Identification and Characterization of Phytocyanin Family Genes in Cotton Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification, Sequence Alignment, and Phylogenetic Tree
2.2. Chromosomal Mapping and Collinearity Analysis
2.3. Gene Structure, Conserved Motifs, and Cis-Regulatory Elements
2.4. Selection Pressure
2.5. Transcriptomic Profiling of the Phytocyanin Gene Family in Cotton
3. Results
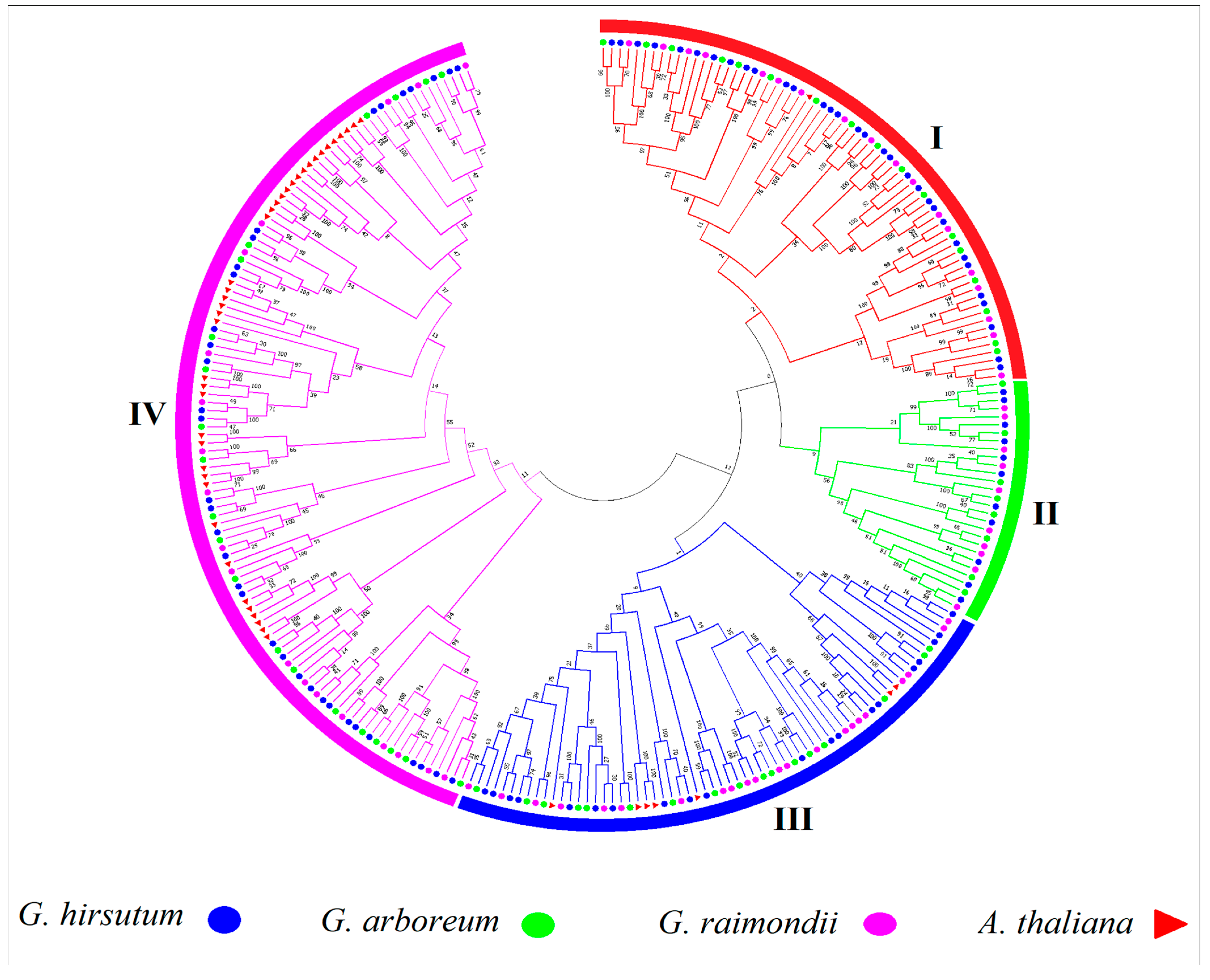
3.1. Genome-Wide Identification, Sequence Alignment, and Phylogenetic Tree
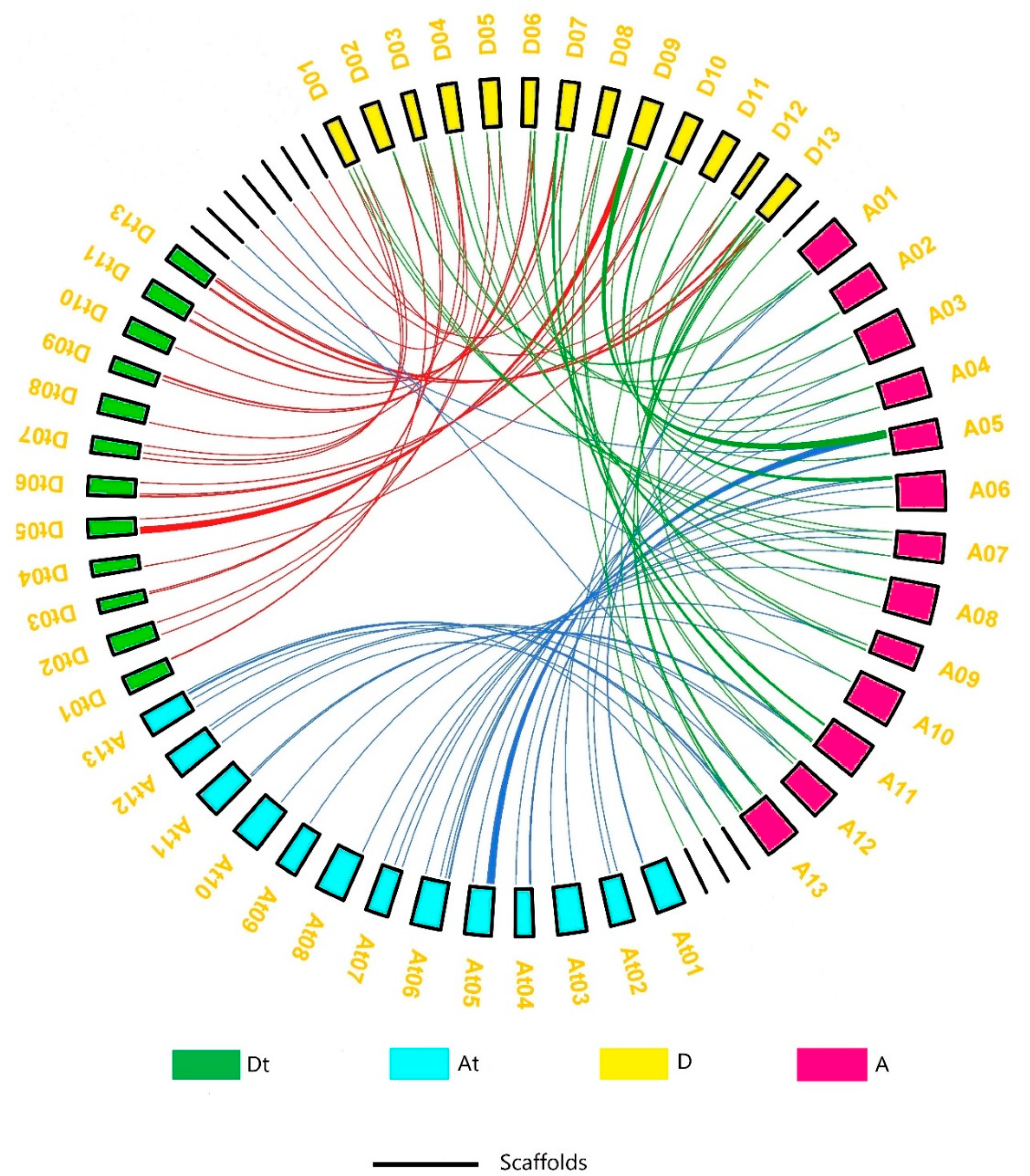
3.2. Chromosomal Mapping and Collinearity Analysis
3.3. Gene Structure, Conserved Motifs, and Cis-Regulatory Elements
3.4. Selection Pressure
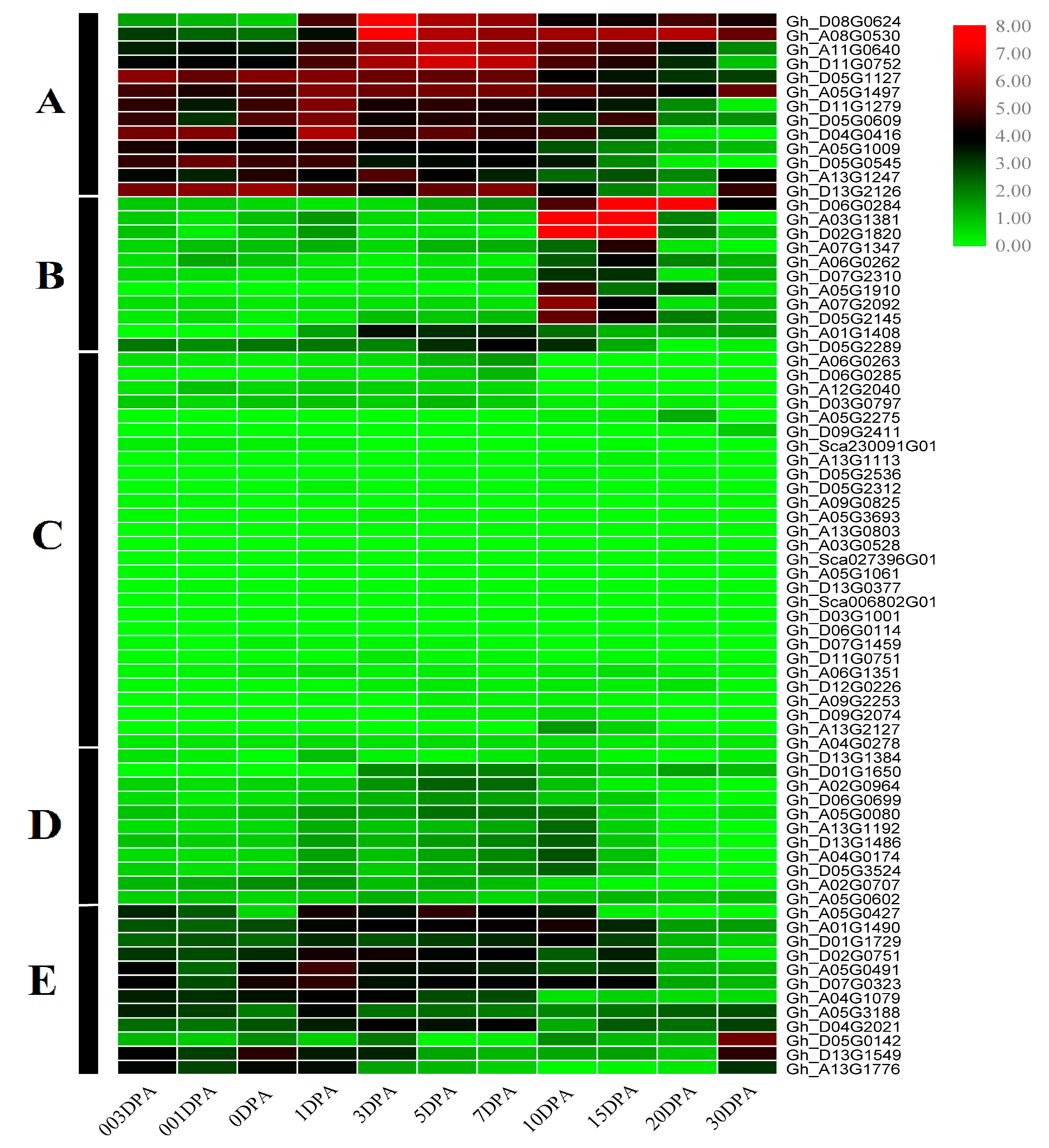
3.5. Transcriptome Profiling of PCs in Cotton
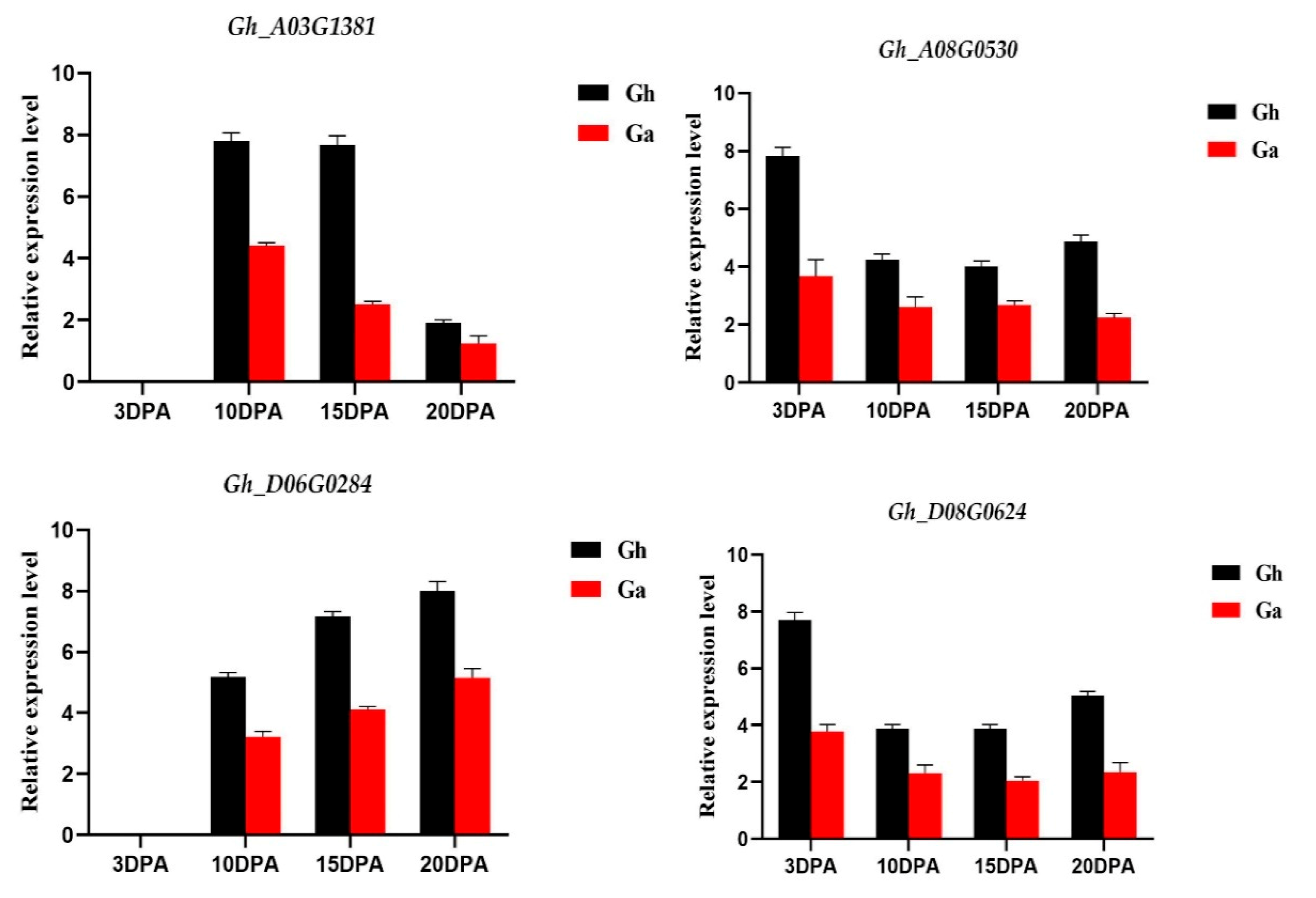
3.6. qRT-PCR Amplification of Four Homologous PC Gene Pairs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rydén, L.G.; Hunt, L.T. Evolution of Protein Complexity: The Blue Copper-Containing Oxidases and Related Proteins. J. Mol. Evol. 1993, 36, 41–66. [Google Scholar] [CrossRef] [PubMed]
- De Rienzo, F.; Gabdoulline, R.R.; Menziani, M.C.; Wade, R.C. Blue Copper Proteins: A Comparative Analysis of Their Molecular Interaction Properties. Protein Sci. 2000, 9, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.-M.; Luo, F.; Li, D.-D.; Zhang, J.; Liu, Z.-H.; Xu, W.-L.; Huang, G.-Q.; Li, X.-B. Cotton BCP Genes Encoding Putative Blue Copper-Binding Proteins Are Functionally Expressed in Fiber Development and Involved in Response to High-Salinity and Heavy Metal Stresses. Physiol. Plant. 2011, 141, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Garrett, T.P.; Clingeleffer, D.J.; Guss, J.M.; Rogers, S.J.; Freeman, H.C. The Crystal Structure of Poplar Apoplastocyanin at 1.8-A Resolution. The Geometry of the Copper-Binding Site Is Created by the Polypeptide. J. Biol. Chem. 1984, 259, 2822–2825. [Google Scholar] [CrossRef]
- Hart, P.J.; Eisenberg, D.; Nersissian, A.M.; Valentine, J.S.; Herrmann, R.G.; Nalbandyan, R.M. A Missing Link in Cupredoxins: Crystal Structure of Cucumber Stellacyanin at 1.6 Å Resolution. Protein Sci. 1996, 5, 2175–2183. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Asami, T.; Suzuki, Y. Genome-Wide Identification, Structure and Expression Studies, and Mutant Collection of 22 Early Nodulin-Like Protein Genes in Arabidopsis. Biosci. Biotechnol. Biochem. 2009, 73, 2452–2459. [Google Scholar] [CrossRef]
- Ma, H.; Zhao, H.; Liu, Z.; Zhao, J. The Phytocyanin Gene Family in Rice (Oryza Sativa L.): Genome-Wide Identification, Classification and Transcriptional Analysis. PLoS ONE 2011, 6, e25184. [Google Scholar] [CrossRef]
- Li, J.; Gao, G.; Zhang, T.; Wu, X. The Putative Phytocyanin Genes in Chinese Cabbage (Brassica Rapa L.): Genome-Wide Identification, Classification and Expression Analysis. Mol. Genet. Genom. 2013, 288, 1–20. [Google Scholar] [CrossRef]
- Nersissian, A.M.; Valentine, J.S.; Immoos, C.; Hill, M.G.; Hart, P.J.; Williams, G.; Herrmann, R.G. Uclacyanins, Stellacyanins, and Plantacyanins Are Distinct Subfamilies of Phytocyanins: Plant-Specific Mononuclear Blue Copper Proteins. Protein Sci. 1998, 7, 1915–1929. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Yamaguchi, I.; Suzuki, Y. Isolation and Identification of Glycosylphosphatidylinositol-Anchored Arabinogalactan Proteins and Novel β-Glucosyl Yariv-Reactive Proteins from Seeds of Rice (Oryza Sativa). Plant Cell Physiol. 2004, 45, 1817–1829. [Google Scholar] [CrossRef]
- Mann, K.; Schäfer, W.; Thoenes, U.; Messerschmidt, A.; Mehrabian, Z.; Nalbandyan, R. The Amino Acid Sequence of a Type I Copper Protein with an Unusual Serine- and Hydroxyproline-Rich C-terminal Domain Isolated from Cucumber Peelings. FEBS Lett. 1992, 314, 220–223. [Google Scholar] [CrossRef]
- Driessche, G.V.; Beeumen, J.V.; Dennison, C.; Geoffrey Sykes, A. Heterogeneity of the Covalent Structure of the Blue Copper Protein Umecyanin from Horseradish Roots. Protein Sci. 1995, 4, 209–227. [Google Scholar] [CrossRef]
- Greene, E.A.; Erard, M.; Dedieu, A.; Barker, D.G. MtENOD16 and 20 Are Members of a Family of Phytocyanin-Related Early Nodulins. Plant Mol. Biol. 1998, 36, 775–783. [Google Scholar] [CrossRef]
- Scheres, B.; van Engelen, F.; van der Knaap, E.; Clemens van de, W.; van Kammen, A.; Bisseling, T. Sequential Lnduction of Nodulin Gene Expression in the Developing Pea Nodule. Plant Cell 1990, 2, 687–700. [Google Scholar]
- Vijn, I.; Yang, W.-C.; Pallisgård, N.; Jensen, E.Ø.; van Kammen, A.; Bisseling, T. VsENOD5, VsENOD12 and VsENOD40 Expression during Rhizobium-Induced Nodule Formation on Vicia Sativa Roots. Plant Mol. Biol. 1995, 28, 1111–1119. [Google Scholar] [CrossRef]
- Denancé, N.; Szurek, B.; Noël, L.D. Emerging Functions of Nodulin-Like Proteins in Non-Nodulating Plant Species. Plant Cell Physiol. 2014, 55, 469–474. [Google Scholar] [CrossRef]
- Pugh, D.A.; Offler, C.E.; Talbot, M.J.; Ruan, Y.-L. Evidence for the Role of Transfer Cells in the Evolutionary Increase in Seed and Fiber Biomass Yield in Cotton. Mol. Plant 2010, 3, 1075–1086. [Google Scholar] [CrossRef]
- Serna, L.; Martin, C. Trichomes: Different Regulatory Networks Lead to Convergent Structures. Trends Plant Sci. 2006, 11, 274–280. [Google Scholar] [CrossRef]
- Fryxell, P.A. A Revised Taxonomic Interpretation of Gossypium L. (Malvaceae). Rheedea 1992, 2, 108–165. [Google Scholar]
- Zhang, D.-Y.; Zhang, T.-Z.; Sang, Z.-Q.; Guo, W.-Z. Comparative Development of Lint and Fuzz Using Different Cotton Fiber-Specific Developmental Mutants in Gossypium Hirsutum. J. Integr. Plant Biol. 2007, 49, 1038–1046. [Google Scholar] [CrossRef]
- Turley, R.B.; Kloth, R.H. Identification of a Third Fuzzless Seed Locus in Upland Cotton (Gossypium Hirsutum L.). J. Hered. 2002, 93, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Wakelyn, P.J.; Bertoniere, N.R.; French, A.D.; Thibodeaux, D.P.; Triplett, B.A.; Rousselle, M.-A.; Goynes, W.R., Jr.; Edwards, J.V.; Hunter, L.; McAlister, D.D.; et al. Cotton Fiber Chemistry and Technology, 1st ed.; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Stewart, J. McD. Fiber initiation on the cotton ovule (Gossypium hirsutum). Am. J. Bot. 1975, 62, 723–730. [Google Scholar] [CrossRef]
- Seagull, R.W.; Giavalis, S. Pre-and post-anthesis application of exogenous hormones alters fiber production in Gossypium hirsutum L. cultivar Maxxa GTO. Mol. Biol. Evol. 2004, 8, 7. [Google Scholar]
- Li, C.; Guo, W.; Zhang, T. Fiber Initiation Development in Upland Cotton (Gossypium Hirsutum L.) Cultivars Varying in Lint Percentage. Euphytica 2009, 165, 223. [Google Scholar] [CrossRef]
- Butterworth, K.M.; Adams, D.C.; Horner, H.T.; Wendel, J.F. Initiation and Early Development of Fiber in Wild and Cultivated Cotton. Int. J. Plant Sci. 2009, 170, 561–574. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Llewellyn, D.J.; Furbank, R.T. Suppression of Sucrose Synthase Gene Expression Represses Cotton Fiber Cell Initiation, Elongation, and Seed Development. Plant Cell 2003, 15, 952–964. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-Wide Comparative Analysis of Long-Chain Acyl-CoA Synthetases (LACSs) Gene Family: A Focus on Identification, Evolution and Expression Profiling Related to Lipid Synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An Integrated Functional Genomics Database for Cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a Core Collection of Upland Cotton Identifies Genomic Variation and Loci Influencing Fiber Quality and Yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Li, D.; Zaman, W.; Lu, J.; Niu, Q.; Zhang, X.; Ayaz, A.; Saqib, S.; Yang, B.; Zhang, J.; Zhao, H.; et al. Natural Lupeol Level Variation among Castor Accessions and the Upregulation of Lupeol Synthesis in Response to Light. Ind. Crops Prod. 2023, 192, 116090. [Google Scholar] [CrossRef]
- Rychlik, W. OLIGO 7 Primer Analysis Software. PCR Primer Des. 2007, 25, 35–39. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hafeez, A.; Gě, Q.; Zhāng, Q.; Lǐ, J.; Gōng, J.; Liú, R.; Shí, Y.; Shāng, H.; Liú, À.; Iqbal, M.S.; et al. Multi-Responses of O-Methyltransferase Genes to Salt Stress and Fiber Development of Gossypium Species. BMC Plant Biol. 2021, 21, 37. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The Draft Genome of a Diploid Cotton Gossypium Raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 Diploid Cotton Accessions Based on an Updated A Genome Identifies the Genetic Basis of Key Agronomic Traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium Barbadense and Gossypium Hirsutum Genomes Provide Insights into the Origin and Evolution of Allotetraploid Cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef]
- Gan, X.; Stegle, O.; Behr, J.; Steffen, J.G.; Drewe, P.; Hildebrand, K.L.; Lyngsoe, R.; Schultheiss, S.J.; Osborne, E.J.; Sreedharan, V.T.; et al. Multiple Reference Genomes and Transcriptomes for Arabidopsis Thaliana. Nature 2011, 477, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P. The Evolutionary Consequences of Polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the Bread Wheat Genome Using Whole-Genome Shotgun Sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Lin, K.; Bonnema, G.; Wang, X. Biased Gene Fractionation and Dominant Gene Expression among the Subgenomes of Brassica Rapa. PLoS ONE 2012, 7, e36442. [Google Scholar] [CrossRef]
- Grover, C.E.; Gallagher, J.P.; Szadkowski, E.P.; Yoo, M.J.; Flagel, L.E.; Wendel, J.F. Homoeolog Expression Bias and Expression Level Dominance in Allopolyploids. New Phytol. 2012, 196, 966–971. [Google Scholar] [CrossRef]
- Leach, L.J.; Belfield, E.J.; Jiang, C.; Brown, C.; Mithani, A.; Harberd, N.P. Patterns of Homoeologous Gene Expression Shown by RNA Sequencing in Hexaploid Bread Wheat. BMC Genom. 2014, 15, 276. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of Allotetraploid Cotton (Gossypium Hirsutum L. Acc. TM-1) Provides a Resource for Fiber Improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Gě, Q.; Cūi, Y.; Lǐ, J.; Gōng, J.; Lú, Q.; Lǐ, P.; Shí, Y.; Shāng, H.; Liú, À.; Dèng, X.; et al. Disequilibrium Evolution of the Fructose-1,6-Bisphosphatase Gene Family Leads to Their Functional Biodiversity in Gossypium Species. BMC Genom. 2020, 21, 379. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Madhava Rao, K.V.; Raghavendra, A.S.; Janardhan Reddy, K. (Eds.) Physiology and Molecular Biology of Stress Tolerance in Plants; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2006; ISBN 978-1-4020-4224-9. [Google Scholar]
- Xu, P.; Liu, Z.; Fan, X.; Gao, J.; Zhang, X.; Zhang, X.; Shen, X. De Novo Transcriptome Sequencing and Comparative Analysis of Differentially Expressed Genes in Gossypium Aridum under Salt Stress. Gene 2013, 525, 26–34. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY Superfamily of Plant Transcription Factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Hara, K.; Yagi, M.; Kusano, T.; Sano, H. Rapid Systemic Accumulation of Transcripts Encoding a Tobacco WRKY Transcription Factor upon Wounding. Mol. Gen. Genet. 2000, 263, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Maleck, K.; Levine, A.; Eulgem, T.; Morgan, A.; Schmid, J.; Lawton, K.A.; Dangl, J.L.; Dietrich, R.A. The Transcriptome of Arabidopsis Thaliana during Systemic Acquired Resistance. Nat. Genet. 2000, 26, 403–410. [Google Scholar] [CrossRef]
- Huang, T.; Nicodemus, J.; Zarka, D.G.; Thomashow, M.F.; Duman, J.G. Expression of an Insect (Dendroides Canadensis) Antifreeze Protein in Arabidopsis Thaliana Results in a Decrease in Plant Freezing Temperature. Plant Mol. Biol. 2002, 50, 333–344. [Google Scholar] [CrossRef]
- Pnueli, L.; Hallak-Herr, E.; Rozenberg, M.; Cohen, M.; Goloubinoff, P.; Kaplan, A.; Mittler, R. Molecular and Biochemical Mechanisms Associated with Dormancy and Drought Tolerance in the Desert Legume Retama Raetam: Dormancy and Drought Tolerance in Retama Raetam. Plant J. 2002, 31, 319–330. [Google Scholar] [CrossRef]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the Expression Pattern of around 7,000 Arabidopsis Genes under ABA Treatments Using a Full-Length CDNA Microarray. Funct. Integr. Genom. 2002, 2, 282–291. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When Defense Pathways Collide. The Response of Arabidopsis to a Combination of Drought and Heat Stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Bastian, R.; Donaldson, L.; Murray, S.; Bajic, V.; Gehring, C. Co-Expression and Promoter Content Analyses Assign a Role in Biotic and Abiotic Stress Responses to Plant Natriuretic Peptides. BMC Plant Biol. 2008, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Kazuko, Y.-S.; Urao, T.; Iwasak, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB Homologs in Drought- and Abscisic Acid-Regulated Gene Expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Zhang, Y.; Sun, T.; Liu, S.; Dong, L.; Liu, C.; Song, W.; Liu, J.; Gai, S. MYC Cis-Elements in PsMPT Promoter Is Involved in Chilling Response of Paeonia Suffruticosa. PLoS ONE 2016, 11, e0155780. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bilal Tufail, M.; Yasir, M.; Zuo, D.; Cheng, H.; Ali, M.; Hafeez, A.; Soomro, M.; Song, G. Identification and Characterization of Phytocyanin Family Genes in Cotton Genomes. Genes 2023, 14, 611. https://doi.org/10.3390/genes14030611
Bilal Tufail M, Yasir M, Zuo D, Cheng H, Ali M, Hafeez A, Soomro M, Song G. Identification and Characterization of Phytocyanin Family Genes in Cotton Genomes. Genes. 2023; 14(3):611. https://doi.org/10.3390/genes14030611
Chicago/Turabian StyleBilal Tufail, Muhammad, Muhammad Yasir, Dongyun Zuo, Hailiang Cheng, Mushtaque Ali, Abdul Hafeez, Mahtab Soomro, and Guoli Song. 2023. "Identification and Characterization of Phytocyanin Family Genes in Cotton Genomes" Genes 14, no. 3: 611. https://doi.org/10.3390/genes14030611
APA StyleBilal Tufail, M., Yasir, M., Zuo, D., Cheng, H., Ali, M., Hafeez, A., Soomro, M., & Song, G. (2023). Identification and Characterization of Phytocyanin Family Genes in Cotton Genomes. Genes, 14(3), 611. https://doi.org/10.3390/genes14030611