Identification and Expression Analysis of UPS Gene Family in Potato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growing Conditions
2.2. Identification of Potato UPS Family Members
2.3. Physicochemical Properties, Transmembrane Structure, and Subcellular Localization of Potato UPS Proteins
2.4. Phylogenetic Analysis of UPS Proteins
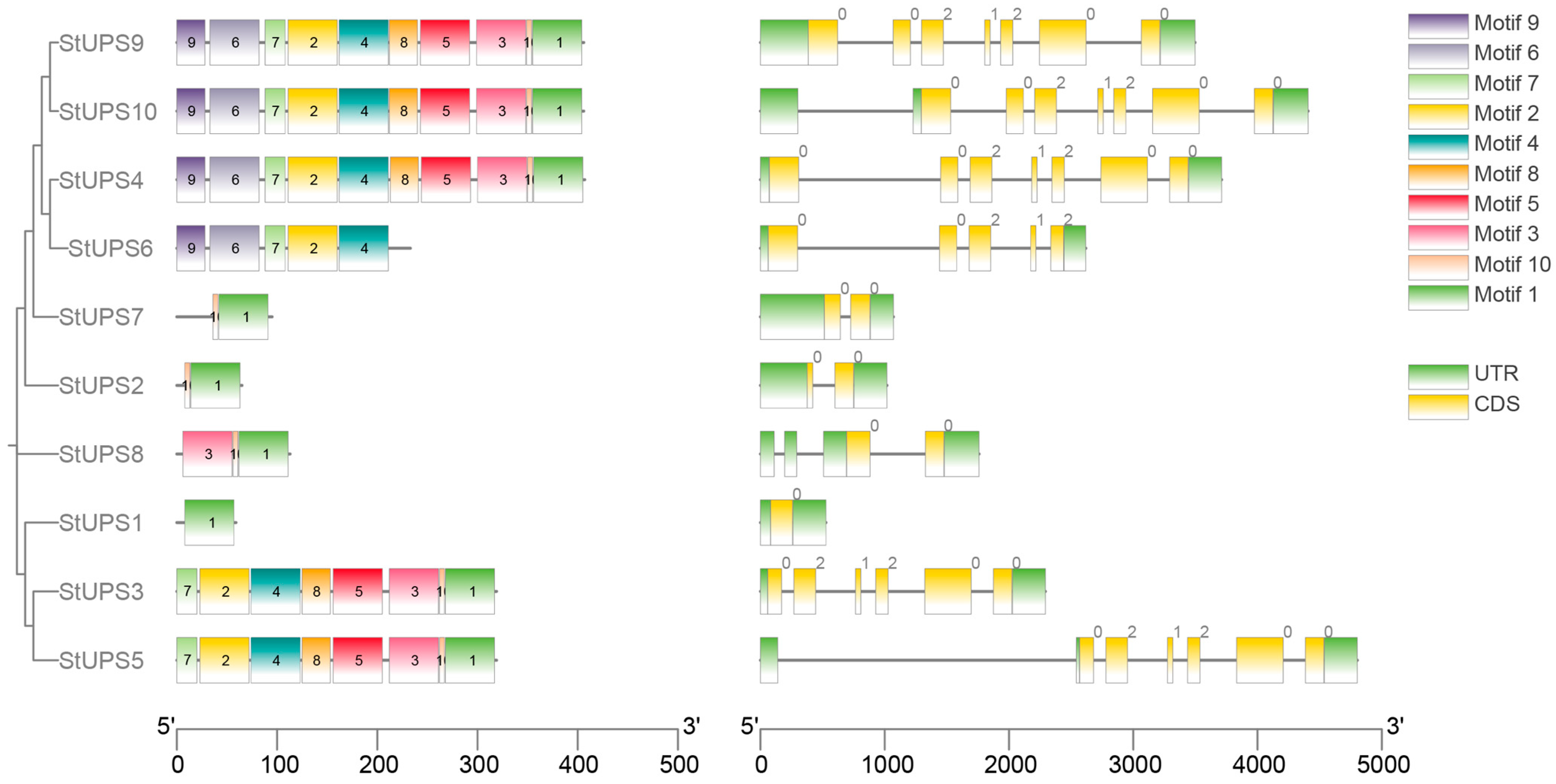
2.5. Analysis of UPS Motifs and Gene Structures in Potato
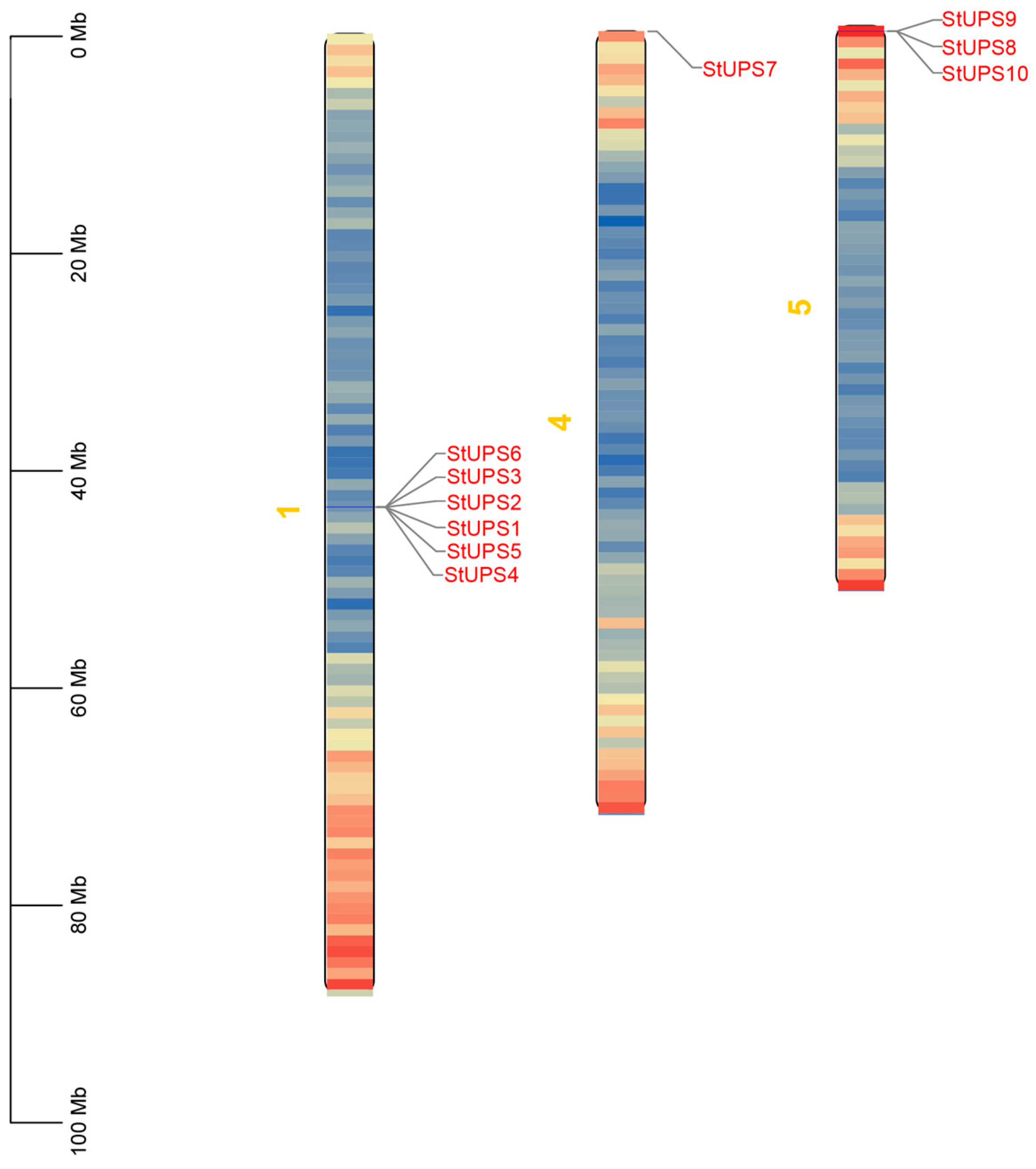
2.6. Analysis of Chromosomal Location
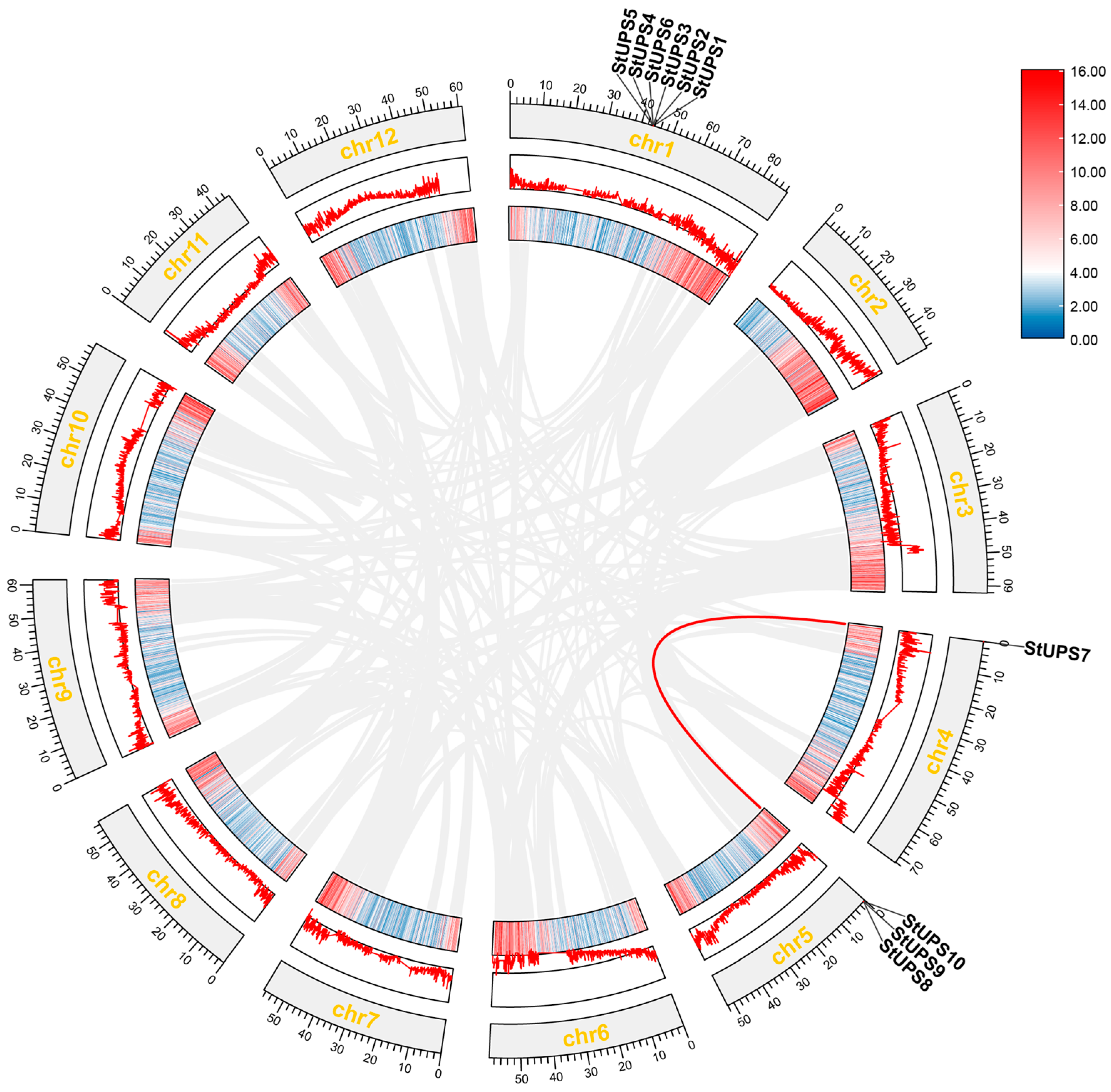
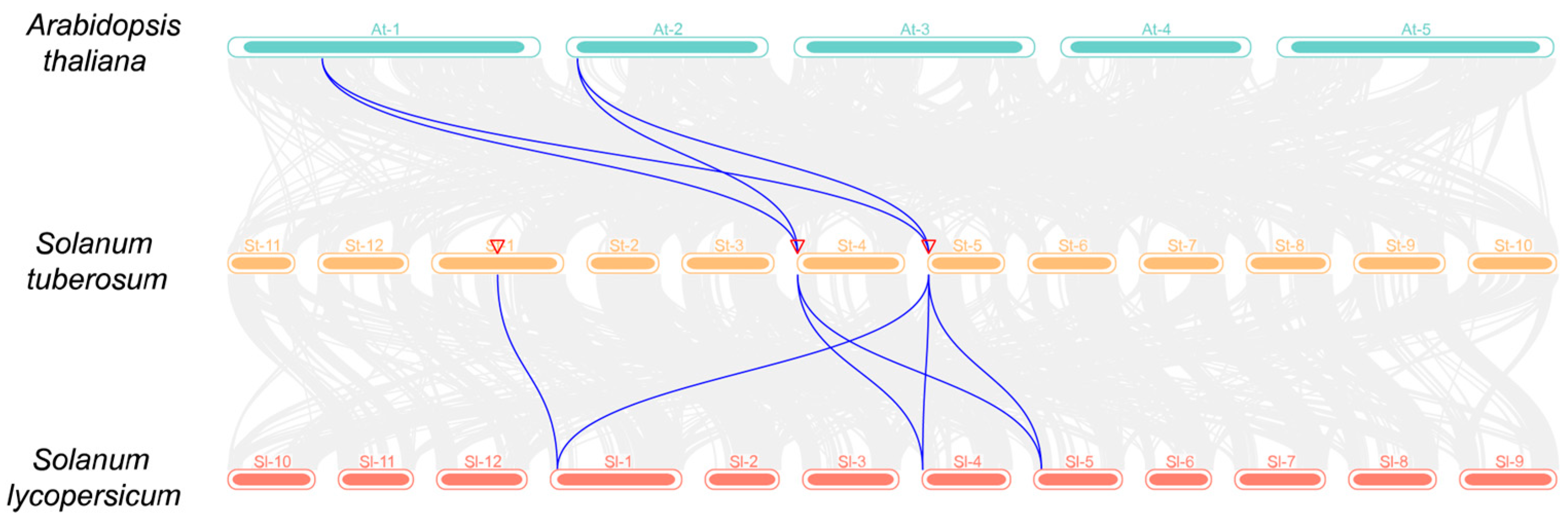
2.7. Collinearity of UPS Family in Potato
2.8. Expression Pattern Analysis of the UPS Gene Family
2.9. Expression Verification of Screened UPS Genes
3. Results
3.1. Identification of the Potato UPS Family Members
3.2. Physicochemical Properties, Transmembrane Structure, and Subcellular Localization of UPS Proteins
3.3. Phylogenetic Analysis of UPS Proteins
3.4. Analysis of UPS Motifs and Gene Structures in Potato
3.5. The Chromosomal Distribution of StUPS Genes
3.6. Collinearity Analysis of UPS Gene Family
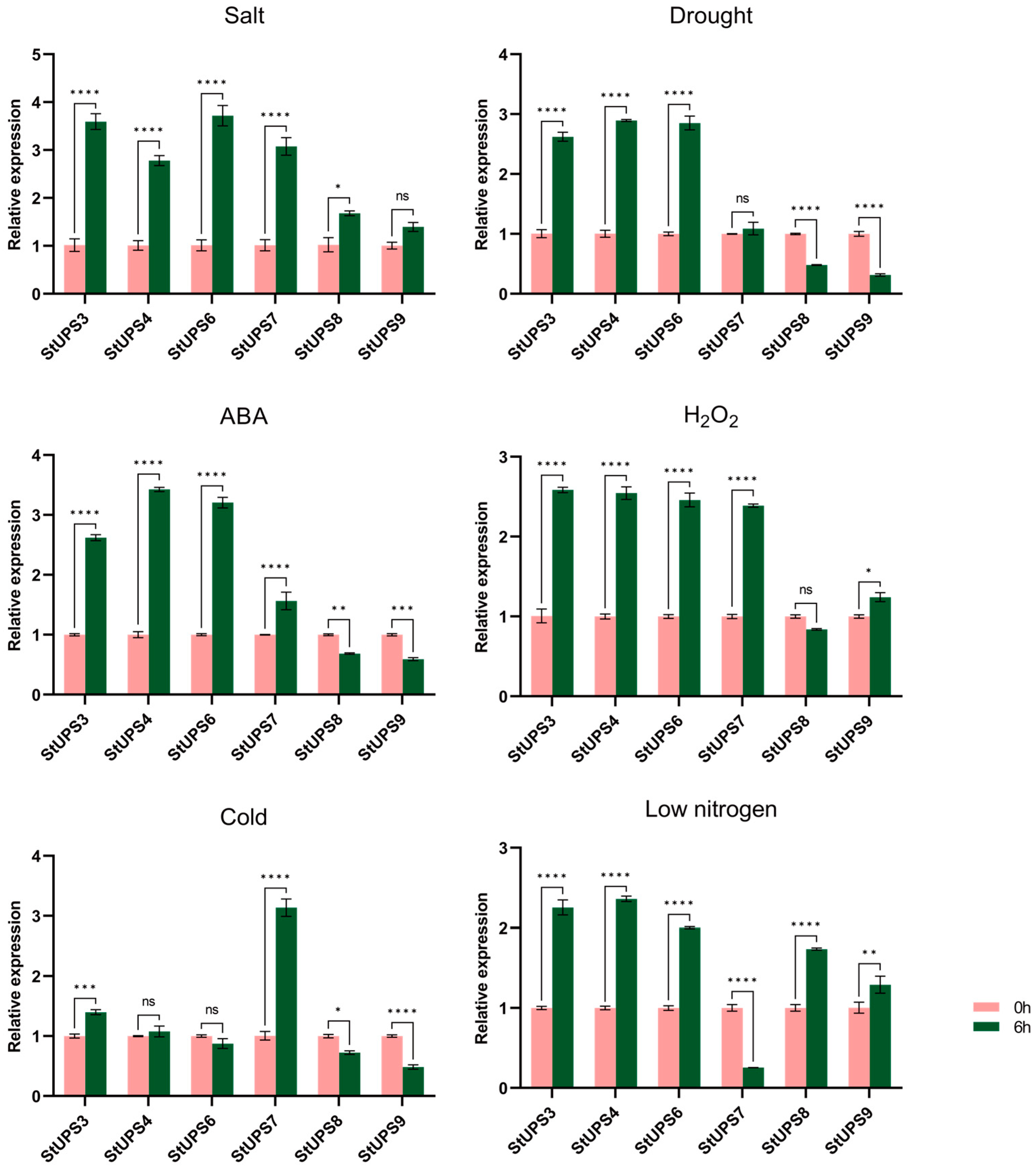
3.7. Expression Pattern Analysis of UPS Genes in Potato
3.8. Expression Verification of Potato UPS Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Havé, M.; Marmagne, A. Nitrogen remobilisation during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef]
- Desimone, M.; Catoni, E. A Novel Superfamily of Transporters for Allantoin and Other Oxo Derivatives of Nitrogen Heterocyclic Compounds in Arabidopsis. Plant Cell 2002, 14, 847–856. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmidt, A.; Su, Y.H. UPS1 and UPS2 from Arabidopsis Mediate High Affinity Transport of Uracil and 5-Fluorouracil. J. Biol. Chem. 2004, 279, 44817–44824. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Baumann, N. Comparative studies on Ureide Permeases in Arabidopsis thaliana and analysis of two alternative splice variants of AtUPS5. Planta 2006, 224, 1329–1340. [Google Scholar] [CrossRef]
- Pélissier, H.C.; Frerich, A. PvUPS1, an Allantoin Transporter in Nodulated Roots of French Bean. Plant Physiol. 2004, 134, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Tegeder, M. Soybean ureide transporters play a critical role in nodule development, function and nitrogen export. Plant J. 2012, 72, 355–367. [Google Scholar] [CrossRef]
- Witte, C.P.; Herde, M. Nucleotide Metabolism in Plants. Plant Physiol. 2020, 182, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 2014, 65, 1865–1878. [Google Scholar] [CrossRef]
- Pélissier, H.C.; Tegeder, M. PvUPS1 plays a role in source-sink transport of allantoin in French bean (Phaseolus vulgaris). Funct. Plant Biol. 2007, 34, 282–291. [Google Scholar] [CrossRef]
- Lu, M.Z.; Carter, A.M. Altering ureide transport in nodulated soybean results in whole-plant adjustments of metabolism, assimilate partitioning, and sink strength. J. Plant Physiol. 2022, 269, 153613. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M.; Tegeder, M. Increasing Nitrogen Fixation and Seed Development in Soybean Requires Complex Adjustments of Nodule Nitrogen Metabolism and Partitioning Processes. Curr. Biol. 2016, 26, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Herridge, D.; Atkins, C. Allantoin and Allantoic Acid in the Nitrogen Economy of the Cowpea (Vigna unguiculata [L.] Walp.). Plant Physiol. 1978, 62, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Casartelli, A.; Melino, V.J. Opposite fates of the purine metabolite allantoin under water and nitrogen limitations in bread wheat. Plant Mol. Biol. 2019, 99, 477–497. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Redillas, M.C.F.R. A Nitrogen Molecular Sensing System, Comprised of the ALLANTOINASE and UREIDE PERMEASE 1 Genes, Can Be Used to Monitor N Status in Rice. Front Plant Sci. 2018, 9, 444. [Google Scholar] [CrossRef] [PubMed]
- Brychkova, G.; Alikulov, Z. A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 2008, 54, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, L. A New Perspective on the Role of Glutamine Synthetase in Nitrogen Remobilization in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 11083. [Google Scholar] [CrossRef] [PubMed]
- Lescano, I.; Bogino, M.F. Ureide Permease 5 (AtUPS5) Connects Cell Compartments Involved in Ureide Metabolism. Plant Physiol. 2020, 182, 1310–1325. [Google Scholar] [CrossRef] [PubMed]
- Nourimand, M.; Todd, C.D. Allantoin Increases Cadmium Tolerance in Arabidopsis via Activation of Antioxidant Mechanisms. Plant Cell Physiol. 2016, 57, 2485–2496. [Google Scholar] [CrossRef]
- Watanabe, S.; Matsumoto, M. The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. 2014, 37, 1022–1036. [Google Scholar] [CrossRef]
- Lescano, C.I.; Martini, C. Allantoin accumulation mediated by allantoinase downregulation and transport by Ureide Permease 5 confers salt stress tolerance to Arabidopsis plants. Plant Mol. Biol. 2016, 91, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Kanehisa, M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 1992, 14, 897–911. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 13, 268. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Amode, M.R. Ensembl 2024. Nucleic Acids Res. 2024, 52, 891–899. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [PubMed]
- Nicot, N.; Hausman, J.F. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Thu, S.W.; Lu, M.Z. Role of ureides in source-to-sink transport of photoassimilates in non-fixing soybean. J. Exp. Bot. 2020, 71, 4495–4511. [Google Scholar] [CrossRef] [PubMed]
- Redillas, M.C.F.R.; Bang, S.W. Allantoin accumulation through overexpression of ureide permease1 improves rice growth under limited nitrogen conditions. Plant Biotechnol. J. 2019, 17, 1289–1301. [Google Scholar] [CrossRef]
- Werner, A.K.; Romeis, T. Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat. Chem. Biol. 2010, 6, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, Z. Multi-scale analysis provides insights into the roles of ureide permeases in wheat nitrogen use efficiency. J. Exp. Bot. 2023, 74, 5564–5590. [Google Scholar] [CrossRef]
- Tang, C.; Zhu, X. Characterization of the pectin methyl-esterase gene family and its function in controlling pollen tube growth in pear (Pyrus bretschneideri). Genomics 2020, 112, 2467–2477. [Google Scholar]
- De Grassi, A.; Lanave, C. Genome duplication and gene-family evolution: The case of three OXPHOS gene families. Gene 2008, 421, 1–6. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X. Evolutionary Origin, Gradual Accumulation and Functional Divergence of Heat Shock Factor Gene Family with Plant Evolution. Front Plant Sci. 2018, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Richard, C.M.; Michael, D.P. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar]
- Kong, H.; Landherr, L.L. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Bowne, J.B.; Erwin, T.A. Drought Responses of Leaf Tissues from Wheat Cultivars of Differing Drought Tolerance at the Metabolite Level. Mol. Plant 2012, 5, 418–429. [Google Scholar] [CrossRef]
- Oliver, M.J.; Guo, L. A Sister Group Contrast Using Untargeted Global Metabolomic Analysis Delineates the Biochemical Regulation Underlying Desiccation Tolerance in Sporobolus stapfianus. Plant Cell 2011, 23, 1231–1248. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Silvente, S. Metabolite Adjustments in Drought Tolerant and Sensitive Soybean Genotypes in Response to Water Stress. PLoS ONE 2012, 7, e38554. [Google Scholar]
- Pandey, G.K.; Degenkolbe, T. Identification of Drought Tolerance Markers in a Diverse Population of Rice Cultivars by Expression and Metabolite Profiling. PLoS ONE 2013, 8, e63637. [Google Scholar]
- Yobi, A.; Wone, B.W.M. Metabolomic Profiling in Selaginella lepidophylla at Various Hydration States Provides New Insights into the Mechanistic Basis of Desiccation Tolerance. Mol. Plant 2013, 6, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Casartelli, A.; Riewe, D. Exploring traditional aus-type rice for metabolites conferring drought tolerance. Rice 2018, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Kanani, H.; Dutta, B. Individual vs. combinatorial effect of elevated CO2 conditions and salinity stress on Arabidopsis thaliana liquid cultures: Comparing the early molecular response using time-series transcriptomic and metabolomic analyses. BMC Syst. Biol. 2010, 4, 177. [Google Scholar] [CrossRef] [PubMed]
- Nam, M.H.; Bang, E. Metabolite Profiling of Diverse Rice Germplasm and Identification of Conserved Metabolic Markers of Rice Roots in Response to Long-Term Mild Salinity Stress. Int. J. Mol. Sci. 2015, 16, 21959–21974. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.S.; Zhao, X.Q. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J. Exp. Bot. 2016, 67, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Kopka, J. Exploring the Temperature-Stress Metabolome of Arabidopsis. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, V.J.; Kopka, J. Systems Rebalancing of Metabolism in Response to Sulfur Deprivation, as Revealed by Metabolome Analysis of Arabidopsis Plants. Plant Physiol. 2005, 138, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Irani, S.; Todd, C.D. Ureide metabolism under abiotic stress in Arabidopsis thaliana. J. Plant Physiol. 2016, 199, 87–95. [Google Scholar] [CrossRef]
- Irani, S.; Todd, C.D. Exogenous allantoin increases Arabidopsis seedlings tolerance to NaCl stress and regulates expression of oxidative stress response genes. J. Plant Physiol. 2018, 221, 43–50. [Google Scholar] [CrossRef]




| ID | Sequence ID | Number of Amino Acid | Molecular Weight | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Localization Prediction |
|---|---|---|---|---|---|---|---|---|
| PGSC0003DMT400025459 | StUPS1 | 59 | 6671.11 | 10.55 | 40.63 | 108.98 | 0.731 | Extracellular |
| PGSC0003DMT400025460 | StUPS2 | 65 | 6942.19 | 10.16 | 39.34 | 96.15 | 0.598 | Endoplasmic reticulum |
| PGSC0003DMT400025462 | StUPS3 | 319 | 34,322.81 | 9.09 | 33.03 | 103.1 | 0.443 | Vacuole |
| PGSC0003DMT400025463 | StUPS4 | 407 | 44,147.34 | 8.87 | 32.33 | 105.5 | 0.465 | Plasma membrane |
| PGSC0003DMT400025464 | StUPS5 | 319 | 34,322.81 | 9.09 | 33.03 | 103.1 | 0.443 | Vacuole |
| PGSC0003DMT400025465 | StUPS6 | 233 | 25,250.13 | 6.13 | 32.77 | 106.78 | 0.302 | Vacuole |
| PGSC0003DMT400045883 | StUPS7 | 95 | 10,056.74 | 9.43 | 30.23 | 96.74 | 0.537 | Chloroplast |
| PGSC0003DMT400072929 | StUPS8 | 113 | 12,247.4 | 9.84 | 35.1 | 99.47 | 0.477 | Extracellular |
| PGSC0003DMT400072930 | StUPS9 | 406 | 44,175.16 | 8.04 | 31.08 | 95.44 | 0.385 | Plasma membrane |
| PGSC0003DMT400072931 | StUPS10 | 406 | 44,175.16 | 8.04 | 31.08 | 95.44 | 0.385 | Plasma membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Lu, Y.; Ren, B.; Zeng, F.; Liu, Y.; Lu, L.; Li, L. Identification and Expression Analysis of UPS Gene Family in Potato. Genes 2024, 15, 870. https://doi.org/10.3390/genes15070870
Huang W, Lu Y, Ren B, Zeng F, Liu Y, Lu L, Li L. Identification and Expression Analysis of UPS Gene Family in Potato. Genes. 2024; 15(7):870. https://doi.org/10.3390/genes15070870
Chicago/Turabian StyleHuang, Wenyue, Yifei Lu, Bi Ren, Fuchun Zeng, Yongjian Liu, Liming Lu, and Liqin Li. 2024. "Identification and Expression Analysis of UPS Gene Family in Potato" Genes 15, no. 7: 870. https://doi.org/10.3390/genes15070870
APA StyleHuang, W., Lu, Y., Ren, B., Zeng, F., Liu, Y., Lu, L., & Li, L. (2024). Identification and Expression Analysis of UPS Gene Family in Potato. Genes, 15(7), 870. https://doi.org/10.3390/genes15070870





