Abstract
Intracranial aneurysm (IA) is a relatively common vascular malformation of an intracranial artery. In most cases, its presence is asymptomatic, but IA rupture causing subarachnoid hemorrhage is a life-threating condition with very high mortality and disability rates. Despite intensive studies, molecular mechanisms underlying the pathophysiology of IA formation, growth, and rupture remain poorly understood. There are no specific biomarkers of IA presence or rupture. Analysis of expression of mRNA and other RNA types offers a deeper insight into IA pathobiology. Here, we present results of published human studies on IA-focused transcriptomics.
1. Introduction
The most common cause of spontaneous subarachnoid hemorrhage (SAH) is a rupture of an intracranial aneurysm (IA). This form of hemorrhagic stroke comprises about 5% of all strokes. Despite overall improvements in patients’ care, SAH is burdened with high mortality (approximately 50%) and disability rates—only 25% of patients who survived are likely to live independently. Most SAH patients have permanent neurological and cognitive deficits and remain dependent [1,2]. The prognosis is heavily influenced by the development of vasospasm and delayed cerebral ischemia (DCI). Vasospasm, which can be detected in approximately two thirds of SAH patients, may lead to DCI and subsequent neurological impairment. The incidence of spontaneous SAH is 8 persons in 100,000 person-years. The prevalence of unruptured IAs (UAs) in the general population is estimated at 3%. Most of the UAs remain unruptured since the risk of aneurysmal rupture is about 1% per year [3,4]. Unfortunately, it is still impossible to predict the fate of a particular IA. So far, only some risk factors of IA presence, growth, and rupture have been identified (for instance: female sex, hypertension, smoking, IA location, IA size), and based on them, a risk of an IA rupture can be estimated [4].
Molecular mechanisms underlying IA formation and rupture remain not fully recognized. Similarly, the knowledge about molecular drivers of systemic responses to the rupture of an IA is incomplete.
One of the approaches to investigate these aspects of IA pathobiology is to analyze alterations in RNA expression associated with the presence of IAs, their status (ruptured vs. unruptured), and sequels of SAH. The first studies focused on mRNA as a molecule containing the genetic information which is translated into proteins. However, over time, non-coding RNAs and RNA regulatory networks drew attention as well. Depending on the underlying question, RNA expression was analyzed in various samples, such as: IA wall, peripheral blood cells, and serum/plasma. The first broad gene expression profiling was performed by Peters et al. by means of the SAGE-Lite method in a single patient [5]. Afterwards, a microarray approach was used, subsequently replaced by RNA sequencing (RNAseq). In addition, by developing new bioinformatics tools, there is an increasing number of studies in which available original data are re-analyzed and/or existing datasets are combined.
In this review, we will focus on transcriptomics studies conducted on human-derived samples obtained from patients with IA. Firstly, a concise overview of original transcriptomics studies will be presented. Then, a brief summary of studies in which existing datasets were used (secondary studies) will be provided. A literature review was performed using PubMed and Web of Science. The search terms were “intracranial aneurysms”, “cerebral aneurysm”, “brain aneurysm” AND “gene expression”, and “RNA expression”. The identified reports were manually checked to select only transcriptomics studies on human-derived samples.
2. Original Studies
We identified 27 original studies which investigated RNA expression in the aneurysmal wall. Seven studies were focused on the mechanisms associated with IA rupture, eighteen on aneurysm formation, and in two reports alterations in RNA expression were analyzed both in present IAs and after their rupture. In studies on blood-derived samples, the corresponding numbers were the following: 24 studies, among them: 9 focused on the rupture-related changes (and complications of SAH in 2 studies), 10 on the IA presence, and in 5 studies, markers of IA formation and rupture were investigated.
2.1. Transcriptomics in IA Samples
These studies can be divided into two subgroups. The first one utilizes aneurysmal tissue and is focused on mechanisms involved in IA formation and rupture. IA samples and control arteries were obtained during neurosurgical procedures, except a study published by Weinsheimer et al. [6], where samples came from autopsies. In two other studies, expression data from available datasets served as controls [7,8]. In general, control vessels served as superficial temporal arteries [5,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23] or middle meningeal arteries [11,23,24,25,26], and in single reports as arteriovenous malformation (AVM) feders [27] or cortical arteries [28]. One group did not specify which vessel was used as a control [29]. Numbers of analyzed samples vary from 3 [7,9,10] to 70 [26] per group. In two studies, in addition to vessels, peripheral blood samples were analyzed to investigate potential similarities between aneurysmal expression profiles and blood, searching for biomarkers of IA presence and/or rupture [22,30]. Three other studies comprised in vitro parts, which allowed to verify some findings from the expression analyses in vascular smooth-muscle cells [23,26] or endothelial cells’ cultures [31].
The first published analysis of global gene expression profiles in aneurysmal tissue was performed using the SAGE-Lite technique. Samples were obtained from a single patient—a 3-year-old girl with SAH: walls of a ruptured IA (RA) and a control vessel—the superficial temporal artery (STA). The analysis comprised 4924 and 3552 genes in the RA and STA samples, respectively, and revealed an overexpression of genes related to extracellular matrix, cell adhesion, and cell migration [5]. In subsequent studies, these two elements, i.e., differential expression of RNAs and their functional annotation, remained the core of the performed analyses. From non-coding RNAs, miRNAs were the most investigated class with or without a concomitant profiling of mRNAs [8,14,15,16,25,26]. In four studies, expression of lncRNAs was analyzed [16,17,20,30], and in two, circular RNA (circRNA) [22,31]. When mRNA expression was not directly measured, mRNA target prediction analysis was provided. Attempts to compare results of expression data on a single RNA molecule level are rather disappointing. For instance, Roder et al. in their meta-analysis of 5 microarray-based IA studies found that only 57 out of 507 reported differentially expressed genes (DEGs) were identified in more than 2 studies [32]. However, while looking at the functional annotations of differentially expressed RNAs, categories related to inflammatory reaction, immune system, cellular adhesion, extracellular matrix, muscles, apoptosis, and cellular signaling were identified as key players in the pathophysiology of IAs. Details are summarized in Table 1.

Table 1.
Original studies on RNA expression in the intracranial aneurysm wall.
2.2. Transcriptomics in Blood-Derived Samples
In expression studies in blood samples, RNAs were isolated from whole blood [33,34,35,36,37], blood cells as a whole [38,39,40,41,42,43,44], or specifically from leukocytes [24], mononuclear cells [45,46,47], or neutrophils [48,49,50]. Circulating RNAs were isolated from plasma [51,52,53,54,55], serum [56], or circulating exosomes [57]. The main goals of this group of studies were: (i) search of biomarkers of IAs or their categories (RAs, UAs), and (ii) investigation of systemic consequences of IA rupture, including clinical status of SAH patients or SAH complications such as vasospasm [33,44] or DCI [38]. Circulating blood cells are notably sensitive to pathologic processes affecting the body. Only in one study was gene expression examined in intracranial, not peripheral, vessels—blood samples were obtained from IA lumen and IA proximal parent vessels [37]. The range of cohort sizes was from 3 [46] to 130 patients [24]. Korostynski et al. [41,42] and Morga et al. [43] analyzed differences in RNA expression profiles between acute and chronic phase of RA, whereas van’t Hof et al. searched for potential biomarkers of past aSAH (at least 2 years after RA) [40]. Similar to tissue-based studies, in most of the blood-based studies, mRNA expression was examined (cell-derived or circulating) [24,34,37,38,39,40,41,44,45,47,48,49,50,53]. However, non-coding RNAs were also studied—mainly miRNAs [33,42,51,52,54,55,56,57] and lncRNAs [24,35,53]. CircRNAs were investigated in two studies [36,46] and expression of different subtypes of small RNAs (piRNAs, rRNAs, tRNAs, snoRNAs, scRNAs) was presented in one report [43]. Functional analyses and target prediction for non-coding RNAs can be considered as standard approaches. In general, results of functional annotation resemble tissue-based studies. More details of this group of transcriptomics studies are presented in Table 2.

Table 2.
Original studies on RNA expression in blood-derived samples.
3. Studies Based on Existing Datasets
We identified 27 secondary studies which used datasets with RNA expression in the aneurysmal wall. Eighteen studies were focused on the mechanisms associated with IA rupture, twelve on aneurysm formation, and in nine, alterations in RNA expression were analyzed both in present IAs and after their rupture. In studies on datasets with blood-derived samples, the corresponding numbers were following: nine studies, among them: eight focused on the rupture-related changes, and one focused on the IA presence.
3.1. Transcriptomics in IA Samples
Along with the development of bioinformatic tools appeared a new type of study presenting re-analyzed data from available datasets, including expression data from the Gene Expression Omnibus (GEO). Approximately one third of these published secondary analyses utilized a single dataset [58,59,60,61,62,63,64,65,66] and two thirds leveraged data from two to eight datasets [67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85]. These studies did not provide any new additional clinical data but rather aimed to deepen the insight into molecular mechanisms of the IA pathophysiology by revealing key regulatory networks and interactions between investigated molecules. Although differential expression and functional annotation were examined, further analyses of co-expression networks with identification of hub RNA molecules, competing endogenous RNA (ceRNA) networks, or protein–protein interaction (PPI) networks became a standard approach. In some of these studies, specific areas of interest were predefined, such as: epithelial–mesenchymal transition [78], endoplasmic reticulum stress [81], immune environment [79,83], or ferroptosis [84,85]. In three studies, an attempt was made to identify potential therapeutic targets [71,82,83]. Sun et al. investigated expression profiles and networks in various aneurysms, including thoracic and abdominal aorta aneurysms [77]. More detailed information about this group of studies is provided in Table 3.

Table 3.
Studies on RNA expression in the intracranial aneurysm wall utilizing existing datasets.
3.2. Transcriptomics in Blood-Derived Samples
Interestingly, the number of studies utilizing existing blood-based transcriptomics results is smaller than tissue-based studies. This could be explained by the availability of bio-samples. It is easier to design a new study and to obtain blood samples than aneurysmal specimens. In this category of studies, only three out of nine studies used data from at least two datasets [86,87,88], and six analyses were based on a single dataset [89,90,91,92,93,94]. Analytical methods used did not significantly differ when compared to tissue-based studies. Two reports comprised validation cohorts [87,88]. Table 4 shows more details.

Table 4.
Studies on RNA expression in blood-related samples in intracranial aneurysm utilizing existing datasets.
4. Conclusions
In the last decade, the number of studies focused on different aspects of transcriptomics in IAs significantly increased (Figure 1). This is associated with the technology development and bioinformatics allowing to analyze big data.
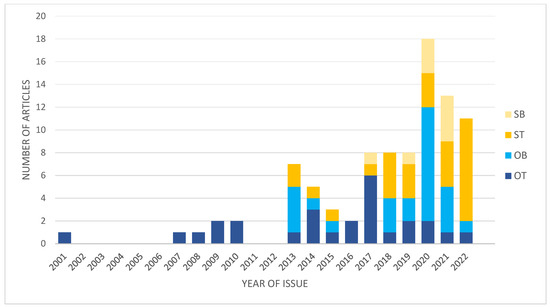
Figure 1.
Graph presenting changes in numbers and types of studies focused on IA transcriptomics. OT, original studies using IA wall; OB, original studies using blood-derived samples; ST, secondary studies using tissue-derived data; SB, secondary studies using blood-derived data.
However, there are so many open questions regarding the pathophysiology of IAs and molecular mechanisms underlying the consequences of IA rupture. After more than 20 years of studies on the expression of coding and non-coding RNAs, it is obvious that there is not one single pathway responsible for IA formation or rupture. However, there are some networks, some groups of genes, that seem to play important role, such as immune/inflammatory response, extracellular matrix- or focal adhesion-related, cellular signaling, regulated cell death, and muscles. These terms are consistently repeated in presented studies, although in studies on blood-derived samples the most common identified pathways are those related to the immune/inflammatory response, cell death, or cellular metabolic processes. Secondary studies based on existing datasets echo these findings.
The existing expression studies are burdened with several limitations. These are human studies and not all factors that can affect gene expression are controllable and comparable between studied groups, including comorbidities, medications, and lifestyle habits. Next, time between sampling and placement of the sample on ice or transportation/storage solutions may impact expression measurements. Furthermore, the quality of the sample is important—what is the composition of the vessel/aneurysmal wall? For instance, there are acellular or hypocellular areas in some ruptured aneurysms. Moreover, the presence of even residual amounts of blood elements on the tissue will influence the results of expression analyses. Another important issue is the choice of the control tissue. In most studies, IAs and controls were obtained from different individuals. Some researchers used intracranial vessels (e.g., cortical arteries or AVM feders), whereas others used extracranial arteries. The anatomical differences between these vessels may affect the results of expression analyses. In 2019, Laarman et al. published results of their search for optimal controls in gene expression studies on IAs [95]. In blood-derived samples, a background cell count may play an important role for the analytical output. All these elements increase the heterogeneity of analyzed samples, including the RNA types. The secondary studies that use the existing datasets rarely pay much attention to clinical variables and focus on raw expression data.
With the progress of our knowledge about the gene expression, the regulatory mechanisms of transcription, and roles played by different classes of RNA, accompanied by the development of available research tools, researchers have started to analyze the alterations in other (not mRNA) types of RNA. However, it seems that we are still at the beginning of understanding the processes underlying the pathophysiology of IAs. Very little is known about the role of small noncoding RNAs other than microRNA. We do not even understand what the significance is of altered expression of gene isoforms. Further studies are needed to explain the role of gene expression and RNA molecules in the pathobiology of IAs and the consequences of their rupture. These studies cannot be limited to a pure transcriptomic analysis. Functional analyses using experimental approaches both in vitro and in vivo are needed to test the results from expression studies in a more complex environment of living cells or whole organisms.
Author Contributions
Conceptualization, J.P.; literature search, R.M.; writing—original draft preparation, R.M. and J.P.; writing—review and editing, J.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nieuwkamp, D.J.; Setz, L.E.; Algra, A.; Linn, F.H.; de Rooij, N.K.; Rinkel, G.J. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex, and region: A meta-analysis. Lancet Neurol. 2009, 8, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Rinkel, G.J.; Algra, A. Long-term outcomes of patients with aneurysmal subarachnoid haemorrhage. Lancet Neurol. 2011, 10, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Lawton, M.T.; Vates, G.E. Subarachnoid Hemorrhage. N. Engl. J. Med. 2017, 377, 257–266. [Google Scholar] [CrossRef]
- Claassen, J.; Park, S. Spontaneous subarachnoid haemorrhage. Lancet 2022, 400, 846–862. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.G.; Kassam, A.B.; Feingold, E.; Heidrich-O’Hare, E.; Yonas, H.; Ferrell, R.E.; Brufsky, A. Molecular anatomy of an intracranial aneurysm: Coordinated expression of genes involved in wound healing and tissue remodeling. Stroke 2001, 32, 1036–1042. [Google Scholar] [CrossRef]
- Weinsheimer, S.; Lenk, G.M.; van der Voet, M.; Land, S.; Ronkainen, A.; Alafuzoff, I.; Kuivaniemi, H.; Tromp, G. Integration of expression profiles and genetic mapping data to identify candidate genes in intracranial aneurysm. Physiol. Genom. 2007, 32, 45–57. [Google Scholar] [CrossRef]
- Li, Z.; Tan, H.; Shi, Y.; Huang, G.; Wang, Z.; Liu, L.; Yin, C.; Wang, Q. Global Gene Expression Patterns and Somatic Mutations in Sporadic Intracranial Aneurysms. World Neurosurg. 2017, 100, 15–21. [Google Scholar] [CrossRef]
- Supriya, M.; Christopher, R.; Devi, B.I.; Bhat, D.I.; Shukla, D.; Kalpana, S.R. Altered MicroRNA Expression in Intracranial Aneurysmal Tissues: Possible Role in TGF-β Signaling Pathway. Cell Mol. Neurobiol. 2022, 42, 2393–2405. [Google Scholar] [CrossRef]
- Li, L.; Yang, X.; Jiang, F.; Dusting, G.J.; Wu, Z. Transcriptome-wide characterization of gene expression associated with unruptured intracranial aneurysms. Eur. Neurol. 2009, 62, 330–337. [Google Scholar] [CrossRef]
- Shi, C.; Awad, I.A.; Jafari, N.; Lin, S.; Du, P.; Hage, Z.A.; Shenkar, R.; Getch, C.C.; Bredel, M.; Batjer, H.H.; et al. Genomics of human intracranial aneurysm wall. Stroke 2009, 40, 1252–1261. [Google Scholar] [CrossRef]
- Marchese, E.; Vignati, A.; Albanese, A.; Nucci, C.G.; Sabatino, G.; Tirpakova, B.; Lofrese, G.; Zelano, G.; Maira, G. Comparative evaluation of genome-wide gene expression profiles in ruptured and unruptured human intracranial aneurysms. J. Biol. Regul. Homeost. Agents 2010, 24, 185–195. [Google Scholar]
- Yu, L.; Fan, J.; Wang, S.; Zhang, D.; Wang, R.; Zhao, Y.; Zhao, J. Gene expression profiles in intracranial aneurysms. Neurosci. Bull 2014, 30, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, H.; Tajima, A.; Yoneyama, T.; Hosomichi, K.; Kasuya, H.; Mizutani, T.; Inoue, I. Gene expression profiling reveals distinct molecular signatures associated with the rupture of intracranial aneurysm. Stroke 2014, 45, 2239–2245. [Google Scholar] [CrossRef]
- Liu, D.; Han, L.; Wu, X.; Yang, X.; Zhang, Q.; Jiang, F. Genome-wide microRNA changes in human intracranial aneurysms. BMC Neurol. 2014, 14, 188. [Google Scholar] [CrossRef]
- Bekelis, K.; Kerley-Hamilton, J.S.; Teegarden, A.; Tomlinson, C.R.; Kuintzle, R.; Simmons, N.; Singer, R.J.; Roberts, D.W.; Kellis, M.; Hendrix, D.A. MicroRNA and gene expression changes in unruptured human cerebral aneurysms. J. Neurosurg. 2016, 125, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, W.; Zhang, L.; Lan, Q.; Wang, J.; Cao, Y.; Zhao, J. Identification of a Long Noncoding RNA-Associated Competing Endogenous RNA Network in Intracranial Aneurysm. World Neurosurg. 2017, 97, 684–692.e4. [Google Scholar] [CrossRef]
- Wang, W.; Li, H.; Yu, L.; Zhao, Z.; Wang, H.; Zhang, D.; Zhang, Y.; Lan, Q.; Wang, J.; Zhao, J. Aberrant expression of lncRNAs and mRNAs in patients with intracranial aneurysm. Oncotarget 2017, 8, 2477–2484. [Google Scholar] [CrossRef]
- Li, H.; Li, H.; Yue, H.; Wang, W.; Yu, L.; Shuo, W.; Cao, Y.; Zhao, J. Comparison between smaller ruptured intracranial aneurysm and larger un-ruptured intracranial aneurysm: Gene expression profile analysis. Neurosurg. Rev. 2017, 40, 419–425. [Google Scholar] [CrossRef]
- Yu, L.; Wang, J.; Wang, S.; Zhang, D.; Zhao, Y.; Wang, R.; Zhao, J. DNA Methylation Regulates Gene Expression in Intracranial Aneurysms. World Neurosurg. 2017, 105, 28–36. [Google Scholar] [CrossRef]
- Li, H.; Yue, H.; Hao, Y.; Li, H.; Wang, S.; Yu, L.; Zhang, D.; Cao, Y.; Zhao, J. Expression profile of long noncoding RNAs in human cerebral aneurysms: A microarray analysis. J. Neurosurg. 2017, 127, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yu, L.; Zhao, J. Comparative transcriptome analysis reveals involvement of TLR-2 signaling in the pathogenesis of intracranial aneurysm. J. Clin. Neurosci. 2018, 47, 258–263. [Google Scholar] [CrossRef]
- Huang, Q.; Huang, Q.Y.; Sun, Y.; Wu, S. High-Throughput Data Reveals Novel Circular RNAs via Competitive Endogenous RNA Networks Associated with Human Intracranial Aneurysms. Med. Sci. Monit. 2019, 25, 4819–4830. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Koseki, H.; Miyata, H.; Itoh, M.; Kawaji, H.; Takizawa, K.; Kawashima, A.; Ujiie, H.; Higa, T.; Minamimura, K.; et al. RNA sequencing analysis revealed the induction of CCL3 expression in human intracranial aneurysms. Sci. Rep. 2019, 9, 10387. [Google Scholar] [CrossRef] [PubMed]
- Pera, J.; Korostynski, M.; Krzyszkowski, T.; Czopek, J.; Slowik, A.; Dziedzic, T.; Piechota, M.; Stachura, K.; Moskala, M.; Przewlocki, R.; et al. Gene expression profiles in human ruptured and unruptured intracranial aneurysms: What is the role of inflammation? Stroke 2010, 41, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, M.; He, H.; Chen, J.; Zeng, H.; Li, J.; Duan, R. MicroRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med. Genom. 2013, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ren, Y.; Wang, Y.; Wang, R.; Zhou, Q.; Peng, Y.; Li, Q.; Yu, M.; Jiang, Y. Regulation of smooth muscle contractility by competing endogenous mRNAs in intracranial aneurysms. J. Neuropathol. Exp. Neurol. 2015, 74, 411–424. [Google Scholar] [CrossRef]
- Krischek, B.; Kasuya, H.; Tajima, A.; Akagawa, H.; Sasaki, T.; Yoneyama, T.; Ujiie, H.; Kubo, O.; Bonin, M.; Takakura, K.; et al. Network-based gene expression analysis of intracranial aneurysm tissue reveals role of antigen presenting cells. Neuroscience 2008, 154, 1398–1407. [Google Scholar] [CrossRef]
- Kleinloog, R.; Verweij, B.H.; van der Vlies, P.; Deelen, P.; Swertz, M.A.; de Muynck, L.; Van Damme, P.; Giuliani, F.; Regli, L.; van der Zwan, A.; et al. RNA Sequencing Analysis of Intracranial Aneurysm Walls Reveals Involvement of Lysosomes and Immunoglobulins in Rupture. Stroke 2016, 47, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Yang, C.; Jia, C.; Xie, X.; Du, L. miR-566 expression and immune changes in patients with intracranial aneurysm. Int. J. Clin. Exp. Pathol. 2020, 13, 685–691. [Google Scholar]
- Sun, Y.; Wen, Y.; Ruan, Q.; Yang, L.; Huang, S.; Xu, X.; Cai, Y.; Li, H.; Wu, S. Exploring the association of long noncoding RNA expression profiles with intracranial aneurysms, based on sequencing and related bioinformatics analysis. BMC Med. Genom. 2020, 13, 147. [Google Scholar] [CrossRef]
- Zhang, Z.; Sui, R.; Ge, L.; Xia, D. CircRNA_0079586 and circRNA_RanGAP1 are involved in the pathogenesis of intracranial aneurysms rupture by regulating the expression of MPO. Sci. Rep. 2021, 11, 19800. [Google Scholar] [CrossRef]
- Roder, C.; Kasuya, H.; Harati, A.; Tatagiba, M.; Inoue, I.; Krischek, B. Meta-analysis of microarray gene expression studies on intracranial aneurysms. Neuroscience 2012, 201, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Lopes, K.P.; Vinasco-Sandoval, T.; Vialle, R.A.; Paschoal, F.M., Jr.; Bastos, V.A.P.A.; Bor-Seng-Shu, E.; Teixeira, M.J.; Yamada, E.S.; Pinto, P.; Vidal, A.F.; et al. Global miRNA expression profile reveals novel molecular players in aneurysmal subarachnoid haemorrhage. Sci. Rep. 2018, 8, 8786. [Google Scholar] [CrossRef]
- Poppenberg, K.E.; Li, L.; Waqas, M.; Paliwal, N.; Jiang, K.; Jarvis, J.N.; Sun, Y.; Snyder, K.V.; Levy, E.I.; Siddiqui, A.H.; et al. Whole blood transcriptome biomarkers of unruptured intracranial aneurysm. PLoS ONE 2020, 15, e0241838. [Google Scholar] [CrossRef]
- Tutino, V.M.; Poppenberg, K.E.; Damiano, R.J.; Patel, T.R.; Waqas, M.; Dmytriw, A.A.; Snyder, K.V.; Siddiqui, A.H.; Jarvis, J.N. Characterization of Long Non-coding RNA Signatures of Intracranial Aneurysm in Circulating Whole Blood. Mol. Diagn. Ther. 2020, 24, 723–736. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, B.; Zhang, D.; Wang, S.; Li, M.; Zhao, J. Differentially Expressed Circular RNA Profile in an Intracranial Aneurysm Group Compared with a Healthy Control Group. Dis. Markers 2021, 2021, 8889569. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.M.; Lu, Y.; Ishii, D.; Poppenberg, K.E.; Rajabzadeh-Oghaz, H.; Siddiqui, A.H.; Hasan, D.M. Aberrant Whole Blood Gene Expression in the Lumen of Human Intracranial Aneurysms. Diagnostics 2021, 11, 1442. [Google Scholar] [CrossRef]
- Baumann, A.; Devaux, Y.; Audibert, G.; Zhang, L.; Bracard, S.; Colnat-Coulbois, S.; Klein, O.; Zannad, F.; Charpentier, C.; Longrois, D.; et al. Gene expression profile of blood cells for the prediction of delayed cerebral ischemia after intracranial aneurysm rupture: A pilot study in humans. Cerebrovasc. Dis. 2013, 36, 236–242. [Google Scholar] [CrossRef]
- Pera, J.; Korostynski, M.; Golda, S.; Piechota, M.; Dzbek, J.; Krzyszkowski, T.; Dziedzic, T.; Moskala, M.; Przewlocki, R.; Szczudlik, A.; et al. Gene expression profiling of blood in ruptured intracranial aneurysms: In search of biomarkers. J. Cereb. Blood Flow Metab. 2013, 33, 1025–1031. [Google Scholar] [CrossRef]
- van ‘t Hof, F.N.; Ruigrok, Y.M.; Medic, J.; Sanjabi, B.; van der Vlies, P.; Rinkel, G.J.; Veldink, J.H. Whole Blood Gene Expression Profiles of Patients with a Past Aneurysmal Subarachnoid Hemorrhage. PLoS ONE 2015, 10, e0139352. [Google Scholar] [CrossRef]
- Korostynski, M.; Piechota, M.; Morga, R.; Hoinkis, D.; Golda, S.; Zygmunt, M.; Dziedzic, T.; Moskala, M.; Slowik, A.; Pera, J. Systemic response to rupture of intracranial aneurysms involves expression of specific gene isoforms. J. Transl. Med. 2019, 17, 141. [Google Scholar] [CrossRef]
- Korostynski, M.; Morga, R.; Piechota, M.; Hoinkis, D.; Golda, S.; Dziedzic, T.; Slowik, A.; Moskala, M.; Pera, J. Inflammatory Responses Induced by the Rupture of Intracranial Aneurysms Are Modulated by miRNAs. Mol. Neurobiol. 2020, 57, 988–996. [Google Scholar] [CrossRef]
- Morga, R.; Borczyk, M.; Korostynski, M.; Piechota, M.; Hoinkis, D.; Golda, S.; Dziedzic, T.; Slowik, A.; Moskala, M.; Pera, J. Opposite regulation of piRNAs, rRNAs and miRNAs in the blood after subarachnoid hemorrhage. J. Mol. Med. 2020, 98, 887–896. [Google Scholar] [CrossRef]
- Xu, H.; Stamova, B.; Ander, B.P.; Waldau, B.; Jickling, G.C.; Sharp, F.R.; Ko, N.U. mRNA Expression Profiles from Whole Blood Associated with Vasospasm in Patients with Subarachnoid Hemorrhage. Neurocrit. Care 2020, 33, 82–89. [Google Scholar] [CrossRef]
- Sabatino, G.; Rigante, L.; Minella, D.; Novelli, G.; Della Pepa, G.M.; Esposito, G.; Albanese, A.; Maira, G.; Marchese, E. Transcriptional profile characterization for the identification of peripheral blood biomarkers in patients with cerebral aneurysms. J. Biol. Regul. Homeost. Agents 2013, 27, 729–738. [Google Scholar] [PubMed]
- Cao, H.; Chen, J.; Lai, X.; Liu, T.; Qiu, P.; Que, S.; Huang, Y. Circular RNA expression profile in human primary multiple intracranial aneurysm. Exp. Ther. Med. 2021, 21, 239. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.M.; Zebraski, H.R.; Rajabzadeh-Oghaz, H.; Waqas, M.; Jarvis, J.N.; Bach, K.; Mokin, M.; Snyder, K.V.; Siddiqui, A.H.; Poppenberg, K.E. Identification of Circulating Gene Expression Signatures of Intracranial Aneurysm in Peripheral Blood Mononuclear Cells. Diagnostics 2021, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.M.; Poppenberg, K.E.; Jiang, K.; Jarvis, J.N.; Sun, Y.; Sonig, A.; Siddiqui, A.H.; Snyder, K.V.; Levy, E.I.; Kolega, J.; et al. Circulating neutrophil transcriptome may reveal intracranial aneurysm signature. PLoS ONE 2018, 13, e0191407. [Google Scholar] [CrossRef]
- Tutino, V.M.; Poppenberg, K.E.; Li, L.; Shallwani, H.; Jiang, K.; Jarvis, J.N.; Sun, Y.; Snyder, K.V.; Levy, E.I.; Siddiqui, A.H.; et al. Biomarkers from circulating neutrophil transcriptomes have potential to detect unruptured intracranial aneurysms. J. Transl. Med. 2018, 16, 373. [Google Scholar] [CrossRef]
- Poppenberg, K.E.; Tutino, V.M.; Li, L.; Waqas, M.; June, A.; Chaves, L.; Jiang, K.; Jarvis, J.N.; Sun, Y.; Snyder, K.V.; et al. Classification models using circulating neutrophil transcripts can detect unruptured intracranial aneurysm. J. Transl. Med. 2020, 18, 392. [Google Scholar] [CrossRef]
- Jin, H.; Li, C.; Ge, H.; Jiang, Y.; Li, Y. Circulating microRNA: A novel potential biomarker for early diagnosis of intracranial aneurysm rupture a case control study. J. Transl. Med. 2013, 11, 296. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Wu, X.; Yang, X.; Zhang, Y.; Li, Y.; Jiang, F. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J. Am. Heart Assoc. 2014, 3, e000972. [Google Scholar] [CrossRef]
- Wu, C.; Song, H.; Wang, Y.; Gao, L.; Cai, Y.; Cheng, Q.; Chen, Y.; Zheng, Z.; Liao, Y.; Lin, J.; et al. Long non-coding RNA TCONS_00000200 as a non-invasive biomarker in patients with intracranial aneurysm. Biosci. Rep. 2019, 39, BSR20182224. [Google Scholar] [CrossRef] [PubMed]
- Supriya, M.; Christopher, R.; Indira Devi, B.; Bhat, D.I.; Shukla, D. Circulating MicroRNAs as Potential Molecular Biomarkers for Intracranial Aneurysmal Rupture. Mol. Diagn. Ther. 2020, 24, 351–364. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, X.; Liu, L.; Pu, Y. Altered Expression of Specific MicroRNAs in Plasma of Aneurysmal Subarachnoid Hemorrhage Patients. Front. Neurol. 2022, 13, 842888. [Google Scholar] [CrossRef]
- Jin, H.; Jiang, Y.; Liu, X.; Meng, X.; Li, Y. Cell-free microRNA-21: Biomarker for intracranial aneurysm rupture. Chin. Neurosurg. J. 2020, 6, 15. [Google Scholar] [CrossRef]
- Liao, B.; Zhou, M.X.; Zhou, F.K.; Luo, X.M.; Zhong, S.X.; Zhou, Y.F.; Qin, Y.S.; Li, P.P.; Qin, C. Exosome-Derived MiRNAs as Biomarkers of the Development and Progression of Intracranial Aneurysms. J. Atheroscler. Thromb. 2020, 27, 545–610. [Google Scholar] [CrossRef]
- Chen, L.; Wan, J.Q.; Zhou, J.P.; Fan, Y.L.; Jiang, J.Y. Gene expression analysis of ruptured and un-ruptured saccular intracranial aneurysm. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 1374–1381. [Google Scholar] [PubMed]
- Wei, L.; Gao, Y.J.; Wei, S.P.; Zhang, Y.F.; Zhang, W.F.; Jiang, J.X.; Sun, Z.Y.; Xu, W. Transcriptome network-based method to identify genes associated with unruptured intracranial aneurysms. Genet. Mol. Res. 2013, 12, 3263–3273. [Google Scholar] [CrossRef]
- Chen, L.; Fan, Y.; Wan, J. Screening of key genes of unruptured intracranial aneurysms by using DNA microarray data analysis techniques. Genet. Mol. Res. 2014, 13, 758–767. [Google Scholar] [CrossRef]
- Wei, L.; Wang, Q.; Zhang, Y.; Yang, C.; Guan, H.; Chen, Y.; Sun, Z. Identification of key genes, transcription factors and microRNAs involved in intracranial aneurysm. Mol. Med. Rep. 2018, 17, 891–897. [Google Scholar] [CrossRef]
- Bo, L.; Wei, B.; Wang, Z.; Li, C.; Gao, Z.; Miao, Z. Bioinformatic analysis of gene expression profiling of intracranial aneurysm. Mol. Med. Rep. 2018, 17, 3473–3480. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Hou, D.; Yu, D. Bioinformatics analysis of gene expression profile data to screen key genes involved in intracranial aneurysms. Mol. Med. Rep. 2019, 20, 4415–4424. [Google Scholar] [CrossRef]
- Pan, Y.B.; Lu, J.; Yang, B.; Lenahan, C.; Zhang, J.; Shao, A. Construction of competitive endogenous RNA network reveals regulatory role of long non-coding RNAs in intracranial aneurysm. BMC Neurosci. 2021, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.M.; Zebraski, H.R.; Rajabzadeh-Oghaz, H.; Chaves, L.; Dmytriw, A.A.; Siddiqui, A.H.; Kolega, J.; Poppenberg, K.E. RNA Sequencing Data from Human Intracranial Aneurysm Tissue Reveals a Complex Inflammatory Environment Associated with Rupture. Mol. Diagn. Ther. 2021, 25, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhong, P.; Yue, P.; Sun, Z. Uncovering of Key Pathways and miRNAs for Intracranial Aneurysm Based on Weighted Gene Co-Expression Network Analysis. Eur. Neurol. 2022, 85, 212–223. [Google Scholar] [CrossRef]
- Zheng, X.; Xue, C.; Luo, G.; Hu, Y.; Luo, W.; Sun, X. Identification of crucial genes in intracranial aneurysm based on weighted gene coexpression network analysis. Cancer Gene Ther. 2015, 22, 238–245. [Google Scholar] [CrossRef]
- Wang, K.; Wang, X.; Lv, H.; Cui, C.; Leng, J.; Xu, K.; Yu, G.; Chen, J.; Cong, P. Identification of the miRNA-target gene regulatory network in intracranial aneurysm based on microarray expression data. Exp. Ther. Med. 2017, 13, 3239–3248. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, Q.; Zhang, Y.; Yang, C.; Guan, H.; Jiang, J.; Sun, Z. Integrated analysis of microarray data to identify the genes critical for the rupture of intracranial aneurysm. Oncol. Lett. 2018, 15, 4951–4957. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, X.; Yi, D.; Song, Y.; Zhao, Y.H.; Luo, Q. Expression profile analysis of differentially expressed genes in ruptured intracranial aneurysms: In search of biomarkers. Biochem. Biophys. Res. Commun. 2018, 506, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Landry, A.P.; Balas, M.; Spears, J.; Zador, Z. Microenvironment of ruptured cerebral aneurysms discovered using data driven analysis of gene expression. PLoS ONE 2019, 14, e0220121. [Google Scholar] [CrossRef]
- Cheng, Q.; Li, Z.; Wang, R.; Zhang, H.; Cao, H.; Chen, F.; Li, H.; Xia, Z.; Feng, S.; Zhang, H.; et al. Genetic Profiles Related to Pathogenesis in Sporadic Intracranial Aneurysm Patients. World Neurosurg. 2019, 131, e23–e31. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, C.; Wang, J.; Li, H.; Yang, B. The potential biomarkers for the formation and development of intracranial aneurysm. J. Clin. Neurosci. 2020, 81, 270–278. [Google Scholar] [CrossRef]
- Chen, S.; Yang, D.; Liu, B.; Wang, L.; Chen, Y.; Ye, W.; Liu, C.; Ni, L.; Zhang, X.; Zheng, Y. Identification and validation of key genes mediating intracranial aneurysm rupture by weighted correlation network analysis. Ann. Transl. Med. 2020, 8, 1407. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Geng, D.; Zhou, K.; Fan, Y.; Su, R.; Zhou, Q.; Liu, B.; Duysenbi, S. Identification of potential key pathways, genes and circulating markers in the development of intracranial aneurysm based on weighted gene co-expression network analysis. Artif. Cells Nanomed. Biotechnol. 2020, 48, 999–1007. [Google Scholar] [CrossRef]
- Zhong, A.; Ding, N.; Zhou, Y.; Yang, G.; Peng, Z.; Zhang, H.; Chai, X. Identification of Hub Genes Associated with the Pathogenesis of Intracranial Aneurysm via Integrated Bioinformatics Analysis. Int. J. Gen. Med. 2021, 14, 4039–4050. [Google Scholar] [CrossRef]
- Sun, R.; Zhou, Y.; Cui, Q. Comparative analysis of aneurysm subtypes associated genes based on protein-protein interaction network. BMC Bioinform. 2021, 22, 587. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Leng, J.; Lin, Q.; Zhou, F. Epithelial-mesenchymal transition related genes in unruptured aneurysms identified through weighted gene coexpression network analysis. Sci. Rep. 2022, 12, 225. [Google Scholar] [CrossRef]
- Zhu, H.; Tan, J.; Zhao, Y.; Wang, Z.; Wu, Z.; Li, M. Potential Role of the Chemotaxis System in Formation and Progression of Intracranial Aneurysms Through Weighted Gene Co-Expression Network Analysis. Int. J. Gen. Med. 2022, 15, 2217–2231. [Google Scholar] [CrossRef]
- Lin, Y.; Ma, H.Y.; Wang, Y.; He, J.; Liu, H.J. Identification of Potential Core Genes for the Rupture of Intracranial Aneurysms by a Bioinformatics Analysis. Front. Genet. 2022, 13, 875007. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhou, H.; Zhou, X.; Yang, L.; Xiong, Y.; Zhang, L. Comprehensive Analysis of Endoplasmic Reticulum Stress in Intracranial Aneurysm. Front. Cell Neurosci. 2022, 16, 865005. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Liu, D. Intracranial Aneurysms Induced by RUNX1 Through Regulation of NFKB1 in Patients with Hypertension-An Integrated Analysis Based on Multiple Datasets and Algorithms. Front. Neurol. 2022, 13, 877801. [Google Scholar] [CrossRef]
- Lu, T.; Liu, Z.; Guo, D.; Ma, C.; Duan, L.; He, Y.; Jia, R.; Guo, C.; Xing, Z.; Liu, Y.; et al. Transcriptome-Based Dissection of Intracranial Aneurysms Unveils an “Immuno-Thermal” Microenvironment and Defines a Pathological Feature-Derived Gene Signature for Risk Estimation. Front. Immunol. 2022, 13, 878195. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ma, Z.; Shang, J.; Cui, X.; Liu, J.; Shi, R.; Wang, S.; Wu, A. Bioinformatics analysis reveals potential biomarkers associated with the occurrence of intracranial aneurysms. Sci. Rep. 2022, 12, 13282. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Q.; Chen, Z.; Huang, Z.; Zhang, L.; Chen, F. Novel insight into ferroptosis-related genes, molecular subtypes, and immune characteristics in intracranial aneurysms. Inflamm. Res. 2022, 71, 1347–1364. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Luo, Q.; Yang, Z.; Zhao, Y.H.; Li, J.; Wang, J.; Piao, J.; Chen, X. Weighted gene co-expression network analysis identified six hub genes associated with rupture of intracranial aneurysms. PLoS ONE 2020, 15, e0229308. [Google Scholar] [CrossRef]
- Niu, S.; Zhao, Y.; Ma, B.; Zhang, R.; Rong, Z.; Ni, L.; Di, X.; Liu, C. Construction and Validation of a New Model for the Prediction of Rupture in Patients with Intracranial Aneurysms. World Neurosurg. 2021, 149, e437–e446. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, J. A Two-Gene-Based Diagnostic Signature for Ruptured Intracranial Aneurysms. Front. Cardiovasc. Med. 2021, 8, 671655. [Google Scholar] [CrossRef] [PubMed]
- Bo, L.; Wei, B.; Wang, Z.; Kong, D.; Gao, Z.; Miao, Z. Screening of Critical Genes and MicroRNAs in Blood Samples of Patients with Ruptured Intracranial Aneurysms by Bioinformatic Analysis of Gene Expression Data. Med. Sci. Monit. 2017, 23, 4518–4525. [Google Scholar] [CrossRef]
- Zhao, H.; Li, S.T.; Zhu, J.; Hua, X.M.; Wan, L. Analysis of Peripheral Blood Cells’ Transcriptome in Patients With Subarachnoid Hemorrhage From Ruptured Aneurysm Reveals Potential Biomarkers. World Neurosurg. 2019, 129, e16–e22. [Google Scholar] [CrossRef]
- Zhao, M.; Xu, L.; Qian, H. Bioinformatics analysis of microRNA profiles and identification of microRNA-mRNA network and biological markers in intracranial aneurysm. Medicine 2020, 99, e21186. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, X.; Chen, Z.; Liu, Y.; Zhang, X. Identification of inflammation-associated circulating long non-coding RNAs and genes in intracranial aneurysm rupture-induced subarachnoid hemorrhage. Mol. Med. Rep. 2020, 22, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Leng, W.; Fan, D.; Ren, Z.; Li, Q. Identification of upregulated NF-κB inhibitor alpha and IRAK3 targeting lncRNA following intracranial aneurysm rupture-induced subarachnoid hemorrhage. BMC Neurol. 2021, 21, 197. [Google Scholar] [CrossRef]
- Yan, Z.; Wu, Q.; Cai, W.; Xiang, H.; Wen, L.; Zhang, A.; Peng, Y.; Zhang, X.; Wang, H. Identifying critical genes associated with aneurysmal subarachnoid hemorrhage by weighted gene co-expression network analysis. Aging 2021, 13, 22345–22360. [Google Scholar] [CrossRef] [PubMed]
- Laarman, M.D.; Kleinloog, R.; Bakker, M.K.; Rinkel, G.J.E.; Bakkers, J.; Ruigrok, Y.M. Assessment of the Most Optimal Control Tissue for Intracranial Aneurysm Gene Expression Studies. Stroke 2019, 50, 2933–2936. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).