Objective Assessments of Smoking and Drinking Outperform Clinical Phenotypes in Predicting Variance in Epigenetic Aging
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Methylation Analyses
2.3. Statistical Analyses
3. Results
3.1. Correlations
3.2. Multiple Regression Models Predicting PC-GrimAge Acceleration
3.3. Multiple Regression Models Predicting PACE
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rodgers, J.L.; Jones, S.I.J.; Bolleddu, S.; Vanthenapalli, L.E.; Rodgers, K.; Karia, S.K.; Panguluri, S.K. Cardiovascular risks associated with gender and aging. J. Cardiovasc. Dev. Dis. 2019, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Oblak, L.; van der, Z.J.; Higgins-Chen, A.T.; Levine, M.E.; Boks, M.P. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res. Rev. 2021, 69, 101348. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, C.; Pirazzini, C.; Sala, C.; Sambati, L.; Yusipov, I.; Kalyakulina, A.; Ravaioli, F.; Kwiatkowska, K.M.; Durso, D.F.; Ivanchenko, M.; et al. A meta-analysis of brain DNA methylation across sex, age, and alzheimer’s disease points for accelerated epigenetic aging in neurodegeneration. Front. Aging Neurosci. 2021, 13, 639428. [Google Scholar] [CrossRef] [PubMed]
- Kresovich, J.K.; Martinez Lopez, A.M.; Garval, E.L.; Xu, Z.; White, A.J.; Sandler, D.P.; Taylor, J.A. Alcohol consumption and methylation-based measures of biological age. J. Gerontol. Ser. A 2021, 76, 2107–2111. [Google Scholar] [CrossRef] [PubMed]
- Nannini, D.R.; Joyce, B.T.; Zheng, Y.; Gao, T.; Wang, J.; Liu, L.; Jacobs, D.R., Jr.; Schreiner, P.J.; Liu, C.; Dai, Q. Alcohol consumption and epigenetic age acceleration in young adults. Aging 2023, 15, 371. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Y.; Lai, M.; Nannini, D.R.; Hou, L.; Joehanes, R.; Huan, T.; Levy, D.; Ma, J.; Liu, C. Alcohol consumption and epigenetic age acceleration across human adulthood. Aging 2023, 15, 10938–10971. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.D.; Robertson, K.D.; Hlady, R.A.; Muench, C.; Lee, J.; Philibert, R.; Horvath, S.; Kaminsky, Z.A.; Lohoff, F.W. DNA methylation age is accelerated in alcohol dependence. Transl. Psychiatry 2018, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Y.; Breitling, L.P.; Brenner, H. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget 2016, 7, 46878–46889. [Google Scholar] [CrossRef]
- Cardenas, A.; Ecker, S.; Fadadu, R.P.; Huen, K.; Orozco, A.; McEwen, L.M.; Engelbrecht, H.-R.; Gladish, N.; Kobor, M.S.; Rosero-Bixby, L. Epigenome-wide association study and epigenetic age acceleration associated with cigarette smoking among costa rican adults. Sci. Rep. 2022, 12, 4277. [Google Scholar] [CrossRef]
- Jatlow, P.; Toll, B.A.; Leary, V.; Krishnan-Sarin, S.; O’Malley, S.S. Comparison of expired carbon monoxide and plasma cotinine as markers of cigarette abstinence. Drug Alcohol. Depend. 2008, 98, 203–209. [Google Scholar] [CrossRef]
- Shipton, D.; Tappin, D.M.; Vadiveloo, T.; Crossley, J.A.; Aitken, D.A.; Chalmers, J. Reliability of self reported smoking status by pregnant women for estimating smoking prevalence: A retrospective, cross sectional study. BMJ 2009, 339, b4347. [Google Scholar] [CrossRef] [PubMed]
- Grüner Nielsen, D.; Andersen, K.; Nielsen, A.S.; Juhl, C.; Mellentin, A. Consistency between self-reported alcohol consumption and biological markers among patients with alcohol use disorder—A systematic review. Neurosci. Biobehav. Rev. 2021, 124, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Wetterling, T.; Dibbelt, L.; Wetterling, G.; Göder, R.; Wurst, F.; Margraf, M.; Junghanns, K. Ethyl glucuronide (etg): Better than breathalyser or self-reports to detect covert short-term relapses into drinking. Alcohol. Alcohol. 2014, 49, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.-K.; Gibbons, F.X.; Gerrard, M.; Beach, S.R.H.; Dawes, K.; Philibert, R. Digital methylation assessments of alcohol and cigarette consumption account for common variance in accelerated epigenetic ageing. Epigenetics 2022, 17, 1991–2005. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.; Milefchik, E.; Papworth, E.; Penaluna, B.; Dawes, K.; Moody, J.; Weeks, G.; Froehlich, E.; deBlois, K.; Long, J.D.; et al. Zscan25 methylation predicts seizures and severe alcohol withdrawal syndrome. Epigenetics 2024, 19, 2298057. [Google Scholar] [CrossRef] [PubMed]
- Dawes, K.; Andersen, A.; Reimer, R.; Mills, J.A.; Hoffman, E.; Long, J.D.; Miller, S.; Philibert, R. The relationship of smoking to cg05575921 methylation in blood and saliva DNA samples from several studies. Sci. Rep. 2021, 11, 21627. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Andersen, A.M.; Philibert, R.; Hancock, D.B. Epigenetic biomarkers for smoking cessation. Addict. Neurosci. 2023, 6, 100079. [Google Scholar] [CrossRef]
- Gelernter, J.; Polimanti, R. Genetics of substance use disorders in the era of big data. Nat. Rev. Genet. 2021, 22, 712–729. [Google Scholar] [CrossRef]
- Monick, M.M.; Beach, S.R.; Plume, J.; Sears, R.; Gerrard, M.; Brody, G.H.; Philibert, R.A. Coordinated changes in ahrr methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012, 159B, 141–151. [Google Scholar] [CrossRef]
- Miller, S.; Mills, J.A.; Long, J.; Philibert, R. A comparison of the predictive power of DNA methylation with carbohydrate deficient transferrin for heavy alcohol consumption. Epigenetics 2020, 16, 969–979. [Google Scholar] [CrossRef]
- Philibert, R.; Miller, S.; Noel, A.; Dawes, K.; Papworth, E.; Black, D.W.; Beach, S.R.H.; Long, J.D.; Mills, J.A.; Dogan, M. A four marker digital PCR toolkit for detecting heavy alcohol consumption and the effectiveness of its treatment. J. Insur. Med. 2019, 48, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Beach, S.R.; Ong, M.L.; Gibbons, F.X.; Gerrard, M.; Lei, M.-K.; Dawes, K.; Philibert, R.A. Epigenetic and proteomic biomarkers of elevated alcohol use predict epigenetic aging and cell-type variation better than self-report. Genes 2022, 13, 1888. [Google Scholar] [CrossRef] [PubMed]
- Philibert, R.A.; Beach, S.R.; Brody, G.H. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics 2012, 7, 1331–1338. [Google Scholar] [CrossRef] [PubMed]
- Dogan, M.V.; Beach, S.R.H.; Philibert, R.A. Genetically contextual effects of smoking on genome wide DNA methylation. Am. J. Med. Genet. Part. B Neuropsychiatr. Genet. 2017, 174, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Philibert, R.; Mills, J.A.; Long, J.D.; Salisbury, S.E.; Comellas, A.; Gerke, A.; Dawes, K.; Weg, M.V.; Hoffman, E.A. The reversion of cg05575921 methylation in smoking cessation: A potential tool for incentivizing healthy aging. Genes 2020, 11, 1415. [Google Scholar] [CrossRef] [PubMed]
- Tsaprouni, L.G.; Yang, T.-P.; Bell, J.; Dick, K.J.; Kanoni, S.; Nisbet, J.; Viñuela, A.; Grundberg, E.; Nelson, C.P.; Meduri, E.; et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 2020, 9, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Belsky, D.W.; Caspi, A.; Corcoran, D.L.; Sugden, K.; Poulton, R.; Arseneault, L.; Baccarelli, A.; Chamarti, K.; Gao, X.; Hannon, E. Dunedinpace, a DNA methylation biomarker of the pace of aging. Elife 2022, 11, e73420. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D. DNA methylation grimage strongly predicts lifespan and healthspan. Aging 2019, 11, 303. [Google Scholar] [CrossRef]
- Lu, A.T.; Binder, A.M.; Zhang, J.; Yan, Q.; Reiner, A.P.; Cox, S.R.; Corley, J.; Harris, S.E.; Kuo, P.L.; Moore, A.Z.; et al. DNA methylation grimage version 2. Aging 2022, 14, 9484–9549. [Google Scholar] [CrossRef]
- Beach, S.R.H.; Barton, A.W.; Lei, M.K.; Brody, G.H.; Kogan, S.M.; Hurt, T.R.; Fincham, F.D.; Stanley, S.M. The effect of communication change on long-term reductions in child exposure to conflict: Impact of the promoting strong african american families (prosaaf) program. Fam. Process 2014, 53, 580–595. [Google Scholar] [CrossRef]
- Faul, J.D.; Kim, J.K.; Levine, M.E.; Thyagarajan, B.; Weir, D.R.; Crimmins, E.M. Epigenetic-based age acceleration in a representative sample of older americans: Associations with aging-related morbidity and mortality. Proc. Natl. Acad. Sci. USA 2023, 120, e2215840120. [Google Scholar] [CrossRef] [PubMed]
- Philibert, R.; Dogan, M.; Beach, S.R.H.; Mills, J.A.; Long, J.D. Ahrr methylation predicts smoking status and smoking intensity in both saliva and blood DNA. Am. J. Genet. 2019, 183, 51–60. [Google Scholar] [CrossRef]
- Davis, S.; Bilke, S. An introduction to the methylumi package. Biocond. Package 2010, 10, B9. [Google Scholar]
- Wong, C.C.; Pidsley, R.; Schalkwyk, L.C. The Watermelon Package. 2013. Available online: https://bioconductor.org/packages/release/bioc/html/wateRmelon.html (accessed on 2 March 2023).
- Illumina. Infinium Human Methylationepic Array Product Files. Available online: https://bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylationEPICanno.ilm10b2.hg19.html (accessed on 2 March 2023).
- Dogan, M.V.; Xiang, J.; Beach, S.R.; Cutrona, C.; Gibbons, F.X.; Simons, R.L.; Brody, G.H.; Stapleton, J.T.; Philibert, R.A. Ethnicity and smoking-associated DNA methylation changes at hiv co-receptor gpr15. Front. Psychiatry 2015, 6, 132. [Google Scholar] [CrossRef] [PubMed]
- Meeks, K.A.C.; Henneman, P.; Venema, A.; Addo, J.; Bahendeka, S.; Burr, T.; Danquah, I.; Galbete, C.; Mannens, M.M.A.M.; Mockenhaupt, F.P.; et al. Epigenome-wide association study in whole blood on type 2 diabetes among sub-saharan african individuals: Findings from the rodam study. Int. J. Epidemiol. 2018, 48, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Nuotio, M.-L.; Pervjakova, N.; Joensuu, A.; Karhunen, V.; Hiekkalinna, T.; Milani, L.; Kettunen, J.; Järvelin, M.-R.; Jousilahti, P.; Metspalu, A.; et al. An epigenome-wide association study of metabolic syndrome and its components. Sci. Rep. 2020, 10, 20567. [Google Scholar] [CrossRef] [PubMed]
- Soriano-Tárraga, C.; Jiménez-Conde, J.; Giralt-Steinhauer, E.; Mola-Caminal, M.; Vivanco-Hidalgo, R.M.; Ois, A.; Rodríguez-Campello, A.; Cuadrado-Godia, E.; Sayols-Baixeras, S.; Elosua, R.; et al. Epigenome-wide association study identifies txnip gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum. Mol. Genet. 2015, 25, 609–619. [Google Scholar] [CrossRef]
- Dawes, K.; Sampson, L.; Reimer, R.; Miller, S.; Philibert, R.; Andersen, A. Epigenetic analyses of alcohol consumption in combustible and non-combustible nicotine product users. Epigenomes 2021, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. Akaike’s information criterion. In International Encyclopedia of Statistical Science; Springer: Berlin/Heidelberg, Germany, 2011; p. 25. [Google Scholar]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Mills, J.A.; Beach, S.R.; Dogan, M.; Simons, R.L.; Gibbons, F.X.; Long, J.D.; Philibert, R. A direct comparison of the relationship of epigenetic aging and epigenetic substance consumption markers to mortality in the framingham heart study. Genes 2019, 10, 51. [Google Scholar] [CrossRef]
- Marsh, O. Life cycle of a star: Carl sagan and the circulation of reputation. Br. J. Hist. Sci. 2019, 52, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Keenan, N.L.; Rosendorf, K.A. Prevalence of hypertension and controlled hypertension-united states, 2005–2008. Morb. Mortal. Wkly. Rep. 2011, 60, 94–97. [Google Scholar]
- Lu, Y.; Kwong, K.; Wells, J.; Edwards, A.; Chen, Z.; Tseng, T.-S.; Zhang, K. Quitting smoking after a cancer diagnosis is associated with high-risk neutrophil-to-lymphocyte ratio among tobacco use-related cancer survivors. Sci. Rep. 2023, 13, 2745. [Google Scholar] [CrossRef] [PubMed]

| Female | Male | |
|---|---|---|
| Sex | 164 | 114 |
| Age (chronological) | 46.1 ± 7.1 | 49.6 ± 9.3 |
| Physiologic Parameters | ||
| BMI ** | 35.4 ± 8.1 | 32 ± 8.2 |
| Systolic BP ** | 127 ± 21 mm Hg | 140 ± 24 mm Hg |
| Diastolic BP ** | 87 ± 12 mm Hg | 88 ± 14 mm Hg |
| Cholesterol | 182 ± 40 mg/dL | 179 ± 41 mg/dL |
| LDL ** | 106 ± 35 mg/dL | 102 ± 38 mg/dL |
| HDL ** | 55 ± 22 mg/dL | 49 ± 14 mg/dL |
| HbA1c ** | 5.9 ± 1.5% | 6.4 ± 1.8% |
| Triglycerides ** | 111 ± 60 mg/dL | 138 ± 79 mg/dL |
| Self-Reported Behaviors and Conditions | ||
| Smoking * | 25 (15%) | 34 (30%) |
| Binge Drinking * | 33 (20%) | 42 (37%) |
| Heart Disease | 15 (9%) | 17 (15%) |
| Hypertension | 89 (54%) | 64 (56%) |
| Diabetes | 29 (18%) | 25 (22%) |
| Arthritis | 50 (31%) | 24 (21%) |
| Cancer | 2 (1%) | 3 (3%) |
| Liver Disease | 3 (2%) | 1 (1%) |
| Kidney Disease | 6 (4%) | 5 (4%) |
| Cataracts | 7 (4%) | 8 (7%) |
| Exposure to Community Crime | ||
| Crime | 0.103 ± 0.25 | 0.174 ± 0.30 |
| Epigenetic Measures of Age and Aging | ||
| GrimAge | 52.8 ± 8.8 years | 58.9 ± 8.4 years |
| GrimAge2 | 59.0 ± 7.2 years | 64.4 ± 8.7 years |
| PCGrimAge | 60.1 ± 6.2 years | 66.2 ± 7.9 years |
| GrimAgeAcc * | 6.7 ± 7.7 years | 9.35 ± 5.7 years |
| GrimAge2Acc * | 13.0 ± 5.7 years | 14.9 ± 6.5 years |
| PCGrimAge Acc * | 14.1 ± 3.9 years | 16.6 ± 4.3 years |
| PACE | 1.07 ± 0.17 | 1.08 ± 0.14 |
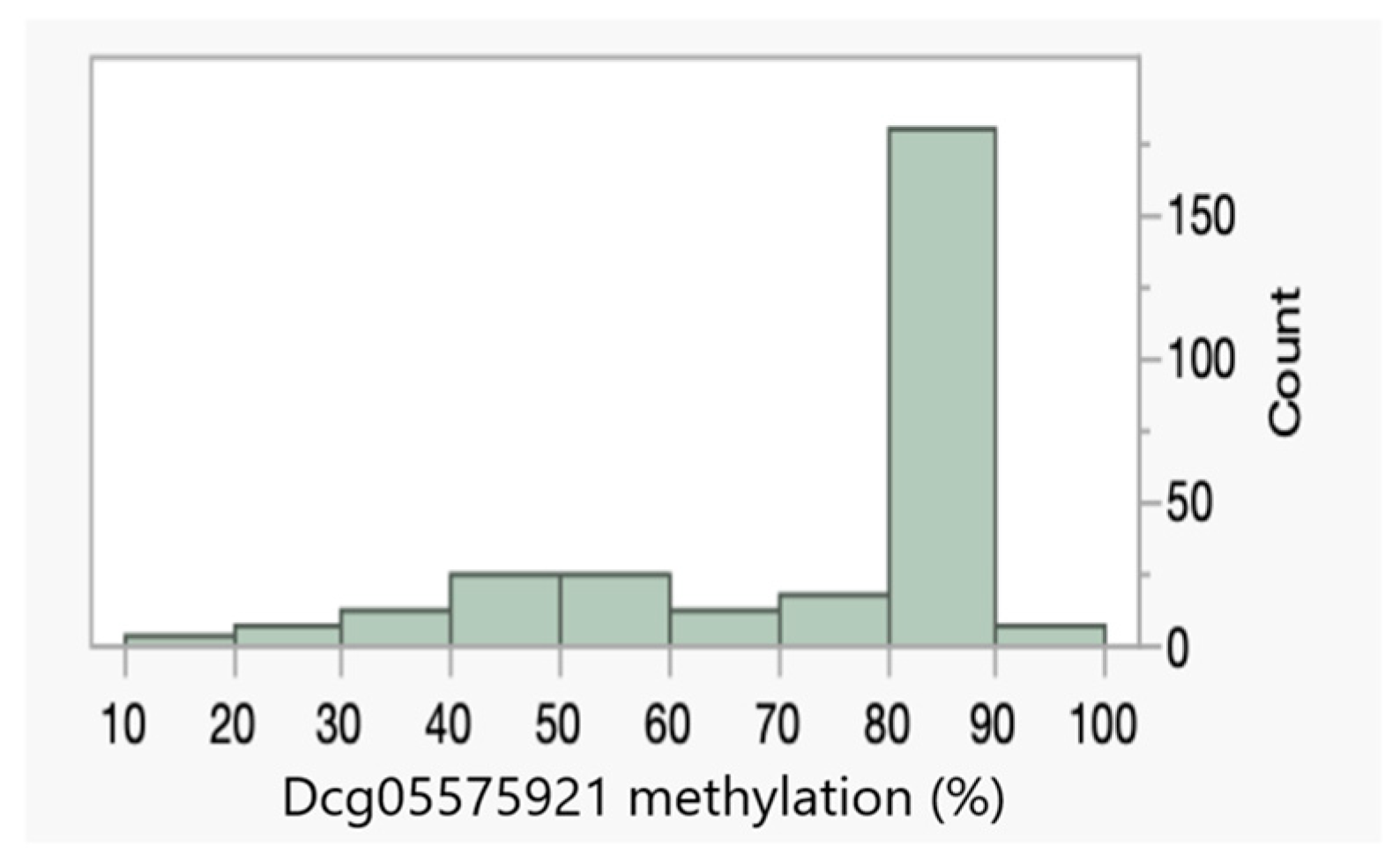
| Dcg05575921 | 79 ± 15% | 68 ± 21% |
| ATS | 1.5 ± 2.9 | 3.2 ± 3.7 |
| Cg19693031 | 78.3% | 73.1% |
| Adj. R2 | AIC | BIC | ||
|---|---|---|---|---|
| Demographic | Age | 0.168 | 1548 | 1555 |
| Sex | 0.085 | 1574 | 1581 | |
| Epigenetic | Dcg05575921 (Dcg055) | 0.459 | 1428 | 1435 |
| ATS | 0.285 | 1505 | 1513 | |
| cg19693031 | 0.037 | 1588 | 1596 | |
| Vitals | BMI | 0.015 | 1594 | 1602 |
| Systolic | 0.004 | 1598 | 1605 | |
| Diastolic | −0.003 | 1600 | 1607 | |
| Serum | HbA1c | −0.001 | 1599 | 1606 |
| Cholesterol | 0.004 | 1598 | 1604 | |
| LDL | 0.006 | 1597 | 1604 | |
| HDL | 0.005 | 1597 | 1604 | |
| Triglycerides | 0.015 | 1595 | 1601 | |
| Med History | Smoking | 0.257 | 1516 | 1524 |
| Binge Drinking | 0.055 | 1583 | 1591 | |
| Heart Disease | −0.002 | 1599 | 1606 | |
| Hypertension | 0.043 | 1587 | 1593 | |
| Diabetes | −0.003 | 1600 | 1607 | |
| Arthritis | 0.024 | 1592 | 1599 | |
| Cancer | −0.002 | 1599 | 1606 | |
| Liver Disease | 0.013 | 1595 | 1602 | |
| Kidney Disease | 0.004 | 1598 | 1605 | |
| Cataracts | 0.005 | 1597 | 1605 | |
| Crime | Crime | 0.025 | 1592 | 1599 |
| Model | ||||
| 1 | Age + Sex | 0.321 | 1492 | 1503 |
| 2 | Dcg055 + ATS | 0.486 | 1415 | 1426 |
| 3 | Age + Sex + Dcg055 + ATS | 0.744 | 1223 | 1241 |
| 4 | Model 3 + cg19693031 | 0.747 | 1221 | 1242 |
| 5 | Model 3 + BMI | 0.744 | 1224 | 1246 |
| 6 | Model 3 + Triglycerides | 0.745 | 1223 | 1245 |
| 7 | Model 3 + Smoking | 0.743 | 1225 | 1247 |
| 8 | Model 3 + Binge Drinking | 0.743 | 1225 | 1247 |
| 9 | Model 3 + Hypertension | 0.743 | 1225 | 1247 |
| 10 | Model 3 + Arthritis | 0.744 | 1225 | 1246 |
| 11 | Model 3 + Liver Disease | 0.743 | 1225 | 1247 |
| 12 | Model 3 + Crime | 0.748 | 1220 | 1242 |
| 13 | Model 3 + All Significant Predictors | 0.748 | 1216 | 1241 |
| 14 | Model 3 + All Significant Predictors + PACE | 0.752 | 1217 | 1246 |
| 15 | Model 3 + PACE | 0.744 | 1224 | 1246 |
| Adj. R2 | AIC | BIC | ||
|---|---|---|---|---|
| Demographic | Age | 0.022 | −248 | −241 |
| Sex | −0.002 | −241 | −234 | |
| Epigenetic | Dcg05575921 (Dcg055) | 0.036 | −252 | −245 |
| ATS | 0.103 | −272 | −265 | |
| cg19693031 | 0.003 | −243 | −235 | |
| Vitals | BMI | 0.043 | −254 | −247 |
| Systolic | 0.006 | −243 | −236 | |
| Diastolic | 0.003 | −242 | −235 | |
| Serum | HbA1c | 0.039 | −253 | −246 |
| Cholesterol | 0.008 | −244 | −237 | |
| LDL | −0.004 | −241 | −234 | |
| HDL | 0.079 | −265 | −257 | |
| Triglycerides | 0.007 | −244 | −236 | |
| Med History | Smoking | 0.014 | −246 | −239 |
| Binge Drinking | −0.000 | −242 | −234 | |
| Heart Disease | 0.047 | −255 | −248 | |
| Hypertension | 0.012 | −245 | −238 | |
| Diabetes | 0.022 | −248 | −241 | |
| Arthritis | 0.004 | −243 | −236 | |
| Cancer | −0.001 | −241 | −234 | |
| Liver Disease | −0.003 | −241 | −233 | |
| Kidney Disease | 0.006 | −243 | −236 | |
| Cataracts | −0.003 | −241 | −233 | |
| Crime | Crime | 0.003 | −243 | −235 |
| Model | ||||
| 1 | Age | 0.022 | −248 | −241 |
| 2 | Dcg055 + ATS | 0.100 | −270 | −259 |
| 3 | Age + Dcg055 + ATS | 0.104 | −270 | −256 |
| 4 | Model 3 + BMI | 0.207 | −303 | −285 |
| 5 | Model 3 + HbA1c | 0.129 | −277 | −259 |
| 6 | Model 3 + HDL | 0.179 | −294 | −276 |
| 7 | Model 3 + Smoking | 0.101 | −268 | −250 |
| 8 | Model 3 + Heart Disease | 0.126 | −276 | −258 |
| 9 | Model 3 + Hypertension | 0.111 | −271 | −253 |
| 10 | Model 3 + Diabetes | 0.116 | −273 | −255 |
| 11 | Model 3 + All Significant Predictors * | 0.255 | −318 | −285 |
| 12 | Model 3 + All Significant + PCGrimAgeAcc | 0.258 | −317 | −280 |
| 13 | Model 3 + PCGrimAgeAcc | 0.118 | −269 | −250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Philibert, R.; Lei, M.-K.; Ong, M.L.; Beach, S.R.H. Objective Assessments of Smoking and Drinking Outperform Clinical Phenotypes in Predicting Variance in Epigenetic Aging. Genes 2024, 15, 869. https://doi.org/10.3390/genes15070869
Philibert R, Lei M-K, Ong ML, Beach SRH. Objective Assessments of Smoking and Drinking Outperform Clinical Phenotypes in Predicting Variance in Epigenetic Aging. Genes. 2024; 15(7):869. https://doi.org/10.3390/genes15070869
Chicago/Turabian StylePhilibert, Robert, Man-Kit Lei, Mei Ling Ong, and Steven R. H. Beach. 2024. "Objective Assessments of Smoking and Drinking Outperform Clinical Phenotypes in Predicting Variance in Epigenetic Aging" Genes 15, no. 7: 869. https://doi.org/10.3390/genes15070869
APA StylePhilibert, R., Lei, M.-K., Ong, M. L., & Beach, S. R. H. (2024). Objective Assessments of Smoking and Drinking Outperform Clinical Phenotypes in Predicting Variance in Epigenetic Aging. Genes, 15(7), 869. https://doi.org/10.3390/genes15070869






