Application of OpenArray Technology to Assess Changes in the Expression of Functionally Significant Genes in the Substantia Nigra of Mice in a Model of Parkinson’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Procedures
2.2. Collection of Biological Material
2.3. High-Performance Liquid Chromatography with Electrochemical Detection
2.4. Reverse Transcription Quantitative Real-Time Polymerase Chain Reaction
2.5. Statistics
3. Results and Discussion
3.1. Characteristics of the Parkinson’s Disease Model
3.2. Selection of Stably Expressed Housekeeping Genes Based on Analysis of Their Expression in Normal Conditions and in Modeling Parkinson’s Disease
3.3. Assessing the Gene Expression Using OpenArray and Developing a Panel of Highly Expressed Genes
- DA synthesis and degradation: Tyrosine hydroxylase (Th), dopa decarboxylase (Ddc), dopamine β-hydroxylase (Dbh), phenylethanolamine N-methyltransferase (Pnmt), monoamine oxidase A and B (Maoa and Maob), and catechol-O-methyltransferase (Comt);
- DA transport, DA receptors, and transcriptional factors: DA transporter (Slc6a3), vesicular monoamine transporter 1 and 2 (Slc18a1 and Slc18a2), plasma membrane monoamine transporter (Slc29a4), DA receptors 1–5 types (Drd1–Drd5), nuclear receptor subfamily 4 group A member 2 (Nurr1, Nr4a2); and paired-like homeodomain 3 (Pitx3);
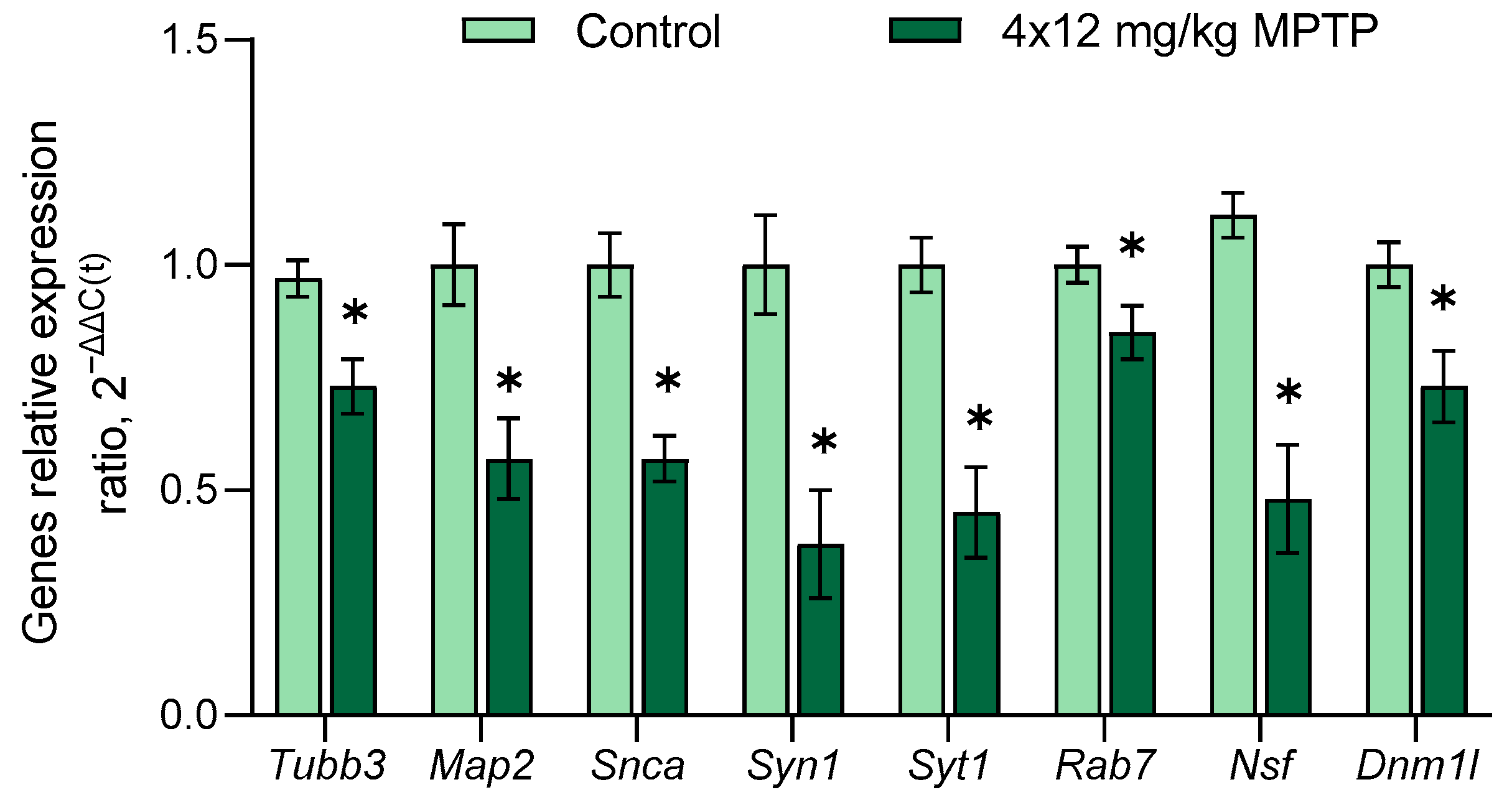
- Axonal transport and microtubules: Kinesin (Kif1a, Kif1b, Kif5a, and Kif2c), dynein (Dync1h1 and Dynll1), dynactin 1 (Dctn1), τau-protein (Mapt), microtubule-associated protein 2 (Map2), MAP/microtubule affinity regulating kinase 2 (Mark2), and tubulin (Tubb3, Tuba1a);
- Vesicle cycle for neurotransmission: α-synuclein (Snca), synapsin 1 (Syn1), syntaxin 1A (Stx1a), synaptotagmin 1 and 11 (Syt1, Syt11), Rab protein 5a и 7 (Rab5a, Rab7), N-ethylmaleimide sensitive fusion protein (Nsf), dynamin 1-like protein (Dnm1l), and vacuolar protein sorting ortholog 35 (Vps35);
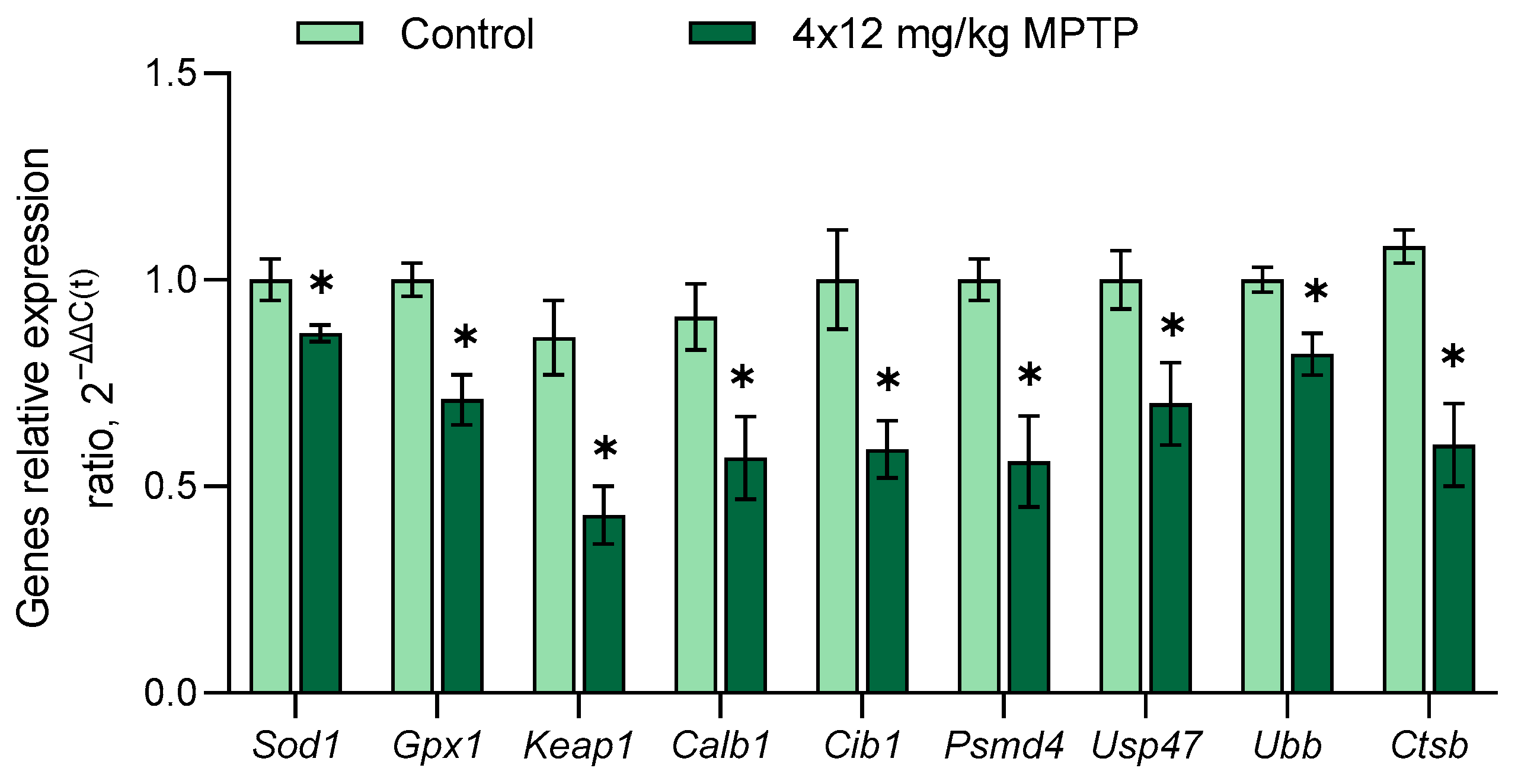
- Neuroprotection: Superoxide dismutase 1 (Sod1), glutathione peroxidase 1 (Gpx1), glutathione reductase (Gsr), thioredoxin reductase 1 (Txnrd1), nitric oxide synthase 1 (Nos1), peroxiredoxin 1 (Prdx1), nuclear factor erythroid 2-related factor 2 (Nfe2l2), angiotensin II receptor type 2 (Agtr2), sigma-1 receptor (Sigmar1), kelch-like ECH-associated protein 1 (Keap1), brain-derived neurotrophic factor (Bdnf), glial cell-derived neurotrophic factor (Gdnf), nerve growth factor (Ngf), vascular endothelial growth factor A (Vegfa), cerebral DA neurotrophic factor (Cdnf), neurotrophic tyrosine kinase receptor types 1 and 2 (Ntrk1 and Ntrk2), nerve growth factor receptor (Ngfr), matrix metalloproteinase-3 (Mmp3), Wnt family member 11 (Wnt11), catenin β-1 (Ctnnb1), and calbindin 1 (Calb1);
- Protein degradation: Calcium channel voltage-dependent L type alpha 1D subunit (Cacna1d), transient receptor potential cation channel subfamily M member 2 (Trpm2), E3 ubiquitin ligase (Parkin) (Park2), ubiquitin-conjugating enzyme E2N (Ube2n), ubiquitin-like modifier activating enzyme 3 (Uba3), proteasome 20S subunit beta 4 (Psmb4), proteasome 26S subunit ATPase 3 (Psmc3), proteasome 26S subunit non-ATPase 4 (Psmd4), ubiquitin-specific peptidase 47 (Usp47), ubiquitin B (Ubb), and cathepsin B (Ctsb);
- Cell death: Caspases 1 and 3 (Casp1 and Casp3), poly [ADP-ribose] polymerase 1 (Parp1), apoptosis-inducing factor mitochondria associated 1 (Aifm1), calcium and integrin binding 1 (Cib1), transformation-related protein 53 (Trp53), Bax protein (Bax), c-Fos protein (Fos), mitogen-activated protein kinase 8 (Mapk8), lysosomal-associated membrane protein 2 (Lamp2), autophagy-related 16-like 1 and 5 (Atg16l1 and Atg5), calpain-1 (Capn1), tumor necrosis factor (Tnf), endoplasmic reticulum to nucleus signaling 2 (Ern2), eukaryotic translation initiation factor 2-alpha kinase 3 (Ef2ak3), and activating transcription factor 6 (Atf6);
- Inflammation and glial activation: Glial fibrillary acidic protein (Gfap), interferon gamma (Ifng), transforming growth factor beta 1 (Tgfb1), protein kinase B alpha (Akt1), cannabinoid receptor 1 (Cnr1), prostaglandin-endoperoxide synthase 2 (Ptgs2), CDC like kinase 1 (Clk1), transforming growth factor beta 1 (Traf1), C-X-C motif chemokine 11 (Cxcl11).
3.4. Evaluation of Gene Expression in the Substantia Nigra in a Mouse Model of Parkinson’s Disease
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3-MT | 3-methoxytyramine |
| DA | dopamine |
| DAergic | dopaminergic |
| DOPAC | 3,4-dihydroxyphenylacetic acid |
| HKG | housekeeping gene |
| HVA | homovanillic acid |
| MPTP | 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine |
| PD | Parkinson’s disease |
| RG | reference gene |
| RT-qPCR | Reverse-transcription quantitative real-time polymerase chain reaction |
| SN | substantia nigra |
References
- Agid, Y. Parkinson’s Disease: Pathophysiology. Lancet 1991, 337, 1321–1324. [Google Scholar] [CrossRef]
- Eriksen, J.L.; Wszolek, Z.; Petrucelli, L. Molecular Pathogenesis of Parkinson Disease. Arch. Neurol. 2005, 62, 353–357. [Google Scholar] [CrossRef] [PubMed]
- GBD 2016 Disease and Injury Incidence and Prevalence Collaborators Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 328 Diseases and Injuries for 195 Countries, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [CrossRef]
- Dorsey, E.R.; Constantinescu, R.; Thompson, J.P.; Biglan, K.M.; Holloway, R.G.; Kieburtz, K.; Marshall, F.J.; Ravina, B.M.; Schifitto, G.; Siderowf, A.; et al. Projected Number of People with Parkinson Disease in the Most Populous Nations, 2005 through 2030. Neurology 2007, 68, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Tatton, W.G. Etiology and Pathogenesis of Parkinson’s Disease. Annu. Rev. Neurosci. 1999, 22, 123–144. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Wood, N.W. Molecular Pathogenesis of Parkinson’s Disease. Hum. Mol. Genet. 2005, 14, 2749–2755. [Google Scholar] [CrossRef]
- Müller, T. Safinamide for Symptoms of Parkinson’s Disease. Drugs Today 2015, 51, 653–659. [Google Scholar] [CrossRef]
- Cacabelos, R. Parkinson’s Disease: From Pathogenesis to Pharmacogenomics. Int. J. Mol. Sci. 2017, 18, 551. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s Disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Postuma, R.B.; Berg, D. Advances in Markers of Prodromal Parkinson Disease. Nat. Rev. Neurol. 2016, 12, 622–634. [Google Scholar] [CrossRef]
- Blesa, J.; Trigo-Damas, I.; Dileone, M.; Del Rey, N.L.-G.; Hernandez, L.F.; Obeso, J.A. Compensatory Mechanisms in Parkinson’s Disease: Circuits Adaptations and Role in Disease Modification. Exp. Neurol. 2017, 298, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Fabbrini, G.; Brotchie, J.M.; Grandas, F.; Nomoto, M.; Goetz, C.G. Levodopa-Induced Dyskinesias. Mov. Disord. 2007, 22, 1379–1389. [Google Scholar] [CrossRef]
- Chotibut, T.; Fields, V.; Salvatore, M.F. Norepinephrine Transporter Inhibition with Desipramine Exacerbates L-DOPA-Induced Dyskinesia: Role for Synaptic Dopamine Regulation in Denervated Nigrostriatal Terminals. Mol. Pharmacol. 2014, 86, 675–685. [Google Scholar] [CrossRef]
- Malek, N. Deep Brain Stimulation in Parkinson’s Disease. Neurol. India 2019, 67, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.K.; Kwatra, M.; Wang, J.; Ko, H.S. Levodopa-Induced Dyskinesia in Parkinson’s Disease: Pathogenesis and Emerging Treatment Strategies. Cells 2022, 11, 3736. [Google Scholar] [CrossRef] [PubMed]
- Ugrumov, M.V.; Khaindrava, V.G.; Kozina, E.A.; Kucheryanu, V.G.; Bocharov, E.V.; Kryzhanovsky, G.N.; Kudrin, V.S.; Narkevich, V.B.; Klodt, P.M.; Rayevsky, K.S.; et al. Modeling of Presymptomatic and Symptomatic Stages of Parkinsonism in Mice. Neuroscience 2011, 181, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Jackson-Lewis, V.; Przedborski, S. Protocol for the MPTP Mouse Model of Parkinson’s Disease. Nat. Protoc. 2007, 2, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Lama, J.; Buhidma, Y.; Fletcher, E.J.R.; Duty, S. Animal Models of Parkinson’s Disease: A Guide to Selecting the Optimal Model for Your Research. Neuronal Signal. 2021, 5, NS20210026. [Google Scholar] [CrossRef]
- Jagmag, S.A.; Tripathi, N.; Shukla, S.D.; Maiti, S.; Khurana, S. Evaluation of Models of Parkinson’s Disease. Front. Neurosci. 2015, 9, 503. [Google Scholar] [CrossRef]
- Bye, C.R.; Jönsson, M.E.; Björklund, A.; Parish, C.L.; Thompson, L.H. Transcriptome Analysis Reveals Transmembrane Targets on Transplantable Midbrain Dopamine Progenitors. Proc. Natl. Acad. Sci. USA 2015, 112, E1946–E1955. [Google Scholar] [CrossRef]
- Agarwal, D.; Sandor, C.; Volpato, V.; Caffrey, T.M.; Monzón-Sandoval, J.; Bowden, R.; Alegre-Abarrategui, J.; Wade-Martins, R.; Webber, C. A Single-Cell Atlas of the Human Substantia Nigra Reveals Cell-Specific Pathways Associated with Neurological Disorders. Nat. Commun. 2020, 11, 4183. [Google Scholar] [CrossRef]
- Troshev, D.; Blokhin, V.; Ukrainskaya, V.; Kolacheva, A.; Ugrumov, M. Isolation of Living Dopaminergic Neurons Labeled with a Fluorescent Ligand of the Dopamine Transporter from Mouse Substantia Nigra as a New Tool for Basic and Applied Research. Front. Mol. Neurosci. 2022, 15, 1020070. [Google Scholar] [CrossRef] [PubMed]
- Mingazov, E.R.; Khakimova, G.R.; Kozina, E.A.; Medvedev, A.E.; Buneeva, O.A.; Bazyan, A.S.; Ugrumov, M.V. MPTP Mouse Model of Preclinical and Clinical Parkinson’s Disease as an Instrument for Translational Medicine. Mol. Neurobiol. 2018, 55, 2991–3006. [Google Scholar] [CrossRef] [PubMed]
- Kolacheva, A.; Alekperova, L.; Pavlova, E.; Bannikova, A.; Ugrumov, M.V. Changes in Tyrosine Hydroxylase Activity and Dopamine Synthesis in the Nigrostriatal System of Mice in an Acute Model of Parkinson’s Disease as a Manifestation of Neurodegeneration and Neuroplasticity. Brain Sci. 2022, 12, 779. [Google Scholar] [CrossRef]
- Coku, I.; Mutez, E.; Eddarkaoui, S.; Carrier, S.; Marchand, A.; Deldycke, C.; Goveas, L.; Baille, G.; Tir, M.; Magnez, R.; et al. Functional Analyses of Two Novel LRRK2 Pathogenic Variants in Familial Parkinson′s Disease. Mov. Disord. 2022, 37, 1761–1767. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, H.; Ohye, T.; Fujita, K.; Pantucek, F.; Lange, K.; Riederer, P.; Nagatsu, T. Quantification of mRNA of Tyrosine Hydroxylase and Aromatic L-Amino Acid Decarboxylase in the Substantia Nigra in Parkinson’s Disease and Schizophrenia. J. Neural Transm. Park. Dis. Dement. Sect. 1994, 8, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Noureddine, M.A.; Li, Y.-J.; van der Walt, J.M.; Walters, R.; Jewett, R.M.; Xu, H.; Wang, T.; Walter, J.W.; Scott, B.L.; Hulette, C.; et al. Genomic Convergence to Identify Candidate Genes for Parkinson Disease: SAGE Analysis of the Substantia Nigra. Mov. Disord. 2005, 20, 1299–1309. [Google Scholar] [CrossRef]
- Duke, D.C.; Moran, L.B.; Pearce, R.K.B.; Graeber, M.B. The Medial and Lateral Substantia Nigra in Parkinson’s Disease: mRNA Profiles Associated with Higher Brain Tissue Vulnerability. Neurogenetics 2007, 8, 83–94. [Google Scholar] [CrossRef]
- Chang, K.-H.; Chen, C.-M. The Role of Oxidative Stress in Parkinson’s Disease. Antioxidants 2020, 9, 597. [Google Scholar] [CrossRef]
- Calabresi, P.; Mechelli, A.; Natale, G.; Volpicelli-Daley, L.; Di Lazzaro, G.; Ghiglieri, V. α-Synuclein in Parkinson’s Disease and Other Synucleinopathies: From Overt Neurodegeneration Back to Early Synaptic Dysfunction. Cell Death Dis. 2023, 14, 176. [Google Scholar] [CrossRef]
- Bondarenko, O.; Saarma, M. Neurotrophic Factors in Parkinson’s Disease: Clinical Trials, Open Challenges and Nanoparticle-Mediated Delivery to the Brain. Front. Cell. Neurosci. 2021, 15, 682597. [Google Scholar] [CrossRef] [PubMed]
- McNaught, K.S.; Jenner, P. Proteasomal Function Is Impaired in Substantia Nigra in Parkinson’s Disease. Neurosci. Lett. 2001, 297, 191–194. [Google Scholar] [CrossRef]
- Abraham, N.A.; Campbell, A.C.; Hirst, W.D.; Nezich, C.L. Optimization of Small-Scale Sample Preparation for High-Throughput OpenArray Analysis. J. Biol. Methods 2021, 8, e143. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Grigorenko, E.; Funari, V.; Enright, E.; Zhang, H.; Kim, H.L. Evaluation of a High-Throughput, Microfluidics Platform for Performing TaqManTM qPCR Using Formalin-Fixed Paraffin-Embedded Tumors. Bioanalysis 2013, 5, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Lamas, A.; Franco, C.M.; Regal, P.; Miranda, J.M.; Vázquez, B.; Cepeda, A.; Lamas, A.; Franco, C.M.; Regal, P.; Miranda, J.M.; et al. High-Throughput Platforms in Real-Time PCR and Applications. In Polymerase Chain Reaction for Biomedical Applications; IntechOpen: London, UK, 2016; ISBN 978-953-51-2796-3. [Google Scholar]
- Rydbirk, R.; Folke, J.; Winge, K.; Aznar, S.; Pakkenberg, B.; Brudek, T. Assessment of Brain Reference Genes for RT-qPCR Studies in Neurodegenerative Diseases. Sci. Rep. 2016, 6, 37116. [Google Scholar] [CrossRef]
- Alieva, A.K.; Filatova, E.V.; Rudenok, M.M.; Slominsky, P.A.; Shadrina, M.I. Housekeeping Genes for Parkinson’s Disease in Humans and Mice. Cells 2021, 10, 2252. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, W.; Akao, Y.; Youdim, M.B.; Davis, B.A.; Naoi, M. Transfection-Enforced Bcl-2 Overexpression and an Anti-Parkinson Drug, Rasagiline, Prevent Nuclear Accumulation of Glyceraldehyde-3-Phosphate Dehydrogenase Induced by an Endogenous Dopaminergic Neurotoxin, N-Methyl(R)Salsolinol. J. Neurochem. 2001, 78, 727–735. [Google Scholar] [CrossRef]
- Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates—5th Edition. Available online: https://shop.elsevier.com/books/paxinos-and-franklins-the-mouse-brain-in-stereotaxic-coordinates/paxinos/978-0-12-816157-9 (accessed on 12 October 2023).
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A Web-Based Tool for Comprehensively Analyzing and Identifying Reference Genes. Funct. Integr. Genomics 2023, 23, 125. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Anglade, P.; Vyas, S.; Javoy-Agid, F.; Herrero, M.T.; Michel, P.P.; Marquez, J.; Mouatt-Prigent, A.; Ruberg, M.; Hirsch, E.C.; Agid, Y. Apoptosis and Autophagy in Nigral Neurons of Patients with Parkinson’s Disease. Histol. Histopathol. 1997, 12, 25–31. [Google Scholar] [PubMed]
- Coulson, D.T.R.; Brockbank, S.; Quinn, J.G.; Murphy, S.; Ravid, R.; Irvine, G.B.; Johnston, J.A. Identification of Valid Reference Genes for the Normalization of RT qPCR Gene Expression Data in Human Brain Tissue. BMC Mol. Biol. 2008, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Penna, I.; Vella, S.; Gigoni, A.; Russo, C.; Cancedda, R.; Pagano, A. Selection of Candidate Housekeeping Genes for Normalization in Human Postmortem Brain Samples. Int. J. Mol. Sci. 2011, 12, 5461–5470. [Google Scholar] [CrossRef]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Kommaddi, R.P.; Gnanabharathi, B.; Hirsch, E.C.; Ravindranath, V. Genes Critical for Development and Differentiation of Dopaminergic Neurons Are Downregulated in Parkinson’s Disease. J. Neural Transm. 2023, 130, 495–512. [Google Scholar] [CrossRef]
- Hoang, V.L.T.; Tom, L.N.; Quek, X.-C.; Tan, J.-M.; Payne, E.J.; Lin, L.L.; Sinnya, S.; Raphael, A.P.; Lambie, D.; Frazer, I.H.; et al. RNA-Seq Reveals More Consistent Reference Genes for Gene Expression Studies in Human Non-Melanoma Skin Cancers. PeerJ 2017, 5, e3631. [Google Scholar] [CrossRef]
- Chen, X.; Hou, H.; Qiao, H.; Fan, H.; Zhao, T.; Dong, M. Identification of Blood-Derived Candidate Gene Markers and a New 7-Gene Diagnostic Model for Multiple Sclerosis. Biol. Res. 2021, 54, 12. [Google Scholar] [CrossRef]
- Simunovic, F.; Yi, M.; Wang, Y.; Macey, L.; Brown, L.T.; Krichevsky, A.M.; Andersen, S.L.; Stephens, R.M.; Benes, F.M.; Sonntag, K.C. Gene Expression Profiling of Substantia Nigra Dopamine Neurons: Further Insights into Parkinson’s Disease Pathology. Brain 2009, 132, 1795–1809. [Google Scholar] [CrossRef]
- Zaccaria, A.; Antinori, P.; Licker, V.; Kövari, E.; Lobrinus, J.A.; Burkhard, P.R. Multiomic Analyses of Dopaminergic Neurons Isolated from Human Substantia Nigra in Parkinson’s Disease: A Descriptive and Exploratory Study. Cell Mol. Neurobiol. 2022, 42, 2805–2818. [Google Scholar] [CrossRef]
- Hauser, M.A.; Li, Y.-J.; Xu, H.; Noureddine, M.A.; Shao, Y.S.; Gullans, S.R.; Scherzer, C.R.; Jensen, R.V.; McLaurin, A.C.; Gibson, J.R.; et al. Expression Profiling of Substantia Nigra in Parkinson Disease, Progressive Supranuclear Palsy, and Frontotemporal Dementia with Parkinsonism. Arch. Neurol. 2005, 62, 917–921. [Google Scholar] [CrossRef]
- Moran, L.B.; Duke, D.C.; Deprez, M.; Dexter, D.T.; Pearce, R.K.B.; Graeber, M.B. Whole Genome Expression Profiling of the Medial and Lateral Substantia Nigra in Parkinson’s Disease. Neurogenetics 2006, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, I.; Keller-McGandy, C.; Bouzou, B.; Asteris, G.; Clark, T.W.; Frosch, M.P.; Standaert, D.G. Effects of Gender on Nigral Gene Expression and Parkinson Disease. Neurobiol. Dis. 2007, 26, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Simunovic, F.; Yi, M.; Wang, Y.; Stephens, R.; Sonntag, K.C. Evidence for Gender-Specific Transcriptional Profiles of Nigral Dopamine Neurons in Parkinson Disease. PLoS ONE 2010, 5, e8856. [Google Scholar] [CrossRef]
- Elstner, M.; Morris, C.M.; Heim, K.; Bender, A.; Mehta, D.; Jaros, E.; Klopstock, T.; Meitinger, T.; Turnbull, D.M.; Prokisch, H. Expression Analysis of Dopaminergic Neurons in Parkinson’s Disease and Aging Links Transcriptional Dysregulation of Energy Metabolism to Cell Death. Acta Neuropathol. 2011, 122, 75–86. [Google Scholar] [CrossRef]
- Glaab, E.; Schneider, R. Comparative Pathway and Network Analysis of Brain Transcriptome Changes during Adult Aging and in Parkinson’s Disease. Neurobiol. Dis. 2015, 74, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Prokopec, S.D.; Watson, J.D.; Waggott, D.M.; Smith, A.B.; Wu, A.H.; Okey, A.B.; Pohjanvirta, R.; Boutros, P.C. Systematic Evaluation of Medium-Throughput mRNA Abundance Platforms. RNA 2013, 19, 51–62. [Google Scholar] [CrossRef]
- Morrison, T.; Hurley, J.; Garcia, J.; Yoder, K.; Katz, A.; Roberts, D.; Cho, J.; Kanigan, T.; Ilyin, S.E.; Horowitz, D.; et al. Nanoliter High Throughput Quantitative PCR. Nucleic Acids Res. 2006, 34, e123. [Google Scholar] [CrossRef]
- Patel, S.N.; Wu, Y.; Bao, Y.; Mancebo, R.; Au-Young, J.; Grigorenko, E. TaqMan® OpenArray® High-Throughput Transcriptional Analysis of Human Embryonic and Induced Pluripotent Stem Cells. Methods Mol. Biol. 2013, 997, 191–201. [Google Scholar] [CrossRef]
- Dalgard, C.L.; Polston, K.F.; Sukumar, G.; Mallon, C.T.M.; Wilkerson, M.D.; Pollard, H.B. MicroRNA Expression Profiling of the Armed Forces Health Surveillance Branch Cohort for Identification of “Enviro-miRs” Associated With Deployment-Based Environmental Exposure. J. Occup. Environ. Med. 2016, 58, S97–S103. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.D.; Schafer, M.K.; Bonner, T.I.; Eiden, L.E.; Weihe, E. Distinct Pharmacological Properties and Distribution in Neurons and Endocrine Cells of Two Isoforms of the Human Vesicular Monoamine Transporter. Proc. Natl. Acad. Sci. USA 1996, 93, 5166–5171. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Du, Y.; Wang, X.; Liao, T.; Zhang, Z.; Wang, N.; Li, X.; Shen, Y.; Shi, L.; Luo, J.; et al. KIF2C Regulates Synaptic Plasticity and Cognition in Mice through Dynamic Microtubule Depolymerization. eLife 2022, 11, e72483. [Google Scholar] [CrossRef] [PubMed]
- Kozina, E.A.; Khakimova, G.R.; Khaindrava, V.G.; Kucheryanu, V.G.; Vorobyeva, N.E.; Krasnov, A.N.; Georgieva, S.G.; Kerkerian-Le Goff, L.; Ugrumov, M.V. Tyrosine Hydroxylase Expression and Activity in Nigrostriatal Dopaminergic Neurons of MPTP-Treated Mice at the Presymptomatic and Symptomatic Stages of Parkinsonism. J. Neurol. Sci. 2014, 340, 198–207. [Google Scholar] [CrossRef]
- Alieva, A.K.; Filatova, E.V.; Kolacheva, A.A.; Rudenok, M.M.; Slominsky, P.A.; Ugrumov, M.V.; Shadrina, M.I. Transcriptome Profile Changes in Mice with MPTP-Induced Early Stages of Parkinson’s Disease. Mol. Neurobiol. 2017, 54, 6775–6784. [Google Scholar] [CrossRef] [PubMed]
- Castillo, S.O.; Baffi, J.S.; Palkovits, M.; Goldstein, D.S.; Kopin, I.J.; Witta, J.; Magnuson, M.A.; Nikodem, V.M. Dopamine Biosynthesis Is Selectively Abolished in Substantia Nigra/Ventral Tegmental Area but Not in Hypothalamic Neurons in Mice with Targeted Disruption of the Nurr1 Gene. Mol. Cell. Neurosci. 1998, 11, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Kompoliti, K.; Cochran, E.J.; Mufson, E.J.; Kordower, J.H. Age-Related Decreases in Nurr1 Immunoreactivity in the Human Substantia Nigra. J. Comp. Neurol. 2002, 450, 203–214. [Google Scholar] [CrossRef]
- Iwawaki, T.; Kohno, K.; Kobayashi, K. Identification of a Potential Nurr1 Response Element That Activates the Tyrosine Hydroxylase Gene Promoter in Cultured Cells. Biochem. Biophys. Res. Commun. 2000, 274, 590–595. [Google Scholar] [CrossRef]
- Kim, K.-S.; Kim, C.-H.; Hwang, D.-Y.; Seo, H.; Chung, S.; Hong, S.J.; Lim, J.-K.; Anderson, T.; Isacson, O. Orphan Nuclear Receptor Nurr1 Directly Transactivates the Promoter Activity of the Tyrosine Hydroxylase Gene in a Cell-Specific Manner. J. Neurochem. 2003, 85, 622–634. [Google Scholar] [CrossRef]
- López-Pérez, S.J.; Morales-Villagrán, A.; Medina-Ceja, L. Effect of Perinatal Asphyxia and Carbamazepine Treatment on Cortical Dopamine and DOPAC Levels. J. Biomed. Sci. 2015, 22, 14. [Google Scholar] [CrossRef]
- Juárez Olguín, H.; Calderón Guzmán, D.; Hernández García, E.; Barragán Mejía, G. The Role of Dopamine and Its Dysfunction as a Consequence of Oxidative Stress. Oxid. Med. Cell Longev. 2016, 2016, 9730467. [Google Scholar] [CrossRef]
- Levitt, P.; Maxwell, G.D.; Pintar, J.E. Specific Cellular Expression of Monoamine Oxidase B during Early Stages of Quail Embryogenesis. Dev. Biol. 1985, 110, 346–361. [Google Scholar] [CrossRef]
- D’Andrea, M.R.; Ilyin, S.; Plata-Salaman, C.R. Abnormal Patterns of Microtubule-Associated Protein-2 (MAP-2) Immunolabeling in Neuronal Nuclei and Lewy Bodies in Parkinson’s Disease Substantia Nigra Brain Tissues. Neurosci. Lett. 2001, 306, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Dehmelt, L.; Halpain, S. The MAP2/Tau Family of Microtubule-Associated Proteins. Genome Biol. 2005, 6, 204. [Google Scholar] [CrossRef]
- Nishida, K.; Matsumura, K.; Tamura, M.; Nakamichi, T.; Shimamori, K.; Kuragano, M.; Kabir, A.M.R.; Kakugo, A.; Kotani, S.; Nishishita, N.; et al. Effects of Three Microtubule-Associated Proteins (MAP2, MAP4, and Tau) on Microtubules’ Physical Properties and Neurite Morphology. Sci. Rep. 2023, 13, 8870. [Google Scholar] [CrossRef] [PubMed]
- Poirier, K.; Saillour, Y.; Bahi-Buisson, N.; Jaglin, X.H.; Fallet-Bianco, C.; Nabbout, R.; Castelnau-Ptakhine, L.; Roubertie, A.; Attie-Bitach, T.; Desguerre, I.; et al. Mutations in the Neuronal SS-Tubulin Subunit TUBB3 Result in Malformation of Cortical Development and Neuronal Migration Defects. Hum. Mol. Genet. 2010, 19, 4462–4473. [Google Scholar] [CrossRef] [PubMed]
- Salama, M.; Shalash, A.; Magdy, A.; Makar, M.; Roushdy, T.; Elbalkimy, M.; Elrassas, H.; Elkafrawy, P.; Mohamed, W.; Abou Donia, M.B. Tubulin and Tau: Possible Targets for Diagnosis of Parkinson’s and Alzheimer’s Diseases. PLoS ONE 2018, 13, e0196436. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Stoklund Dittlau, K.; Van Den Bosch, L. Axonal Transport Defects and Neurodegeneration: Molecular Mechanisms and Therapeutic Implications. Semin. Cell Dev. Biol. 2020, 99, 133–150. [Google Scholar] [CrossRef]
- Yang, X.; Ma, Z.; Lian, P.; Xu, Y.; Cao, X. Common Mechanisms Underlying Axonal Transport Deficits in Neurodegenerative Diseases: A Mini Review. Front. Mol. Neurosci. 2023, 16, 1172197. [Google Scholar] [CrossRef]
- Stefanis, L. α-Synuclein in Parkinson’s Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009399. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Higgins, J.J.; Golbe, L.I.; Johnson, W.G.; Ide, S.E.; Di Iorio, G.; Sanges, G.; Stenroos, E.S.; Pho, L.T.; Schaffer, A.A.; et al. Mapping of a Gene for Parkinson’s Disease to Chromosome 4q21-Q23. Science 1996, 274, 1197–1199. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the α-Synuclein Gene Identified in Families with Parkinson’s Disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Ingelsson, M. α-Synuclein Oligomers-Neurotoxic Molecules in Parkinson’s Disease and Other Lewy Body Disorders. Front. Neurosci. 2016, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Cascella, R.; Chen, S.W.; Bigi, A.; Camino, J.D.; Xu, C.K.; Dobson, C.M.; Chiti, F.; Cremades, N.; Cecchi, C. The Release of Toxic Oligomers from α-Synuclein Fibrils Induces Dysfunction in Neuronal Cells. Nat. Commun. 2021, 12, 1814. [Google Scholar] [CrossRef] [PubMed]
- Rudenok, M.M.; Shadrina, M.I.; Filatova, E.V.; Rybolovlev, I.N.; Nesterov, M.S.; Abaimov, D.A.; Ageldinov, R.A.; Kolacheva, A.A.; Ugrumov, M.V.; Slominsky, P.A.; et al. Expression Analysis of Genes Involved in Transport Processes in Mice with MPTP-Induced Model of Parkinson’s Disease. Life 2022, 12, 751. [Google Scholar] [CrossRef] [PubMed]
- Mirza, F.J.; Zahid, S. The Role of Synapsins in Neurological Disorders. Neurosci. Bull. 2017, 34, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Kolacheva, A.; Pavlova, E.; Bannikova, A.; Bogdanov, V.; Troshev, D.; Ugrumov, M. The Gene Expression of Proteins Involved in Intercellular Signaling and Neurodegeneration in the Substantia Nigra in a Mouse Subchronic Model of Parkinson’s Disease. Int. J. Mol. Sci. 2023, 24, 3027. [Google Scholar] [CrossRef] [PubMed]
- Kershberg, L.; Banerjee, A.; Kaeser, P.S. Protein Composition of Axonal Dopamine Release Sites in the Striatum. eLife 2022, 11, e83018. [Google Scholar] [CrossRef]
- Dinter, E.; Saridaki, T.; Nippold, M.; Plum, S.; Diederichs, L.; Komnig, D.; Fensky, L.; May, C.; Marcus, K.; Voigt, A.; et al. Rab7 Induces Clearance of α-Synuclein Aggregates. J. Neurochem. 2016, 138, 758–774. [Google Scholar] [CrossRef]
- Szegö, E.M.; Van den Haute, C.; Höfs, L.; Baekelandt, V.; Van der Perren, A.; Falkenburger, B.H. Rab7 Reduces α-Synuclein Toxicity in Rats and Primary Neurons. Exp. Neurol. 2022, 347, 113900. [Google Scholar] [CrossRef]
- Hollerman, J.R.; Grace, A.A. The Effects of Dopamine-Depleting Brain Lesions on the Electrophysiological Activity of Rat Substantia Nigra Dopamine Neurons. Brain Res. 1990, 533, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, J.; Mørk, A.; Brennum, L.T.; Sager, T.N. Striatal Extracellular Dopamine Levels and Behavioural Reversal in MPTP-Lesioned Mice. Neuroreport 2009, 20, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Belluzzi, E.; Gonnelli, A.; Cirnaru, M.-D.; Marte, A.; Plotegher, N.; Russo, I.; Civiero, L.; Cogo, S.; Carrion, M.P.; Franchin, C.; et al. LRRK2 Phosphorylates Pre-Synaptic N-Ethylmaleimide Sensitive Fusion (NSF) Protein Enhancing Its ATPase Activity and SNARE Complex Disassembling Rate. Mol. Neurodegener. 2016, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Pischedda, F.; Cirnaru, M.D.; Ponzoni, L.; Sandre, M.; Biosa, A.; Carrion, M.P.; Marin, O.; Morari, M.; Pan, L.; Greggio, E.; et al. LRRK2 G2019S Kinase Activity Triggers Neurotoxic NSF Aggregation. Brain 2021, 144, 1509–1525. [Google Scholar] [CrossRef]
- Staal, R.G.W.; Mosharov, E.V.; Sulzer, D. Dopamine Neurons Release Transmitter via a Flickering Fusion Pore. Nat. Neurosci. 2004, 7, 341–346. [Google Scholar] [CrossRef]
- Sulzer, D.; Cragg, S.J.; Rice, M.E. Striatal Dopamine Neurotransmission: Regulation of Release and Uptake. Basal Ganglia 2016, 6, 123–148. [Google Scholar] [CrossRef]
- Feng, S.-T.; Wang, Z.-Z.; Yuan, Y.-H.; Wang, X.-L.; Sun, H.-M.; Chen, N.-H.; Zhang, Y. Dynamin-Related Protein 1: A Protein Critical for Mitochondrial Fission, Mitophagy, and Neuronal Death in Parkinson’s Disease. Pharmacol. Res. 2020, 151, 104553. [Google Scholar] [CrossRef]
- Berthet, A.; Margolis, E.B.; Zhang, J.; Hsieh, I.; Zhang, J.; Hnasko, T.S.; Ahmad, J.; Edwards, R.H.; Sesaki, H.; Huang, E.J.; et al. Loss of Mitochondrial Fission Depletes Axonal Mitochondria in Midbrain Dopamine Neurons. J. Neurosci. 2014, 34, 14304–14317. [Google Scholar] [CrossRef]
- Martínez, J.H.; Fuentes, F.; Vanasco, V.; Alvarez, S.; Alaimo, A.; Cassina, A.; Coluccio Leskow, F.; Velazquez, F. α-Synuclein Mitochondrial Interaction Leads to Irreversible Translocation and Complex I Impairment. Arch. Biochem. Biophys. 2018, 651, 1–12. [Google Scholar] [CrossRef]
- Rappold, P.M.; Cui, M.; Grima, J.C.; Fan, R.Z.; de Mesy-Bentley, K.L.; Chen, L.; Zhuang, X.; Bowers, W.J.; Tieu, K. Drp1 Inhibition Attenuates Neurotoxicity and Dopamine Release Deficits in Vivo. Nat. Commun. 2014, 5, 5244. [Google Scholar] [CrossRef]
- Dias, V.; Junn, E.; Mouradian, M.M. The Role of Oxidative Stress in Parkinson’s Disease. J. Parkinsons Dis. 2013, 3, 461–491. [Google Scholar] [CrossRef] [PubMed]
- Wypijewska, A.; Galazka-Friedman, J.; Bauminger, E.R.; Wszolek, Z.K.; Schweitzer, K.J.; Dickson, D.W.; Jaklewicz, A.; Elbaum, D.; Friedman, A. Iron and Reactive Oxygen Species Activity in Parkinsonian Substantia Nigra. Park. Relat. Disord. 2010, 16, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Toulorge, D.; Schapira, A.H.V.; Hajj, R. Molecular Changes in the Postmortem Parkinsonian Brain. J. Neurochem. 2016, 139 (Suppl. S1), 27–58. [Google Scholar] [CrossRef] [PubMed]
- Perry, T.L.; Yong, V.W. Idiopathic Parkinson’s Disease, Progressive Supranuclear Palsy and Glutathione Metabolism in the Substantia Nigra of Patients. Neurosci. Lett. 1986, 67, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Jenner, P.; Dexter, D.T.; Sian, J.; Schapira, A.H.; Marsden, C.D. Oxidative Stress as a Cause of Nigral Cell Death in Parkinson’s Disease and Incidental Lewy Body Disease. The Royal Kings and Queens Parkinson’s Disease Research Group. Ann. Neurol. 1992, 32, S82–S87. [Google Scholar] [CrossRef] [PubMed]
- Sian, J.; Dexter, D.T.; Lees, A.J.; Daniel, S.; Agid, Y.; Javoy-Agid, F.; Jenner, P.; Marsden, C.D. Alterations in Glutathione Levels in Parkinson’s Disease and Other Neurodegenerative Disorders Affecting Basal Ganglia. Ann. Neurol. 1994, 36, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.-L. The Keap1-Nrf2 Pathway: Mechanisms of Activation and Dysregulation in Cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Taguchi, K.; Motohashi, H.; Yamamoto, M. Molecular Mechanisms of the Keap1–Nrf2 Pathway in Stress Response and Cancer Evolution. Genes Cells 2011, 16, 123–140. [Google Scholar] [CrossRef]
- Rintoul, G.L.; Raymond, L.A.; Baimbridge, K.G. Calcium Buffering and Protection from Excitotoxic Cell Death by Exogenous Calbindin-D28k in HEK 293 Cells. Cell Calcium 2001, 29, 277–287. [Google Scholar] [CrossRef]
- Brimblecombe, K.R.; Vietti-Michelina, S.; Platt, N.J.; Kastli, R.; Hnieno, A.; Gracie, C.J.; Cragg, S.J. Calbindin-D28K Limits Dopamine Release in Ventral but Not Dorsal Striatum by Regulating Ca2+ Availability and Dopamine Transporter Function. ACS Chem. Neurosci. 2019, 10, 3419–3426. [Google Scholar] [CrossRef] [PubMed]
- Naik, M.U.; Naik, U.P. Calcium- and Integrin-Binding Protein 1 Regulates Microtubule Organization and Centrosome Segregation through Polo like Kinase 3 during Cell Cycle Progression. Int. J. Biochem. Cell Biol. 2011, 43, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.W.; Hori, Y.; Ebinuma, I.; Sato, H.; Hara, N.; Ikeuchi, T.; Tomita, T. Identification of Calcium and Integrin-Binding Protein 1 as a Novel Regulator of Production of Amyloid β Peptide Using CRISPR/Cas9-Based Screening System. FASEB J. 2020, 34, 7661–7674. [Google Scholar] [CrossRef]
- Yoon, K.W.; Cho, J.-H.; Lee, J.K.; Kang, Y.-H.; Chae, J.S.; Kim, Y.M.; Kim, J.; Kim, E.K.; Kim, S.E.; Baik, J.-H.; et al. CIB1 Functions as a Ca2+-Sensitive Modulator of Stress-Induced Signaling by Targeting ASK1. Proc. Natl. Acad. Sci. USA 2009, 106, 17389–17394. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.W.; Yang, H.-S.; Kim, Y.M.; Kim, Y.; Kang, S.; Sun, W.; Naik, U.P.; Parise, L.V.; Choi, E.-J. CIB1 Protects against MPTP-Induced Neurotoxicity through Inhibiting ASK1. Sci. Rep. 2017, 7, 12178. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Obeso, J.A.; Stocchi, F. Continuous Dopamine-Receptor Treatment of Parkinson’s Disease: Scientific Rationale and Clinical Implications. Lancet Neurol. 2006, 5, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Mandel, S.; Grünblatt, E.; Maor, G.; Youdim, M.B.H. Early and Late Gene Changes in MPTP Mice Model of Parkinson’s Disease Employing cDNA Microarray. Neurochem. Res. 2002, 27, 1231–1243. [Google Scholar] [CrossRef]
- Lin, Q.-S.; Chen, P.; Wang, W.-X.; Lin, C.-C.; Zhou, Y.; Yu, L.-H.; Lin, Y.-X.; Xu, Y.-F.; Kang, D.-Z. RIP1/RIP3/MLKL Mediates Dopaminergic Neuron Necroptosis in a Mouse Model of Parkinson Disease. Lab. Investig. 2020, 100, 503–511. [Google Scholar] [CrossRef]
- Oliveira, S.R.; Dionísio, P.A.; Gaspar, M.M.; Ferreira, M.B.T.; Rodrigues, C.A.B.; Pereira, R.G.; Estevão, M.S.; Perry, M.J.; Moreira, R.; Afonso, C.A.M.; et al. Discovery of a Necroptosis Inhibitor Improving Dopaminergic Neuronal Loss after MPTP Exposure in Mice. Int. J. Mol. Sci. 2021, 22, 5289. [Google Scholar] [CrossRef]
- Meredith, G.E.; Totterdell, S.; Beales, M.; Meshul, C.K. Impaired Glutamate Homeostasis and Programmed Cell Death in a Chronic MPTP Mouse Model of Parkinson’s Disease. Exp. Neurol. 2009, 219, 334–340. [Google Scholar] [CrossRef]
- Jackson-Lewis, V.; Jakowec, M.; Burke, R.E.; Przedborski, S. Time Course and Morphology of Dopaminergic Neuronal Death Caused by the Neurotoxin 1-Methyl-4-Phenyl-l,2,3,6-Tetrahydropyridine. Neurodegeneration 1995, 4, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Kolacheva, A.A.; Kozina, E.A.; Volina, E.V.; Ugryumov, M.V. Time Course of Degeneration of Dopaminergic Neurons and Respective Compensatory Processes in the Nigrostriatal System in Mice. Dokl. Biol. Sci. 2014, 456, 160–164. [Google Scholar] [CrossRef] [PubMed]


| Gene Rank | Control | 4 × 12 mg/kg MPTP | Summary Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Comprehensive Ranking | Delta CT | BestKeeper | NormFinder | GeNorm | Comprehensive Ranking | Delta CT | BestKeeper | NormFinder | GeNorm | ||
| HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | HKG/Score | |
| 1 | Rps27a 1.41 | Sdha 0.57 | Rps27a 0.333 | Sdha 0.253 | Rps27a 0.389 | Rps27a 1.68 | Cyc1 2.95 | Rpl13 0.333 | Ube2d2a 0.226 | Rps27a 0.452 | Rps27a 3.09 |
| 2 | Sdha 2.45 | Rps27a 0.57 | Gapdh 0.417 | Rps27a 0.267 | Ube2d2a 0.389 | Ube2d2a 1.97 | Rps27a 3.01 | Rps27a 0.417 | Rps27a 0.226 | Ube2d2a 0.452 | Ube2d2a 5.95 |
| 3 | Hprt 3.72 | Gapvd1 0.59 | Aars 0.456 | Gapvd1 0.328 | Hprt 0.402 | Rpl13 2.28 | Rpl13 3.05 | Ube2d2a 0.583 | Rpl13 0.311 | Rpl13 0.583 | Hprt 8.15 |
| 4 | Gapvd1 3.87 | Hprt 0.62 | Hprt 0.5 | Hprt 0.390 | Sdha 0.437 | Cyc1 3.16 | Hprt 3.07 | Hprt 0.889 | Cyc1 1.197 | Hprt 0.836 | Cyc1 8.85 |
| 5 | Ube2d2a 3.98 | Cyc1 0.64 | Gapvd1 0.583 | Cyc1 0.415 | Gapvd1 0.471 | Hprt 4.43 | Ube2d2a 3.1 | Cyc1 1.056 | Xpnpep1 1.655 | Cyc1 0.979 | Sdha 8.93 |
| 6 | Cyc1 5.69 | Ube2d2a 0.65 | Cyc1 0.583 | Ube2d2a 0.431 | Rpl13 0.504 | Xpnpep1 6.19 | Xpnpep1 3.14 | Sdha 1.306 | Hprt 1.729 | Sdha 1.084 | Rpl13 9.24 |
| 7 | Aars 6.26 | Rpl13 0.67 | Ube2d2a 0.583 | Rpl13 0.476 | Cyc1 0.530 | Sdha 6.48 | Sdha 3.21 | Xpnpep1 1.403 | Sdha 2.189 | Xpnpep1 1.163 | Gapvd1 13.87 |
| 8 | Gapdh 6.69 | Aars 0.71 | Rpl13 0.611 | Aars 0.538 | Aars 0.560 | Gapdh 8 | Gapdh 3.33 | Gapdh 1.667 | Gapdh 2.216 | Gapdh 1.259 | Gapdh 14.69 |
| 9 | Rpl13 6.96 | Xpnpep1 0.73 | Sdha 0.625 | Xpnpep1 0.571 | Xpnpep1 0.580 | Aars 9 | Aars 3.42 | Aars 1.75 | Aars 2.593 | Aars 1.303 | Aars 15.26 |
| 10 | Xpnpep1 9.24 | Gapdh 0.78 | Xpnpep1 0.625 | Gapdh 0.623 | Gapdh 0.616 | Gapvd1 10 | Gapvd1 8.95 | Gapvd1 4.208 | Gapvd1 8.064 | Gapvd1 2.834 | Xpnpep1 15.43 |
| 11 | Osbp 11 | Osbp 0.98 | Osbp 0.75 | Osbp 0.889 | Osbp 0.682 | Osbp 11 | Osbp 11.72 | Osbp 7.806 | Osbp 11.389 | Osbp 4.449 | Osbp 22 |
| Gene Clusters | Highly Expressed Genes (Can Be Detected with OpenArray) | Genes with Low Expressions (Can Be Detected with OpenArray Following Pre-amplification) | Genes with Very Low Expressions or Genes Not Expressed (Cannot Be Detected with OpenArray) |
|---|---|---|---|
| Synthesis and degradation of dopamine | Th, Ddc, Comt, Maoa, Maob | - | Dbh, Pnmt |
| Dopamine transport, dopamine receptors, and transcription factors of dopaminergic neurons | Slc18a2, Slc6a3, Drd2, Nr4a2 | Slc29a4, Drd1 | Drd3–Drd5, Slc18a1, Pitx3 |
| Axonal transport | Tubb3, Tuba1a, Dynll1, Kif1a, Kif5a, Dctn1, Map2, Mapt | Kif1b, Dync1h1, Mark2 | Kif2c |
| Vesicular cycle and mediator release | Syn1, Syt1, Snca, Syt11, Rab5a, Rab7, Dnm1l, Vps35, Nsf, | Stx1a | - |
| Neuroprotection | Gpx1, Gsr, Sod1, Prdx1, Txnrd1, Nfe2l2, Keap1, Sigmar1, Calb1, Ctnnb1, Ntrk2 | Bdnf, Vegfa, Nos1, Agtr2 | Ngf, Gdnf, Cdnf, Ntrk1, Mmp3, Wnt11, Ngfr |
| Protein degradation | Ubb, Uba3, Ube2n, Psmb4, Psmd4, Psmc3, Usp47, Ctsb | Park2, Cacna1d, Trpm2 | - |
| Cell death | Parp1, Cib1, Aifm1Bax, Trp53, Lamp2, Mapk8, Atg5 | Casp1, Casp3, Map3k5, Fos, Capn1, Eif2ak3, Atf6, Atg16l1 | Tnf, Bcl2l11, Ern2 |
| Inflammation and glia activation | Gfap, Clk1, Akt1, Cnr1 | Tgfb1 | Ifng, Cxcl11, Ptgs2, Traf1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troshev, D.; Kolacheva, A.; Pavlova, E.; Blokhin, V.; Ugrumov, M. Application of OpenArray Technology to Assess Changes in the Expression of Functionally Significant Genes in the Substantia Nigra of Mice in a Model of Parkinson’s Disease. Genes 2023, 14, 2202. https://doi.org/10.3390/genes14122202
Troshev D, Kolacheva A, Pavlova E, Blokhin V, Ugrumov M. Application of OpenArray Technology to Assess Changes in the Expression of Functionally Significant Genes in the Substantia Nigra of Mice in a Model of Parkinson’s Disease. Genes. 2023; 14(12):2202. https://doi.org/10.3390/genes14122202
Chicago/Turabian StyleTroshev, Dmitry, Anna Kolacheva, Ekaterina Pavlova, Victor Blokhin, and Michael Ugrumov. 2023. "Application of OpenArray Technology to Assess Changes in the Expression of Functionally Significant Genes in the Substantia Nigra of Mice in a Model of Parkinson’s Disease" Genes 14, no. 12: 2202. https://doi.org/10.3390/genes14122202
APA StyleTroshev, D., Kolacheva, A., Pavlova, E., Blokhin, V., & Ugrumov, M. (2023). Application of OpenArray Technology to Assess Changes in the Expression of Functionally Significant Genes in the Substantia Nigra of Mice in a Model of Parkinson’s Disease. Genes, 14(12), 2202. https://doi.org/10.3390/genes14122202






