP transcription factors are very important for the metabolism of sucrose in the plant, especially through the control of sugar signaling and expression of the related genes. From the E. fulvus genome, a total of 79 EfbZIP transcription factors were found, and there were differences observed in the number of bZIP genes found in other species. For example, Oryza sativa has (71) [], Setaria italica (92) [], Salvia miltiorrhiza (70) [], Andrographis paniculata (62) [], Gossypium hirsutum L. (207) [], Solanum tuberosum L. (65) [], Ricinus communis L. (49) [], Pyrus spp. (84) [], and Arabidopsis (75) []. These variations may result from species-specific evolutionary events or differences in genome size.
The instability indices of EfbZIP proteins, determined using physicochemical properties, vary between 40.33 and 82.75, which is low. Proteins with indices above 40 are believed to be unstable []. High instability is closely related to the structure and functions of the proteins, probably involving the dynamic biological functions of stress response and signal transduction [].
Based on the information of the Arabidopsis bZIP gene family, the E. fulvus bZIP gene family consists of nine subfamilies (A, B, C, D, E, F, G, I, and S), each having a different number of genes. There are 18 genes belonging to subfamily A, while only two genes are found in subfamily B. These asymmetrical distributions might suggest expansions and contractions within the gene family of E. fulvus [,]. Tandem duplications and segmental duplications are common genetic events that occur within the E. fulvus genome []. The same is true for the bZIP gene family of the sugarcane plant, where the subfamilies that have a large number of genes (A and D, for instance) are believed to have undergone repeated gene duplication events, thus having important functions relating to environmental adaptations. Moreover, the genes of the H subfamily of the Arabidopsis are not related to any of the E. fulvus bZIP genes of the EfbZIP subgroup, having undergone elimination and having been replaced with other functions during the course of E. fulvus evolution []. Studies have also previously shown that the S subfamily of the Arabidopsis bZIP genes is involved in the metabolism of sugars such as sucrose []. We hypothesize that the S subfamily in E. fulvus may have similar functions, although further experimental validation is required.
Motif analysis indicates that each gene contains the same motif, a phenomenon also observed in rice []. Furthermore, motif diversity varies across subfamilies, potentially due to functional differentiation, a phenomenon previously reported in poplar []. Domain examination showed the presence of subfamily-specific domains. This condition has also been found among other species, such as the mangosteen plant and other organisms []. Gene structure analysis indicated relatively conserved gene structures among members of the same subfamily, consistent with findings in the bZIP gene family of Lycium barbarum []. In summary, the conservation within subfamilies and differences among subfamilies in E. fulvus Efbzip genes not only support the evolutionary conservation of the bZIP gene family across species but also demonstrate its functional diversification [,]. The bZIP transcription factor light response element dominates across multiple species, including E. fulvus, rice, and Arabidopsis [,,,,], indicating its potential in regulating light signal transduction.
In an intraspecific collinearity analysis, 28 duplication events were observed, consisting of 26 segmental duplication pairs and 2 tandem duplication pairs. This indicates that gene duplication likely represents the primary mechanism underlying the Efbzip gene family’s expansion, consistent with patterns observed across other plant gene families [,,,,]. The differentiation time of duplicated gene pairs ranges from 5.58 to 170 million years (MYA). Among these, 8 pairs emerged within 60 MYA, and 20 pairs appeared earlier than 60 MYA, indicating multiple gene expansion events over a long evolutionary period. Additionally, all duplicated events appear to have undergone purifying selection pressure, suggesting these genes maintain functional conservation essential for fundamental biological processes [].
In interspecies comparisons, the high homology between Ef and XTT22 as well as Er reflects their close phylogenetic relationships and shared evolutionary histories. In contrast, Ef shares only five collinear gene pairs with At, indicating significant divergence in the evolution of their bZIP gene families. This divergence likely results from the phylogenetic distance between dicotyledons and monocotyledons. The 114 collinear gene pairs between Ef and Ss suggest partial conservation, possibly related to their shared ancestry within the Poaceae family. Collectively, these interspecies differences underscore the dynamic evolution of the bZIP gene family in plants: closely related species (e.g., Ef, Er, and XTT22) maintain high homology through gene duplication, while distantly related species exhibit more divergent gene family structures due to independent evolutionary events [,,].
Efbzip genes display tissue-specific expression patterns consistent with previous studies [,]. Furthermore, existing research indicates that tissue-specific gene expression may correlate with the gene’s biological function [,,,]. Therefore, studying the gene’s expression patterns is important for further functional research.
Phylogenetic analysis revealed that Efbzip13, Efbzip52, Efbzip61, and Efbzip64 clustered with known bZIP genes associated with sucrose metabolism (such as PpybZIP43, TBZ17, and AtbZIP11), supporting the hypothesis that structurally similar genes possess analogous functions []. Subsequent functional validation revealed that transient overexpression of Efbzip52, Efbzip61, and Efbzip64 in Nicotiana benthamiana significantly increased the sucrose content in tobacco leaves, whereas Efbzip13 did not exhibit this effect. This suggests that Efbzip52, Efbzip61, and Efbzip64 may directly or indirectly regulate sucrose metabolism. These findings reinforce the utility of phylogenetic analysis in predicting gene functions and highlight the potential of Efbzip52, Efbzip61, and Efbzip64 as sucrose metabolism regulators. The lack of effect observed with Efbzip13 may result from structural fine-tuning (such as sequence variations outside conserved domains) affecting protein interaction specificity [,] or promoter evolution [].
This study has some inherent limitations. Firstly, gene functions identified in model plants may not necessarily correspond to identical roles in different species. Secondly, there is currently insufficient experimental evidence validating the functions of these genes. To overcome these constraints, future investigations should incorporate advanced genetic methodologies such as overexpression and CRISPR-Cas9-mediated gene editing in both sugarcane and E. fulvus.
5. Conclusions
This study systematically identified a total of 79 Efbzip genes belonging to the Efbzip gene family in the E. fulvus genome, of which the genes are unequally distributed across the 10 chromosomes. Subcellular localization analysis predicted that the members of the Efbzip gene family are nuclear. Phylogenetic analysis classified the 79 identified Efbzip genes into nine subfamilies. The analysis of the structures of the Efbzip genes revealed the widespread distribution of Motif1 and the consensus of each subfamily. The analysis of the cis-active promoter elements further revealed that the Efbzip gene family is often involved in the light and hormone regulatory pathways of E. fulvus. The gene duplication event of the Efbzip gene family in E. fulvus, revealed through the phylogenetic analysis, included the segment and tandem duplication events of the Efbzip gene family. The expression of the Efbzip genes was further confirmed using the qRT-PCR analysis. The expression of the bZIP genes was largely observed in stems and leaves, with relatively low expression of specific genes in the roots. There was a trend of similar expression among members of the same subfamily. Transient overexpression of Efbzip52, Efbzip61, and Efbzip64 led to significantly elevated sucrose content in tobacco leaves (p < 0.0001). Overall, this study represents the first extensive characterization of the bZIP transcription factor gene family in E. fulvus and identifies promising candidate genes for the genetic improvement of sucrose metabolism traits in sugarcane.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes16121434/s1, Table S1: Physicochemical properties analysis of 79 proteins; Table S2: The secondary structure of 79 proteins; Table S3: Cis-acting elements of the promoter region; Table S4: Segmentally and tandemly duplicated gene pairs; Table S5: The number of collinear genes between the E. fulvus Efbzip gene family and other different species; Table S6: RNA-seq data (FPKM values) of 79 genes; Table S7: Primer sequences for qRT-PCR and gene cloning.
Author Contributions
Conceptualization, C.Z.; methodology, C.Z., Z.Q. and W.N.; formal analysis, C.Z., Z.Q. and Y.W.; investigation, F.L.; resources, F.L.; data curation, C.Z., Q.D., Y.W. and W.N.; writing—original draft preparation, C.Z. and W.N.; writing—review and editing, C.Z., Z.Q. and F.L.; visualization, C.Z. and W.N.; supervision, F.L. and L.H.; project administration, F.L. and L.H.; funding acquisition, F.L. and L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding from various sources, including the Major Science and Technology Project of Yunnan Province (grant number 202202AE090021), the Special Project associated with the Key Laboratory for Crop Production and Smart Agriculture in Yunnan Province (grant number 202105AG070007), the National Natural Science Foundation of China (project number 31960451) and the Yunnan Provincial Department of Education Scientific Research Fund Project (grant number 2025Y0490).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting the results of this study are available from the corresponding authors upon reasonable request.
Acknowledgments
We are deeply grateful for the valuable feedback provided by each reviewer.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhao, K.; Liu, L.; Huang, S. Genome-Wide Identification and Functional Analysis of the bZIP Transcription Factor Family in Rice Bakanae Disease Pathogen, Fusarium fujikuroi. Int. J. Mol. Sci. 2022, 23, 6658. [Google Scholar] [CrossRef]
- Niu, S.; Gu, X.; Zhang, Q.; Tian, X.; Chen, Z.; Liu, J.; Wei, X.; Yan, C.; Liu, Z.; Wang, X.; et al. Grapevine bZIP transcription factor bZIP45 regulates VvANN1 and confers drought tolerance in Arabidopsis. Front. Plant Sci. 2023, 14, 1128002. [Google Scholar] [CrossRef]
- Choi, J.; Lim, C.W.; Lee, S.C. Role of pepper bZIP transcription factor CaADBZ1 in abscisic acid signalling and drought stress response. Physiol. Plant. 2025, 177, e70159. [Google Scholar] [CrossRef]
- Siberil, Y.; Doireau, P.; Gantet, P. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 2001, 268, 5655–5666. [Google Scholar] [CrossRef]
- Soucek, L.; Helmer-Citterich, M.; Sacco, A.; Jucker, R.; Cesareni, G.; Nasi, S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene 1998, 17, 2463–2472. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Qiao, Y.; Pan, X.; Chen, X.; Su, W.; Li, A.; Li, X.; Liao, W. Genome-Wide identification and expression analysis of CsABF/AREB gene family in cucumber (Cucumis sativus L.) and in response to phytohormonal and abiotic stresses. Sci. Rep. 2025, 15, 15757. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Zhao, P.; Ye, M.; Wang, R.; Wang, D.; Chen, Q. Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance. Int. J. Biol. Macromol. 2020, 161, 155–167. [Google Scholar] [CrossRef]
- Jin, Z.; Xu, W.; Liu, A. Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 2014, 239, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Y.; Wang, Q.; Tao, X.; Fang, J.; Zheng, W.; Zhu, L.; Jia, B.; Heng, W.; Li, S. Identification of bZIP transcription factors and their responses to brown spot in pear. Genet. Mol. Biol. 2022, 45, e20210175. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Malik, W.A.; Chen, X.; Wang, J.; Wang, D.; Wang, S.; Chen, C.; Guo, L.; Ye, W. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int. J. Biol. Macromol. 2020, 146, 569–578. [Google Scholar] [CrossRef]
- Guan, R.; Xu, S.; Lu, Z.; Su, L.; Zhang, L.; Sun, W.; Zhang, Y.; Jiang, C.; Liu, Z.; Duan, L.; et al. Genomic characterization of bZIP transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Int. J. Biol. Macromol. 2022, 223, 1619–1631. [Google Scholar] [CrossRef]
- Jia, X.; Gao, H.; Zhang, L.; Tang, W.; Wei, G.; Sun, J.; Xiong, W. Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis. Biomolecules 2024, 14, 958. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Ji, A.; Luo, H.; Song, J. Genomic survey of bZIP transcription factor genes related to tanshinone biosynthesis in Salvia miltiorrhiza. Acta Pharm. Sin. B 2018, 8, 295–305. [Google Scholar] [CrossRef]
- Schlogl, P.S.; Nogueira, F.T.; Drummond, R.; Felix, J.M.; De Rosa, V.J.; Vicentini, R.; Leite, A.; Ulian, E.C.; Menossi, M. Identification of new ABA- and MEJA-activated sugarcane bZIP genes by data mining in the SUCEST database. Plant Cell Rep. 2008, 27, 335–345. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Casaretto, J.; Ho, T.D. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 2003, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Wiese, A.; Elzinga, N.; Wobbes, B.; Smeekens, S. Sucrose-induced translational repression of plant bZIP-type transcription factors. Biochem. Soc. Trans. 2005, 33, 272–275. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Liu, Y.; Chen, S.; Shityakov, S. Meta-Analysis of the Effects of Overexpressed bZIP Transcription Factors in Plants under Drought Stress. Plants 2024, 13, 337. [Google Scholar] [CrossRef]
- Hwang, I.; Jung, H.; Park, J.; Yang, T.; Nou, I. Transcriptome analysis of newly classified bZIP transcription factors of Brassica rapa in cold stress response. Genomics 2014, 104, 194–202. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Goncalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Berberich, T.; Tanaka, S.; Nishiyama, M.; Kanayama, Y.; Kojima, S.; Muramoto, K.; Kusano, T. A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant Biotechnol. J. 2016, 14, 1116–1126. [Google Scholar] [CrossRef]
- Meng, D.; Cao, H.; Yang, Q.; Zhang, M.; Borejsza-Wysocka, E.; Wang, H.; Dandekar, A.M.; Fei, Z.; Cheng, L. SnRK1 kinase-mediated phosphorylation of transcription factor bZIP39 regulates sorbitol metabolism in apple. Plant Physiol. 2023, 192, 2123–2142. [Google Scholar] [CrossRef]
- D’Haeseleer, K.; De Keyser, A.; Goormachtig, S.; Holsters, M. Transcription factor MtATB2: About nodulation, sucrose and senescence. Plant Cell Physiol. 2010, 51, 1416–1424. [Google Scholar] [CrossRef]
- Thalor, S.K.; Berberich, T.; Lee, S.S.; Yang, S.H.; Zhu, X.; Imai, R.; Takahashi, Y.; Kusano, T. Deregulation of sucrose-controlled translation of a bZIP-type transcription factor results in sucrose accumulation in leaves. PLoS ONE 2012, 7, e33111. [Google Scholar] [CrossRef]
- Zhang, H.; Tao, X.; Fan, X.; Zhang, S.; Qin, G. PpybZIP43 contributes to sucrose synthesis in pear fruits by activating PpySPS3 expression and interacts with PpySTOP1. Physiol. Plant. 2022, 174, e13732. [Google Scholar] [CrossRef]
- Araujo, M.A.; Melo, A.; Silva, V.M.; Reis, A. Selenium enhances ROS scavenging systems and sugar metabolism increasing growth of sugarcane plants. Plant Physiol. Biochem. 2023, 201, 107798. [Google Scholar] [CrossRef]
- Akbar, S.; Yao, W.; Qin, L.; Yuan, Y.; Powell, C.A.; Chen, B.; Zhang, M. Comparative Analysis of Sugar Metabolites and Their Transporters in Sugarcane Following Sugarcane mosaic virus (SCMV) Infection. Int. J. Mol. Sci. 2021, 22, 13574. [Google Scholar] [CrossRef]
- Sun, N.; Xu, X.; Zhu, Z.; Zhou, X.; Liu, Y.; Li, D.; Cao, F.; Wang, L.; Zhang, H. Tonoplast sugar transporter ZmTST1 positively regulates plant growth, salt and drought tolerance. Plant Physiol. Biochem. 2025, 229, 110380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Li, Y.P.; Gai, P.Z.; Gao, J.; Xu, L. Exogenously applied ABA alleviates dysplasia of maize (Zea mays L.) ear under drought stress by altering photosynthesis and sucrose transport. Plant Signal. Behav. 2025, 20, 2462497. [Google Scholar] [CrossRef]
- Zhao, J.; Li, S.; Xu, Y.; Ahmad, N.; Kuang, B.; Feng, M.; Wei, N.; Yang, X. The subgenome Saccharum spontaneum contributes to sugar accumulation in sugarcane as revealed by full-length transcriptomic analysis. J. Adv. Res. 2023, 54, 1–13. [Google Scholar] [CrossRef]
- Wang, M.; Li, A.; Liao, F.; Chen, Z.; Qin, C.; Zhang, B.; Li, X.; Su, Z.; Pan, Y.; Huang, D. Sugarcane microRNA shy-miR164 regulates sugar metabolism through direct cleavage of the transcription factor ScNAC mRNA. Plant Physiol. 2025, 198, kiaf354. [Google Scholar] [CrossRef]
- Chen, M.; Liu, P.; An, R.; He, X.; Zhao, P.; Huang, D.; Yang, X. Sugarcane Pan-Transcriptome Identifying a Master Gene ScHCT Regulating Lignin and Sugar Traits. J. Agric. Food. Chem. 2025, 73, 1739–1755. [Google Scholar] [CrossRef]
- Ling, K.; Yi-Ning, D.; Majeed, A.; Zi-Jiang, Y.; Jun-Wen, C.; Li-Lian, H.; Xian-Hong, W.; Lu-Feng, L.; Zhen-Feng, Q.; Dan, Z.; et al. Evaluation of genome size and phylogenetic relationships of the Saccharum complex species. 3 Biotech 2022, 12, 327. [Google Scholar] [CrossRef] [PubMed]
- Kui, L.; Majeed, A.; Wang, X.; Yang, Z.; Chen, J.; He, L.; Di, Y.; Li, X.; Qian, Z.; Jiao, Y.; et al. A chromosome-level genome assembly for Erianthus fulvus provides insights into its biofuel potential and facilitates breeding for improvement of sugarcane. Plant Commun. 2023, 4, 100562. [Google Scholar] [CrossRef]
- Qian, Z.; Zhao, C.; Wan, H.; He, L.; Wang, X.; Li, F. Research Progress and Utilization Potential of Saccharum hortensis, a Wild Relative of Sugarcane. J. Plant Genet. Resour. 2025, 26, 1485–1498. [Google Scholar] [CrossRef]
- Qian, Z.; Zhao, C.; Wan, H.; Rao, X.; Luo, Z.; He, L.; Li, F. Genome-wide identification of DREB1 transcription factors in Erianthus fulvus and the functional role of EfDREB1C in regulating cold tolerance of transgenic Arabidopsis and sugarcane. Int. J. Biol. Macromol. 2025, 317, 144859. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lin, W.; He, S. Reflections on Developing and Utilizing Wild Sugarcane Germplasm Resources. Resour. Dev. Mark. 2004, 20, 266–270. [Google Scholar] [CrossRef]
- Zhang, J.; Qi, Y.; Hua, X.; Wang, Y.; Wang, B.; Qi, Y.; Huang, Y.; Yu, Z.; Gao, R.; Zhang, Y.; et al. The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum. Nat. Genet. 2025, 57, 242–253. [Google Scholar] [CrossRef]
- Wang, T.; Wang, B.; Hua, X.; Tang, H.; Zhang, Z.; Gao, R.; Qi, Y.; Zhang, Q.; Wang, G.; Yu, Z.; et al. A complete gap-free diploid genome in Saccharum complex and the genomic footprints of evolution in the highly polyploid Saccharum genus. Nat. Plants 2023, 9, 554–571. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.T.; Jakovlic, I.; Wang, W. In silico characterisation, homology modelling and structure-based functional annotation of blunt snout bream (Megalobrama amblycephala) Hsp70 and Hsc70 proteins. J. Anim. Sci. Technol. 2015, 57, 44. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Zhao, L.; Yang, H.; Yang, X.; Bai, Y.; Xue, X.; Wang, D.; Han, S. Genome-Wide Characterization of Wholly Disordered Proteins in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 1117. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, X.; Li, F.; Li, D.; Dong, Y.; Fan, Y. Bioinformatics Analysis of WRKY Family Genes in Erianthus fulvus Ness. Genes 2022, 13, 2102. [Google Scholar] [CrossRef]
- Qian, Z.; Rao, X.; Zhang, R.; Gu, S.; Shen, Q.; Wu, H.; Lv, S.; Xie, L.; Li, X.; Wang, X.; et al. Genome-Wide Identification, Evolution, and Expression Analyses of AP2/ERF Family Transcription Factors in Erianthus fulvus. Int. J. Mol. Sci. 2023, 24, 7102. [Google Scholar] [CrossRef]
- E, Z.G.; Zhang, Y.P.; Zhou, J.H.; Wang, L. Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 2014, 13, 3025–3036. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Gao, H.; Cao, X.; Ma, Y.; Qin, X.; Bai, X.; Zhang, X.; Xiong, A.; Yin, Y.; Zheng, R. Genome-Wide Identification of bZIP Gene Family in Lycium barbarum and Expression During Fruit Development. Int. J. Mol. Sci. 2025, 26, 4665. [Google Scholar] [CrossRef]
- Li, H.; Li, L.; Shangguan, G.; Jia, C.; Deng, S.; Noman, M.; Liu, Y.; Guo, Y.; Han, L.; Zhang, X.; et al. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci. Rep. 2020, 10, 15521. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Hou, Z.; He, Q.; Zhang, X.; Yan, K.; Han, R.; Liang, Z. Genome-Wide Characterization and Expression Analysis of bZIP Gene Family Under Abiotic Stress in Glycyrrhiza uralensis. Front. Genet. 2021, 12, 754237. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gao, T.; Bian, K.; Meng, C.; Tang, X.; Mao, Y. Genome-wide analysis and expression profile of the bZIP gene family in Neopyropia yezoensis. Front. Plant Sci. 2024, 15, 1461922. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Wang, J.; Gao, T.; Qu, C.; Mo, X.; Zhang, X. Systematic analysis of bZIP gene family in Suaeda australis reveal their roles under salt stress. BMC Plant Biol. 2024, 24, 816. [Google Scholar] [CrossRef]
- Ye, W.; Wang, Y.; Dong, S.; Tyler, B.M.; Wang, Y. Phylogenetic and transcriptional analysis of an expanded bZIP transcription factor family in Phytophthora sojae. BMC Genom. 2013, 14, 839. [Google Scholar] [CrossRef]
- Yang, X.; Gao, C.; Hu, Y.; Ma, Q.; Li, Z.; Wang, J.; Li, Z.; Zhang, L.; Li, D. Identification and expression analysis of bZIP transcription factors in Setaria italica in response to dehydration stress. Front. Genet. 2024, 15, 1466486. [Google Scholar] [CrossRef]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genom. 2019, 20, 483. [Google Scholar] [CrossRef]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and Analysis of bZIP Family Genes in Potato and Their Potential Roles in Stress Responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef]
- Pophaly, S.D.; Tellier, A. Population Level Purifying Selection and Gene Expression Shape Subgenome Evolution in Maize. Mol. Biol. Evol. 2015, 32, 3226–3235. [Google Scholar] [CrossRef]
- Balakrishnan, S.; Bhasker, R.; Ramasamy, Y.; Dev, S.A. Genome-wide analysis of cellulose synthase gene superfamily in Tectona grandis L.f. 3 Biotech 2024, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, T.; Wu, Y.; Tang, H.; Yu, J.; Dai, X.; Zheng, Y.; Wan, X.; Yang, Y.; Tan, X. Genome-wide analysis of the peanut CaM/CML gene family reveals that the AhCML69 gene is associated with resistance to Ralstonia solanacearum. BMC Genom. 2024, 25, 200. [Google Scholar] [CrossRef]
- Gao, Z.; Wu, Y.; Li, M.; Ding, L.; Li, J.; Liu, Y.; Cao, Y.; Hua, Y.; Jia, Q.; Wang, D. The auxin response factor (ARF) gene family in Cyclocarya paliurus: Genome-wide identification and their expression profiling under heat and drought stresses. Physiol. Mol. Biol. Plants 2024, 30, 921–944. [Google Scholar] [CrossRef]
- Feng, Y.; Bakari, A.; Guan, H.; Wang, J.; Zhang, L.; Xu, M.; Nyoni, M.; Cao, S.; Zhang, Z. An Investigation into the Evolutionary Characteristics and Expression Patterns of the Basic Leucine Zipper Gene Family in the Endangered Species Phoebe bournei Under Abiotic Stress Through Bioinformatics. Plants 2025, 14, 2292. [Google Scholar] [CrossRef]
- Ma, F.; Zhou, H.; Xu, Y.; Huang, D.; Wu, B.; Xing, W.; Chen, D.; Xu, B.; Song, S. Comprehensive analysis of bZIP transcription factors in passion fruit. iScience 2023, 26, 106556. [Google Scholar] [CrossRef]
- Sprenger-Haussels, M.; Weisshaar, B. Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 2000, 22, 1–8. [Google Scholar] [CrossRef]
- Rook, F.; Gerrits, N.; Kortstee, A.; van Kampen, M.; Borrias, M.; Weisbeek, P.; Smeekens, S. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998, 15, 253–263. [Google Scholar] [CrossRef]
- Fox, R.M.; Andrew, D.J. Transcriptional regulation of secretory capacity by bZip transcription factors. Front. Biol. 2015, 10, 28–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.W.; Hsueh, A.J. Genomic analyses of the evolution of LGR genes. Change Gung Med. J. 2006, 29, 2–8. [Google Scholar]
- Bizotto, F.M.; Ceratti, R.S.; Braz, A.; Masuda, H.P. Evolutionary history of Mo25 gene in plants, a component of RAM/MOR signaling network. Mech. Dev. 2018, 153, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Nong, Q.; Xie, J.; Wang, Z.; Liang, Q.; Solanki, M.K.; Malviya, M.K.; Liu, X.; Li, Y.; Htun, R.; et al. Molecular Characterization and Co-expression Analysis of the SnRK2 Gene Family in Sugarcane (Saccharum officinarum L.). Sci. Rep. 2017, 7, 17659. [Google Scholar] [CrossRef] [PubMed]
- Babula-Skowronska, D. Functional divergence of Brassica napus BnaABI1 paralogs in the structurally conserved PP2CA gene subfamily of Brassicaceae. Genomics 2021, 113, 3185–3197. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).