Integrative Analysis of Biomarkers for Cancer Stem Cells in Bladder Cancer and Their Therapeutic Potential
Abstract
1. Introduction
2. Methods and Materials
2.1. Data Collection and Study Design
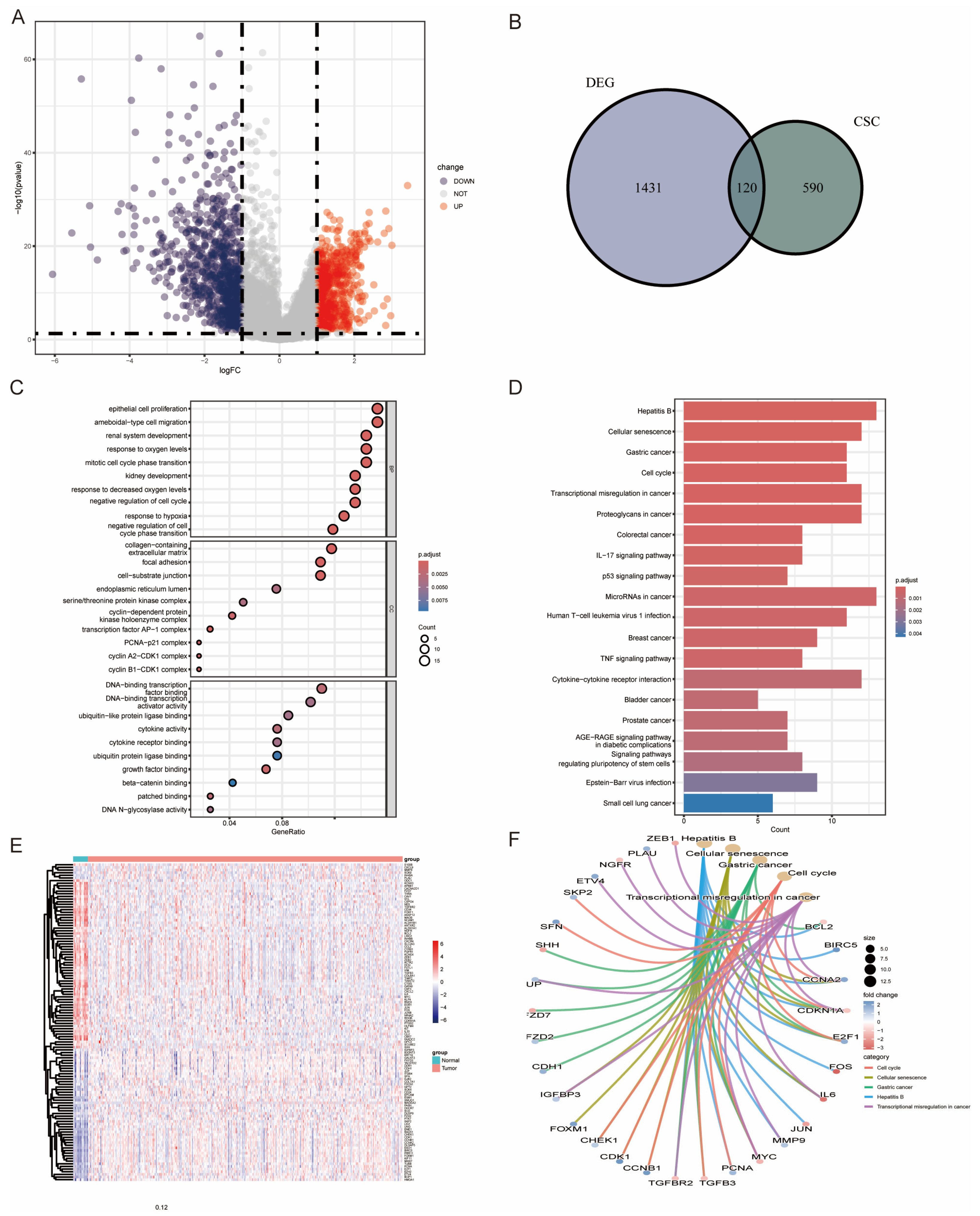
2.2. Identification of Differentially Expressed BCSCs (DE-BCSCs) and Functional Enrichment Analysis
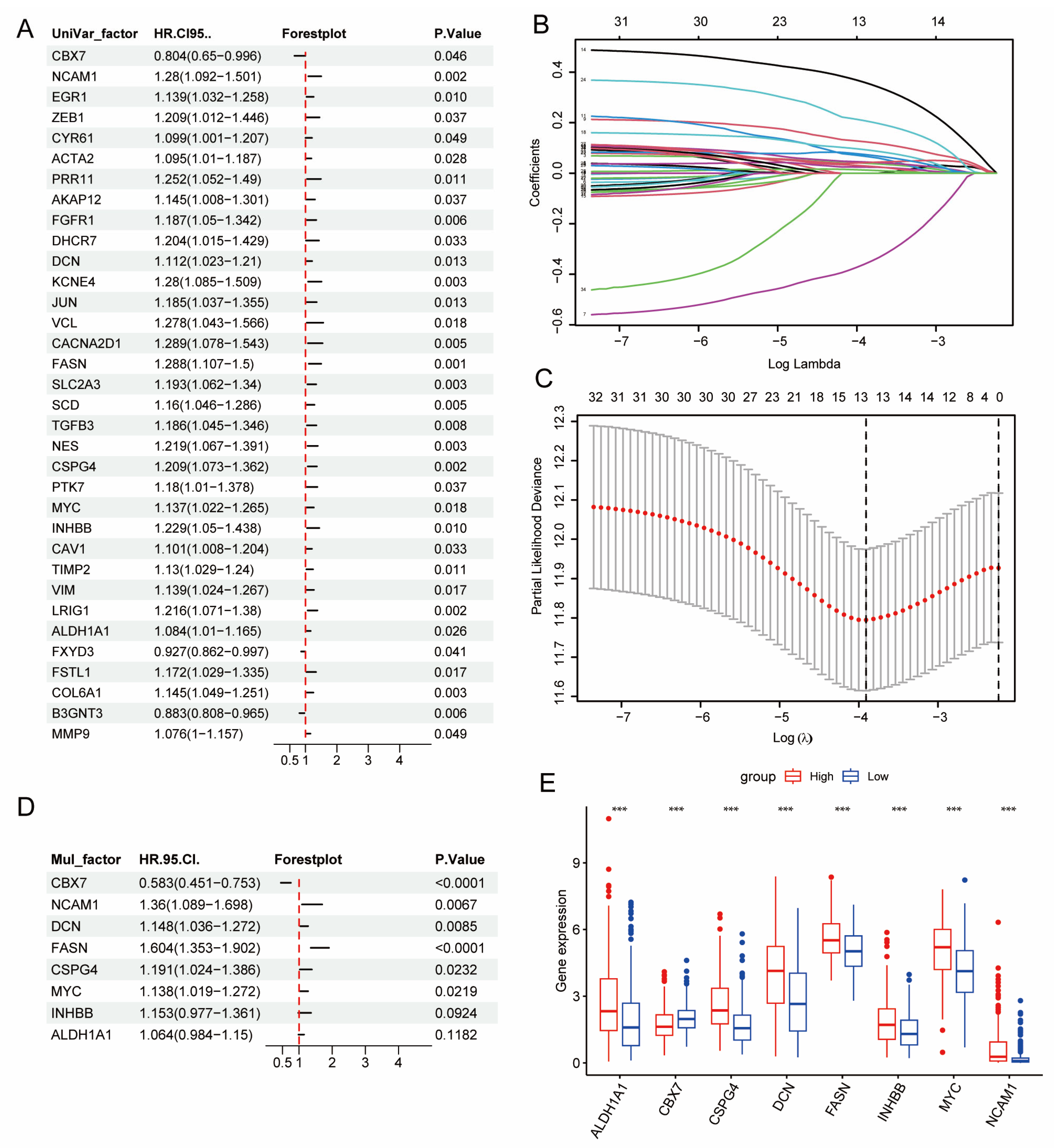
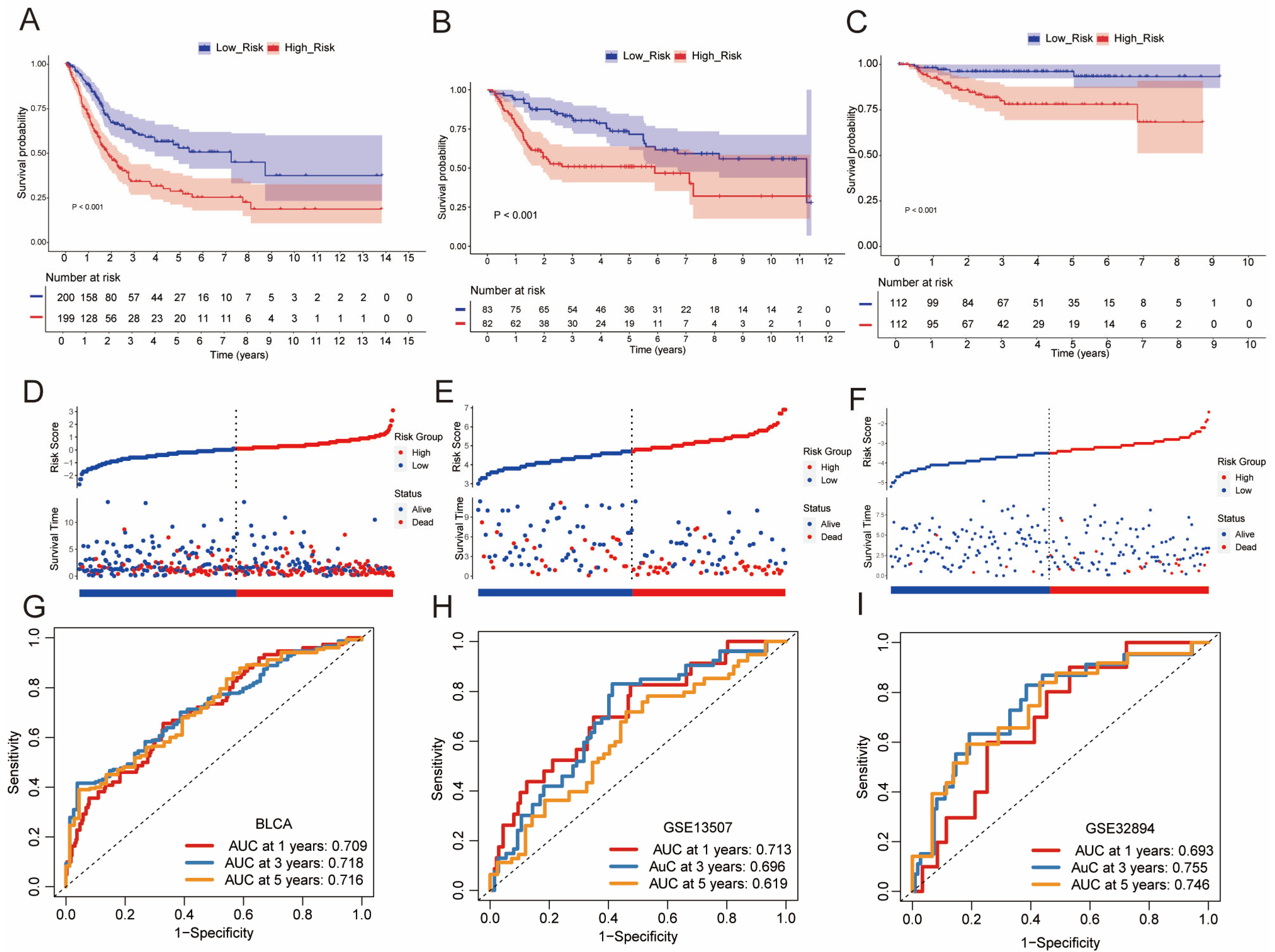
2.3. Construction of BCSCs Prognostic Signature and Risk Model
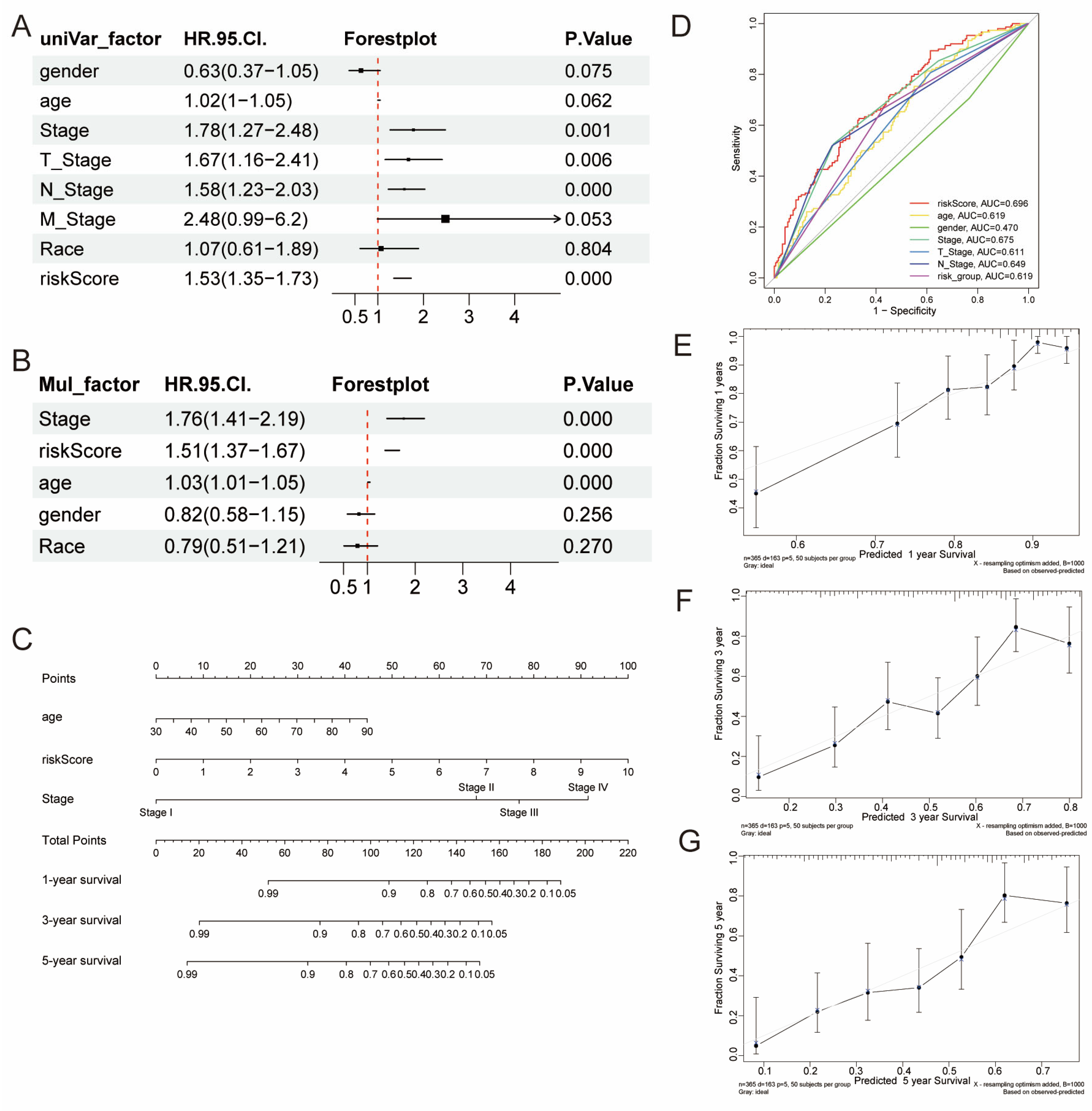
2.4. Development of a Nomogram Model
2.5. Gene Set Enrichment Analysis (GSEA) for Risk Groups in BLCA
2.6. Immune Infiltration Analysis and Gene Mutation Information
2.7. Single Cell RNA-Seq (scRNA) Analysis for GSE222315
2.8. Sensitivity of Antitumor Drugs
2.9. Statistical Analysis
3. Results
3.1. Expression of BCSCs in the TCGA-BLCA Cohort
3.2. Development and Validation of a CSC-Based Signature in BLCA Cohort
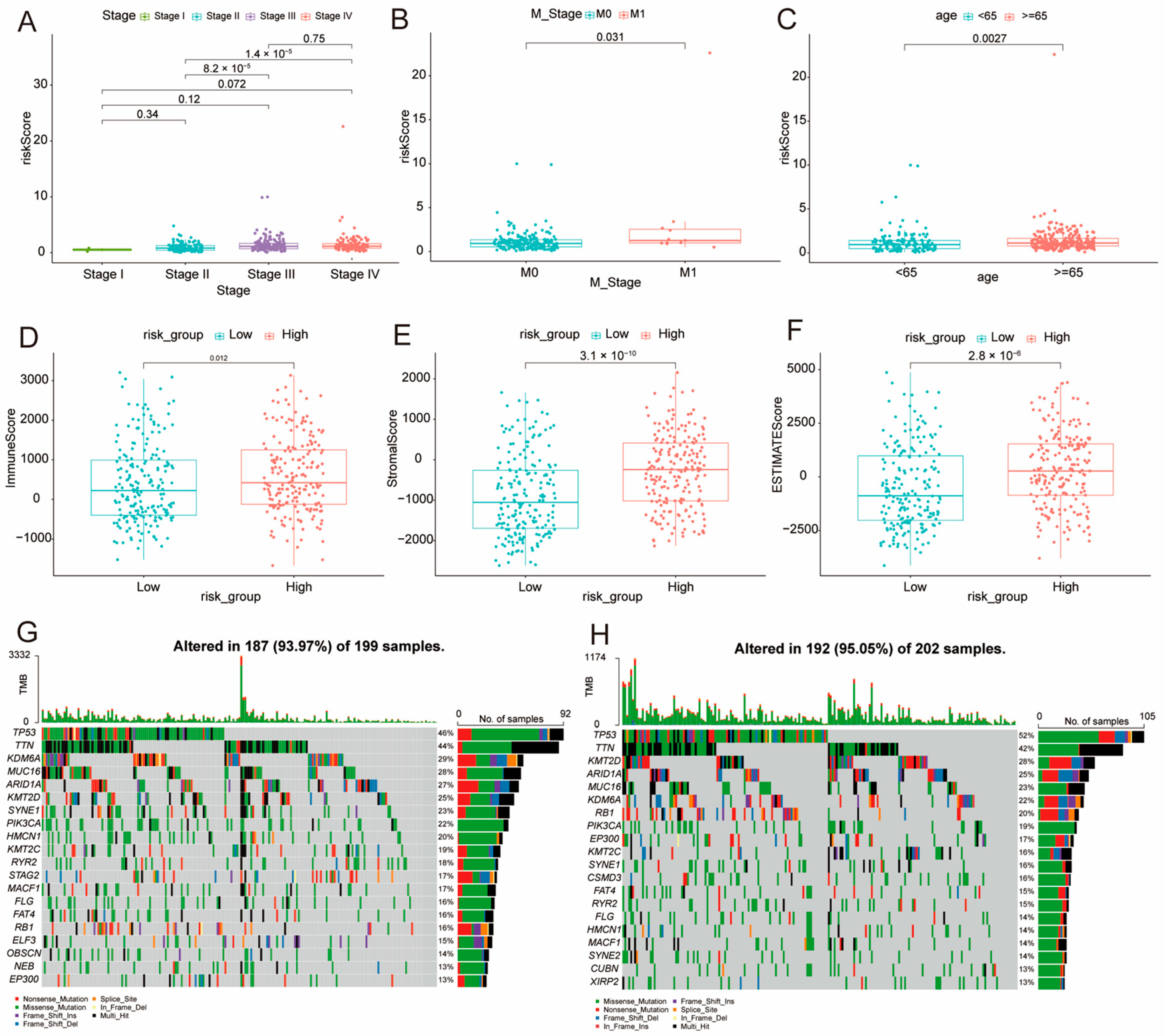
3.3. Clinical Relevance of DE-BCSCs
3.4. Development of a Nomogram
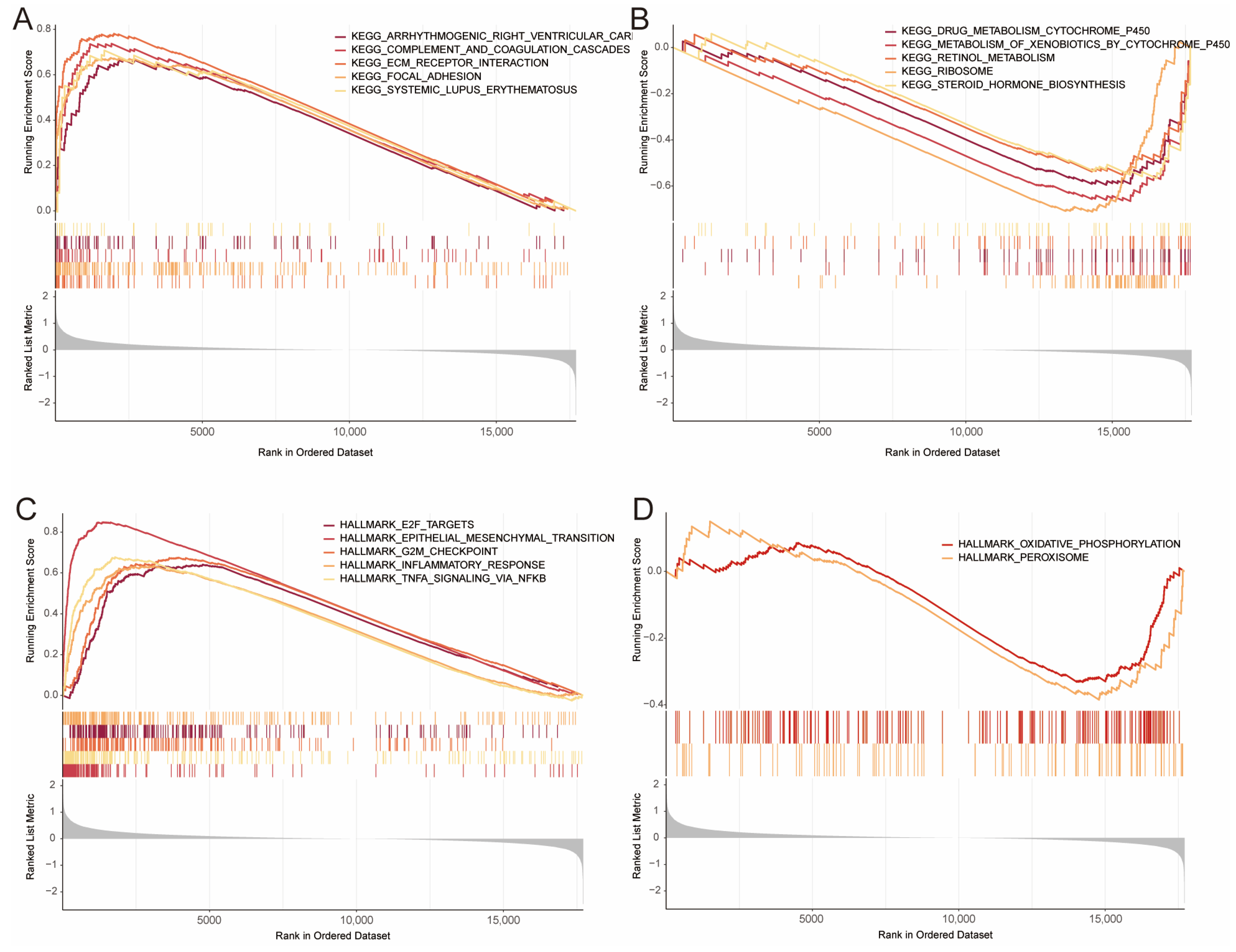
3.5. Gene Set Enrichment Analysis (GSEA)
3.6. Association of Risk Score with Clinical Factors
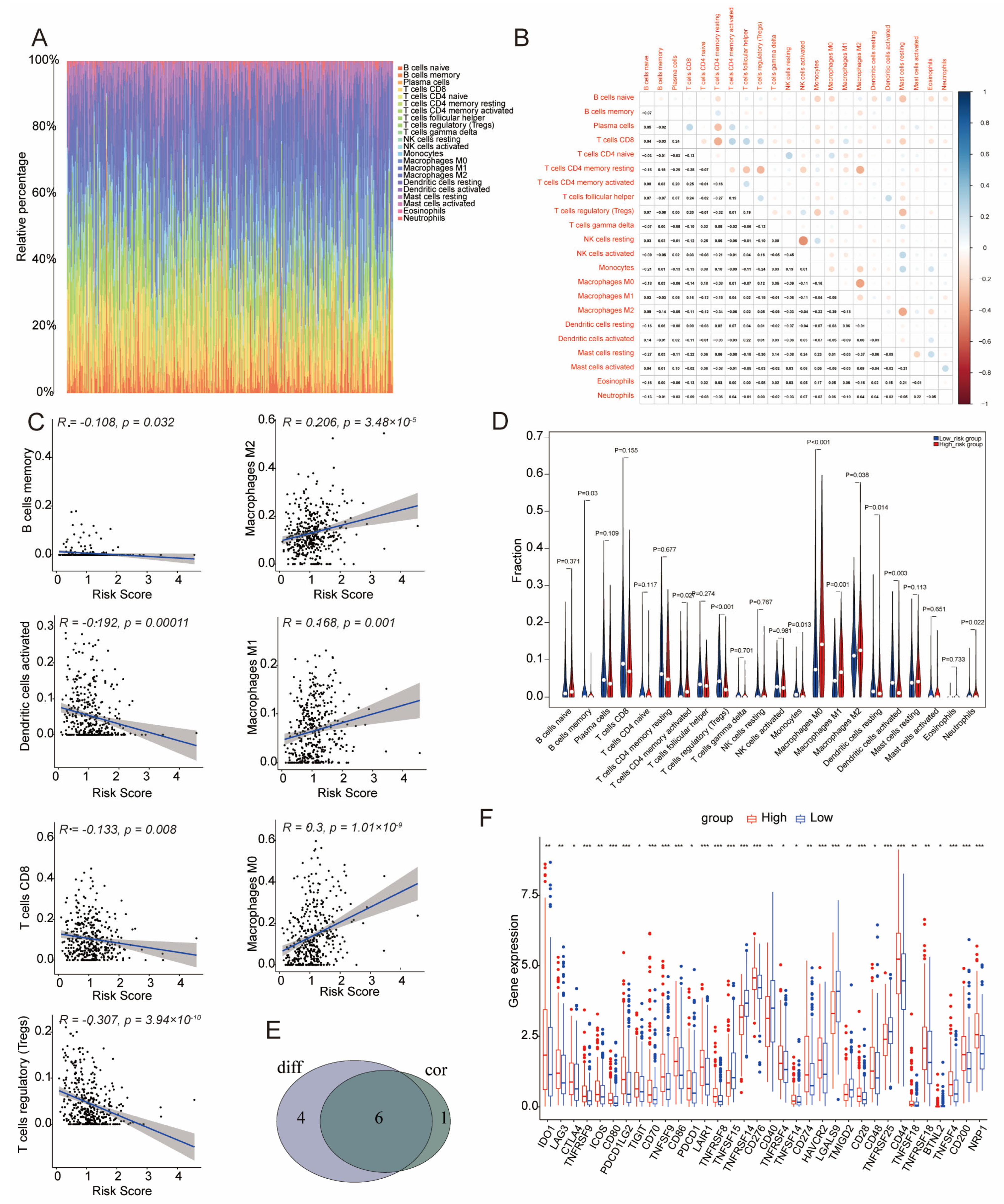
3.7. Immune Infiltration in Low- and High-Risk Groups
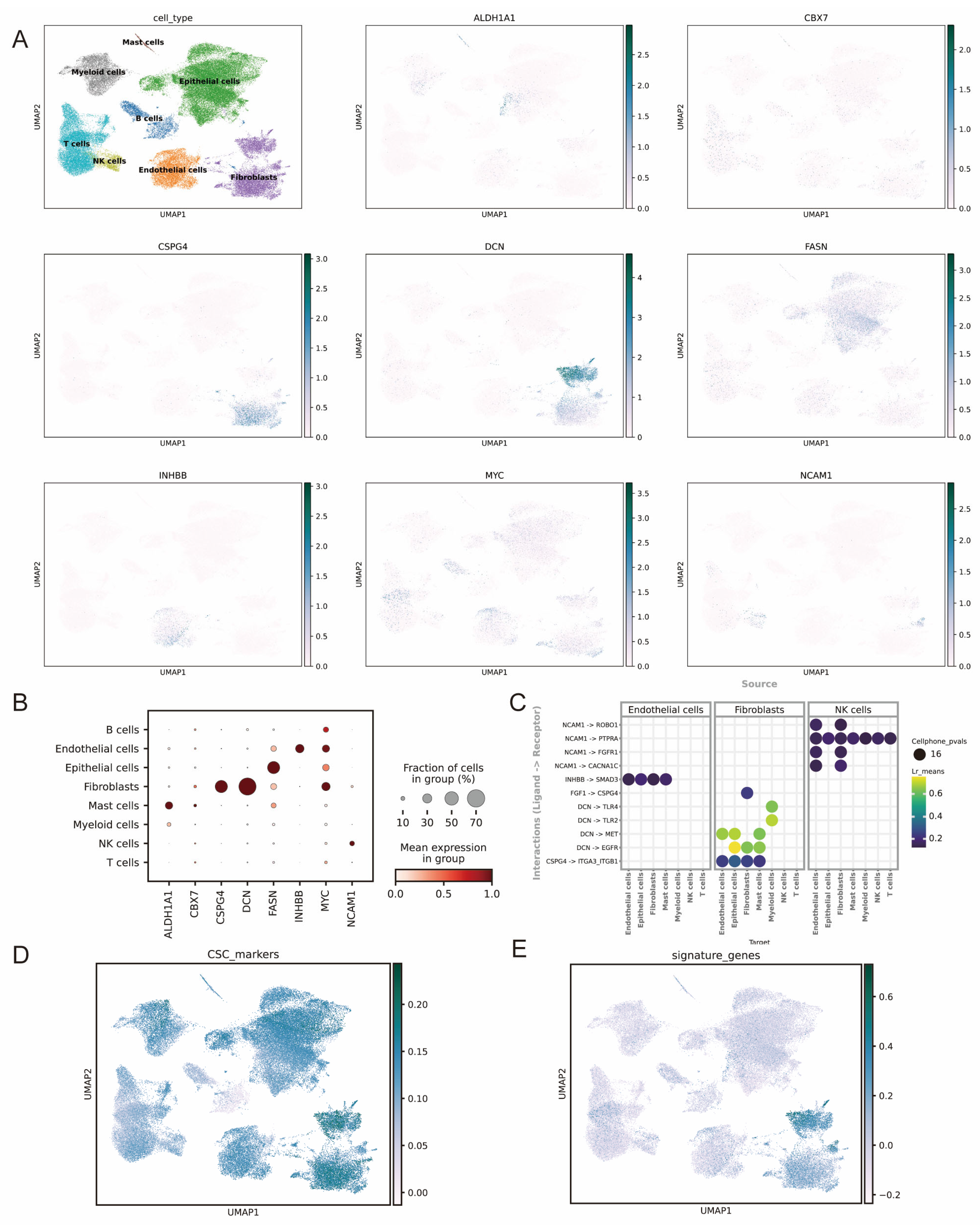
3.8. ScRNA-Seq Analysis of the Signature Genes
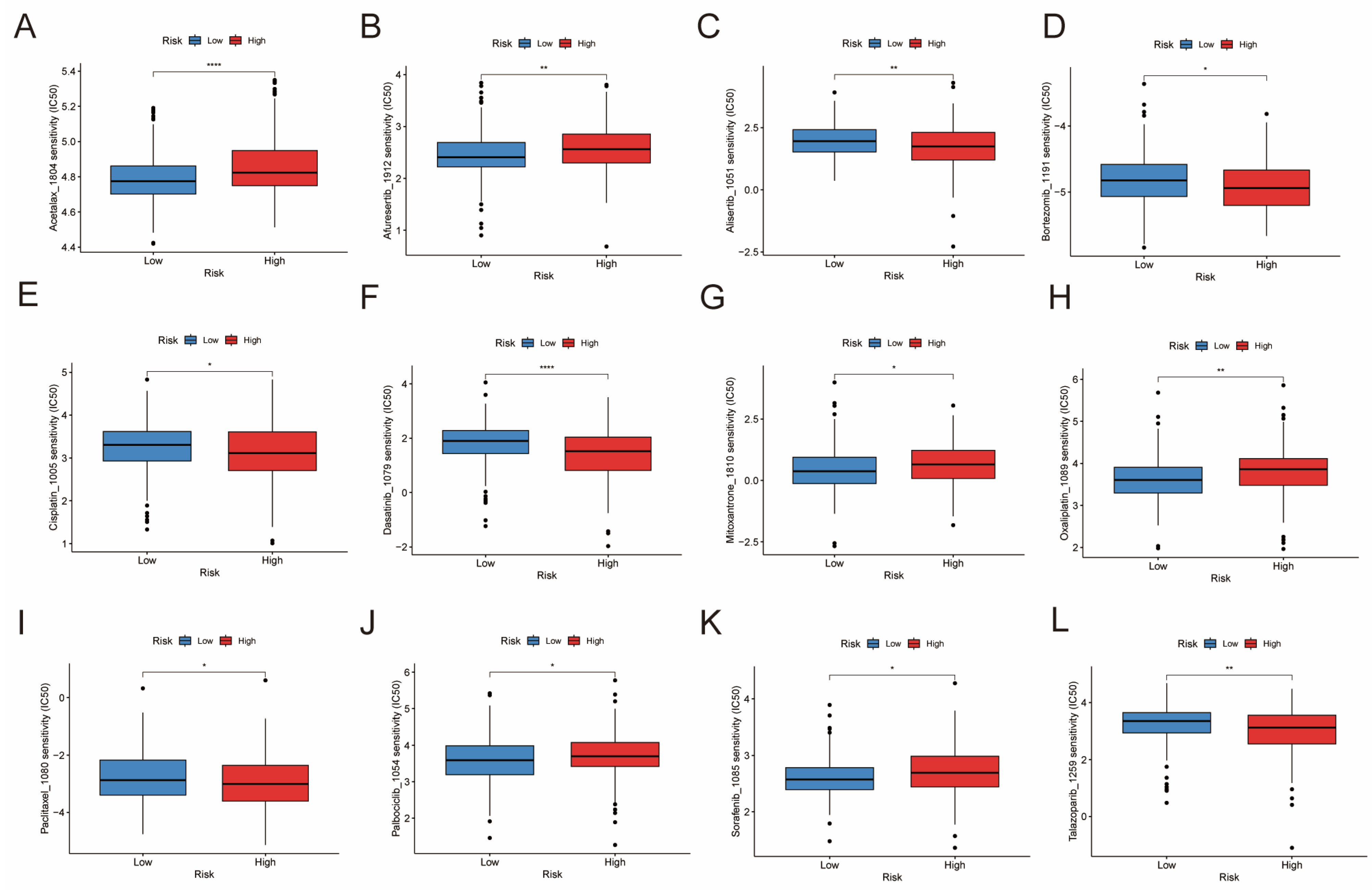
3.9. Drug Sensitivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CSCs | Cancer stem cells |
| BCSCs | Biomarkers of CSCs |
| DE-BCSCs | Differentially expressed BCSCs |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| scRNA-seq | single-cell RNA sequencing |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| OS | Overall Survival |
| TME | Tumor microenvironment |
| BLCA | Bladder cancer |
| EMT | Epithelial–Mesenchymal Transition |
| UMAP | Uniform Manifold Approximation and Projection |
| GSEA | Gene Set Enrichment Analysis |
| LIANA | Ligand–receptor ANalysis frAmework |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Lobo, N.; Afferi, L.; Moschini, M.; Mostafid, H.; Porten, S.; Psutka, S.P.; Gupta, S.; Smith, A.B.; Williams, S.B.; Lotan, Y. Epidemiology, Screening, and Prevention of Bladder Cancer. Eur. Urol. Oncol. 2022, 5, 628–639. [Google Scholar] [CrossRef]
- Zhao, J.; Li, J.; Zhang, R. Off the Fog to Find the Optimal Choice: Research Advances in Biomarkers for Early Diagnosis and Recurrence Monitoring of Bladder Cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188926. [Google Scholar] [CrossRef] [PubMed]
- Maas, M.; Todenhöfer, T.; Black, P.C. Urine Biomarkers in Bladder Cancer—Current Status and Future Perspectives. Nat. Rev. Urol. 2023, 20, 597–614. [Google Scholar] [CrossRef]
- Bazarkin, A.; Morozov, A.; Androsov, A.; Fajkovic, H.; Rivas, J.G.; Singla, N.; Koroleva, S.; Teoh, J.Y.-C.; Zvyagin, A.V.; Shariat, S.F.; et al. Assessment of Prostate and Bladder Cancer Genomic Biomarkers Using Artificial Intelligence: A Systematic Review. Curr. Urol. Rep. 2023, 25, 19–35. [Google Scholar] [CrossRef]
- Easwaran, H.; Tsai, H.-C.; Baylin, S.B. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.S.; Espinosa, I.; Chao, M.; Wong, D.; Ailles, L.; Diehn, M.; Gill, H.; Presti, J., Jr.; Chang, H.Y.; van de Rijn, M.; et al. Identification, Molecular Characterization, Clinical Prognosis, and Therapeutic Targeting of Human Bladder Tumor-Initiating Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 14016–14021. [Google Scholar] [CrossRef]
- Li, Y.; Lin, K.; Yang, Z.; Han, N.; Quan, X.; Guo, X.; Li, C. Bladder Cancer Stem Cells: Clonal Origin and Therapeutic Per-spectives. Oncotarget 2017, 8, 66668–66679. [Google Scholar] [CrossRef]
- Garg, M. Urothelial Cancer Stem Cells and Epithelial Plasticity: Current Concepts and Therapeutic Implications in Bladder Cancer. Cancer Metastasis Rev. 2015, 34, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, C.; Fan, Z.; Liu, H.; Zhang, X.; Cai, Z.; Xu, L.; Luo, J.; Huang, Y.; He, L.; et al. Single-Cell Sequencing Reveals Variants in ARID1A, GPRC5A and MLL2 Driving Self-Renewal of Human Bladder Cancer Stem Cells. Eur. Urol. 2017, 71, 8–12. [Google Scholar] [CrossRef]
- Zheng, H.; An, M.; Luo, Y.; Diao, X.; Zhong, W.; Pang, M.; Lin, Y.; Chen, J.; Li, Y.; Kong, Y.; et al. PDGFRα+ITGA11+ Fi-broblasts Foster Early-Stage Cancer Lymphovascular Invasion and Lymphatic Metastasis via ITGA11-SELE Interplay. Cancer Cell 2024, 42, 682–700.e12. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Firdous, S.; Ghosh, A.; Saha, S. BCSCdb: A Database of Biomarkers of Cancer Stem Cells. Database 2022, 2022, baac082. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A Class Discovery Tool with Confidence Assessments and Item Tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Blanche, P.; Dartigues, J.F.; Jacqmin-Gadda, H. Estimating and Comparing Time-Dependent Areas under Receiver Operating Characteristic Curves for Censored Event Times with Competing Risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture from Expression Data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- He, Y.; Jiang, Z.; Chen, C.; Wang, X. Classification of Triple-Negative Breast Cancers Based on Immunogenomic Profiling. J. Exp. Clin. Cancer Res. 2018, 37, 327. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An Updated Database of Manually Curated Cell Markers in Human/Mouse and Web Tools Based on scRNA-Seq Data. Nucleic Acids Res. 2022, 51, D870–D876. [Google Scholar] [CrossRef]
- Dimitrov, D.; Schäfer, P.S.L.; Farr, E.; Rodriguez-Mier, P.; Lobentanzer, S.; Badia-I-Mompel, P.; Dugourd, A.; Tanevski, J.; Ramirez Flores, R.O.; Saez-Rodriguez, J. LIANA+ provides an all-in-one framework for cell–cell communication inference. Nat. Cell Biol. 2024, 26, 1613–1622. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring Cell-Cell Communication from Combined Expression of Multi-Subunit Ligand-Receptor Complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R Package for Predicting In Vivo or Cancer Patient Drug Response and Biomarkers from Cell Line Screening Data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef]
- Hamid, A.R.A.H.; Syadza, Y.Z.; Yausep, O.E.; Christanto, R.B.I.; Muharia, B.H.R.; Mochtar, C.A. The Expression of Stem Cells Markers and Its Effects on the Propensity for Recurrence and Metastasis in Bladder Cancer: A Systematic Review. PLoS ONE 2023, 18, e0269214. [Google Scholar] [CrossRef]
- Tan, J.; Wang, Y.; Sun, L.; Xu, S.; Li, C.; Jin, X. The Origin and Evolution of Bladder Cancer Stem Cells. Front. Cell Dev. Biol. 2022, 10, 950241. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhu, X.; Wang, X.; Liao, S.; Jin, M.; Zhang, L.; Wu, X.; Zhao, T.; Zhang, J.; Lv, J.; et al. Identification and Validation of the Prognostic Stemness Biomarkers in Bladder Cancer Bone Metastasis. Front. Oncol. 2021, 11, 641184. [Google Scholar] [CrossRef] [PubMed]
- Namekawa, T.; Ikeda, K.; Horie-Inoue, K.; Suzuki, T.; Okamoto, K.; Ichikawa, T.; Yano, A.; Kawakami, S.; Inoue, S. ALDH1A1 in Patient-Derived Bladder Cancer Spheroids Activates Retinoic Acid Signaling Leading to TUBB3 Overexpression and Tumor Progression. Int. J. Cancer 2020, 146, 1099–1113. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Y.; Liang, Z.; Meng, Z.; Zhang, X.; Ma, G.; Chen, Y.; Zhang, M.; Su, Y.; Li, Z.; et al. Modification of Lysine-260 2-Hydroxyisobutyrylation Destabilizes ALDH1A1 Expression to Regulate Bladder Cancer Progression. iScience 2023, 26, 108142. [Google Scholar] [CrossRef]
- Qiu, Y.; Ye, W.; Wang, C.; Zang, J. Prognostic significance and immunoinfiltration analysis of genes associated with epithelial-mesenchymal transition and energy metabolism in bladder urothelial carcinoma. Aging 2023, 15, 13312–13328. [Google Scholar] [CrossRef]
- Song, Y.; Du, Y.; Qin, C.; Liang, H.; Yang, W.; Lin, J.; Ding, M.; Han, J.; Xu, T. Gemcitabine-Resistant Biomarkers in Bladder Cancer Are Associated with Tumor-Immune Microenvironment. Front. Cell Dev. Biol. 2022, 9, 809620. [Google Scholar] [CrossRef]
- Appunni, S.; Anand, V.; Khandelwal, M.; Gupta, N.; Rubens, M.; Sharma, A. Small Leucine Rich Proteoglycans (Decorin, Biglycan and Lumican) in Cancer. Clin. Chim. Acta 2019, 491, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Cao, J.; Li, J.; Yao, Z.; Han, D.; Ying, L.; Wang, Z.; Tian, J. Identification of Prognostic Biomarkers Associated with Stromal Cell Infiltration in Muscle-invasive Bladder Cancer by Bioinformatics Analyses. Cancer Med. 2020, 9, 7253–7267. [Google Scholar] [CrossRef]
- Wu, N.; Wang, J.; Fan, M.; Liang, Y.; Qi, X.W.; Deng, F.; Zeng, F. Non-Glycanated ΔDCN Isoform in Muscle Invasive Bladder Cancer Mediates Cancer Stemness and Gemcitabine Resistance. Cell. Oncol. 2024, 47, 2163–2181. [Google Scholar] [CrossRef]
- Jones, S.F.; Infante, J.R. Molecular Pathways: Fatty Acid Synthase. Clin. Cancer Res. 2015, 21, 5434–5438. [Google Scholar] [CrossRef]
- Peng, R.; Ma, X.; Jiang, Z.; Duan, Y.; Lv, S.; Jing, W. Integrative analysis of Anoikis-related genes reveals that FASN is a novel prognostic biomarker and promotes the malignancy of bladder cancer via Wnt/β-catenin pathway. Heliyon 2024, 10, e34029. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jia, Q.; Zou, Z.; Liu, X.; Li, X.; Chen, H.; Ma, H.; Chen, L. INHBB Promotes Tumor Aggressiveness and Stemness of Glioblastoma via Activating EGFR Signaling. Pathol.-Res. Pract. 2023, 245, 154460. [Google Scholar] [CrossRef]
- Li, H.; Bao, X.; Xiao, Y.; Yin, H.; Han, X.; Kang, S. Identification and Verification of Anoikis-Related Gene Markers to Predict the Prognosis of Patients with Bladder Cancer and Assist in the Diagnosis and Treatment of Bladder Cancer. Transl. Cancer Res. 2024, 13, 579–593. [Google Scholar] [CrossRef]
- Zhuang, C.; Huang, X.; Zhuang, C.; Luo, X.; Zhang, X.; Cai, Z.; Gui, Y. Synthetic Regulatory RNAs Selectively Suppress the Progression of Bladder Cancer. J. Exp. Clin. Cancer Res. 2017, 36, 151. [Google Scholar] [CrossRef]
- Wang, Y.; Ju, L.; Wang, G.; Qian, K.; Jin, W.; Li, M.; Yu, J.; Shi, Y.; Wang, Y.; Zhang, Y.; et al. DNA Polymerase POLD1 Promotes Proliferation and Metastasis of Bladder Cancer by Stabilizing MYC. Nat. Commun. 2023, 14, 2421. [Google Scholar] [CrossRef]
- Wang, H.; Mei, Y.; Luo, C.; Huang, Q.; Wang, Z.; Lu, G.-M.; Qin, L.; Sun, Z.; Huang, C.-W.; Yang, Z.-W.; et al. Single-Cell Analyses Reveal Mechanisms of Cancer Stem Cell Maintenance and Epithelial–Mesenchymal Transition in Recurrent Bladder Cancer. Clin. Cancer Res. 2021, 27, 6265–6278. [Google Scholar] [CrossRef]
- Huang, Z.; Yan, Y.; Zhu, Z.; Liu, J.; He, X.; Dalangood, S.; Li, M.; Tan, M.; Cai, J.; Tang, P.; et al. CBX7 Suppresses Urinary Bladder Cancer Progression via Modulating AKR1B10–ERK Signaling. Cell Death Dis. 2021, 12, 537. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Su, H.; Liu, Z.; Wang, Y.; Lu, Z.; Cheng, S. Integrative Analysis of the Bladder Cancer from a Cell Cycle NCAM1 Perspective at Both Single Cell and Bulk Resolution. Environ. Toxicol. 2024, 40, 445–458. [Google Scholar] [CrossRef]
- Aljagthmi, W.A.; Alasmari, M.A.; Daghestani, M.H.; Al-Kharashi, L.A.; Al-Mohanna, F.H.; Aboussekhra, A. Decorin (DCN) Downregulation Activates Breast Stromal Fibroblasts and Promotes Their Pro-Carcinogenic Effects through the IL-6/STAT3/AUF1 Signaling. Cells 2024, 13, 680. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xia, Q.; Rao, Q.; Shi, S.; Shi, Q.; Ma, H.; Lu, Z.; Chen, H.; Zhou, X. DCN Deficiency Promotes Renal Cell Carcinoma Growth and Metastasis through Downregulation of P21 and E-Cadherin. Tumor Biol. 2016, 37, 5171–5183. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Yang, H.-L.; Zhou, W.-J.; Lai, Z.-Z.; Qiu, X.-M.; Fu, Q.; Zhao, J.-Y.; Wang, J.; Li, D.-J.; Li, M.-Q. Rapamycin Prevents Spontaneous Abortion by Triggering Decidual Stromal Cell Autophagy-Mediated NK Cell Residence. Autophagy 2020, 17, 2511–2527. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Niu, C.; Li, Y.; Gao, B.; Zheng, J.; Guo, X.; Ma, W. Fatty Acid Synthase Expression and Esophageal Cancer. Mol. Biol. Rep. 2012, 39, 9733–9739. [Google Scholar] [CrossRef]
- Chen, K.; Yong, J.; Zauner, R.; Wally, V.; Whitelock, J.; Sajinovic, M.; Kopecki, Z.; Liang, K.; Scott, K.F.; Mellick, A.S. Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma. Cancers 2022, 14, 5564. [Google Scholar] [CrossRef]
- Farrell, A.S.; Joly, M.M.; Allen-Petersen, B.L.; Worth, P.J.; Lanciault, C.; Sauer, D.; Link, J.; Pelz, C.; Heiser, L.M.; Morton, J.P.; et al. MYC Regulates Ductal-Neuroendocrine Lineage Plasticity in Pancreatic Ductal Adenocarcinoma Associated with Poor Outcome and Chemoresistance. Nat. Commun. 2017, 8, 1728. [Google Scholar] [CrossRef]
- Na, Y.; Choi, J.-W.; Kasala, D.; Hong, J.; Oh, E.; Li, Y.; Jung, S.-J.; Kim, S.W.; Yun, C.-O. Potent Antitumor Effect of Neurotensin Receptor-Targeted Oncolytic Adenovirus Co-Expressing Decorin and Wnt Antagonist in an Orthotopic Pancreatic Tumor Model. J. Control. Release 2015, 220, 766–782. [Google Scholar] [CrossRef]
- Macaulay, A.R.K.; Yang, J.; Price, M.A.; Forster, C.L.; Riddle, M.J.; Ebens, C.L.; Albert, F.W.; Giubellino, A.; McCarthy, J.B.; Tolar, J. Chondroitin Sulfate Proteoglycan 4 Increases Invasion of Recessive Dystrophic Epidermolysis Bullosa-Associated Cutaneous Squamous Cell Carcinoma by Modifying Transforming Growth Factor-β Signalling. Br. J. Dermatol. 2024, 192, 104–117. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A. Macrophages, Innate Immunity and Cancer: Balance, Tolerance, and Diversity. Curr. Opin. Immunol. 2010, 22, 231–237. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage Plasticity and Polarization: In Vivo Veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Steiner, R.E.; Parra, E.R.; Vega, F.; Feng, L.; Westin, J.R.; Neelapu, S.S.; Strati, P.; Green, M.R.; Flowers, C.R.; Solis, L.M.; et al. PD-L1+ Macrophages Are Associated with Favorable Features in Primary Mediastinal (Thymic) Large B-Cell Lymphoma. Exp. Hematol. Oncol. 2023, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Orian-Rousseau, V. CD44, a Therapeutic Target for Metastasising Tumours. Eur. J. Cancer 2010, 46, 1271–1277. [Google Scholar] [CrossRef]
- Leblond, M.M.; Zdimerova, H.; Desponds, E.; Verdeil, G. Tumor-Associated Macrophages in Bladder Cancer: Biological Role, Impact on Therapeutic Response and Perspectives for Immunotherapy. Cancers 2021, 13, 4712. [Google Scholar] [CrossRef] [PubMed]
- Dyck, L.; Mills, K.H.G. Immune Checkpoints and Their Inhibition in Cancer and Infectious Diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Meli, V.S.; Rowley, A.T.; Veerasubramanian, P.K.; Heedy, S.E.; Liu, W.F.; Wang, S.-W. Modulation of Stiffness-Dependent Macrophage Inflammatory Responses by Collagen Deposition. ACS Biomater. Sci. Eng. 2024, 10, 2212–2223. [Google Scholar] [CrossRef]
- Kim, Y.K.; Chu, S.; Hsieh, J.Y.; Kamoku, C.M.; Tenner, A.J.; Liu, W.F.; Wang, S. Incorporation of a Ligand Peptide for Immune Inhibitory Receptor LAIR-1 on Biomaterial Surfaces Inhibits Macrophage Inflammatory Responses. Adv. Health Mater. 2017, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Singla, A.K.; Garg, A.; Aggarwal, D. Paclitaxel and Its Formulations. Int. J. Pharm. 2002, 235, 179–192. [Google Scholar] [CrossRef]
- Romani, A.M. Cisplatin in Cancer Treatment. Biochem. Pharmacol. 2022, 206, 115323. [Google Scholar] [CrossRef]
- Chen, X.; Yin, Y.; He, Y.; Meng, F.; Zhao, J.; Liu, F.; Xu, Y.; Wang, G.; Zhu, X.; Ma, S.; et al. The Prognostic Significance and Clinical Relevance of Stem Cell Characteristic in Bladder Cancer. Heliyon 2024, 10, e24858. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Liu, W. Integrative Analysis of Biomarkers for Cancer Stem Cells in Bladder Cancer and Their Therapeutic Potential. Genes 2025, 16, 1146. https://doi.org/10.3390/genes16101146
Wu J, Liu W. Integrative Analysis of Biomarkers for Cancer Stem Cells in Bladder Cancer and Their Therapeutic Potential. Genes. 2025; 16(10):1146. https://doi.org/10.3390/genes16101146
Chicago/Turabian StyleWu, Jing, and Wei Liu. 2025. "Integrative Analysis of Biomarkers for Cancer Stem Cells in Bladder Cancer and Their Therapeutic Potential" Genes 16, no. 10: 1146. https://doi.org/10.3390/genes16101146
APA StyleWu, J., & Liu, W. (2025). Integrative Analysis of Biomarkers for Cancer Stem Cells in Bladder Cancer and Their Therapeutic Potential. Genes, 16(10), 1146. https://doi.org/10.3390/genes16101146




