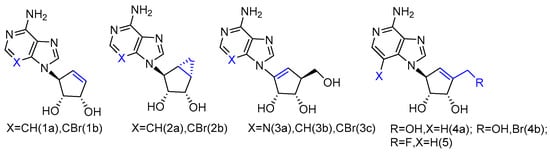
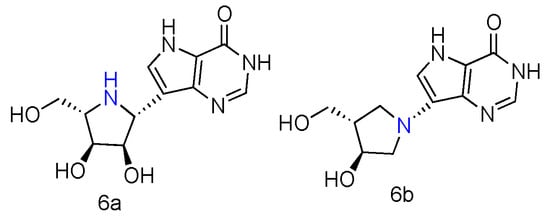
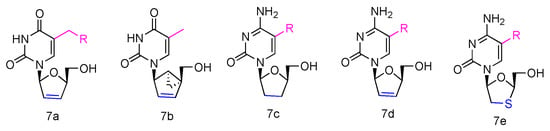
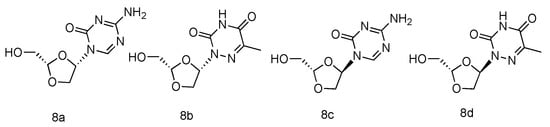
Abstract
Nucleic-acid-based small molecule and oligonucleotide therapies are attractive topics due to their potential for effective target of disease-related modules and specific control of disease gene expression. As the non-naturally occurring biomolecules, modified DNA/RNA nucleoside and oligonucleotide analogues composed of L-(deoxy)riboses, have been designed and applied as innovative therapeutics with superior plasma stability, weakened cytotoxicity, and inexistent immunogenicity. Although all the chiral centers in the backbone are mirror converted from the natural D-nucleic acids, L-nucleic acids are equipped with the same nucleobases (A, G, C and U or T), which are critical to maintain the programmability and form adaptable tertiary structures for target binding. The types of L-nucleic acid drugs are increasingly varied, from chemically modified nucleoside analogues that interact with pathogenic polymerases to nanoparticles containing hundreds of repeating L-nucleotides that circulate durably in vivo. This article mainly reviews three different aspects of L-nucleic acid therapies, including pharmacological L-nucleosides, Spiegelmers as specific target-binding aptamers, and L-nanostructures as effective drug-delivery devices.
1. Introduction
The genetic information of the human genome is stored in nucleic acids, which are constituted by only four major nucleobases (A, G, C, and T). DNA replication and transcription, as well as RNA translation, are specifically regulated by proteins recognizing nucleic acids [1]. In order to preserve the integrated genomic information and secure the accurate nucleic acid-protein recognitions, native DNA and RNA molecules must bear not only the particular base sequence but also the highly ordered structures, including phosphodiester linkages, (deoxy)ribose moieties, and a variety of unique secondary/tertiary geometries [2,3]. Like the phenomenon that most chiral components of natural products occur predominantly in one of the plausible enantiomeric isomers, the biological building blocks of life, including nucleic acids and proteins, are exclusively utilizing only one chirality to function in the metabolism [4]. In particular, native DNA and RNA are only composed of D-configuration, while native proteins only involve L-amino acid residues, except for the rarely arisen D-amino acids and D-peptides [5,6]. In theory, the two enantiomeric DNAs/RNAs should have identical chemical and physical properties; nevertheless, all living organisms on the earth only utilize D-DNAs/RNAs. Meanwhile, only L-proteins can contact D-DNAs/RNAs in vivo to ensure the central dogma [7]. This scenario would be closely related to the molecular evolution of life. There is no substantial solution to this evolutionary mystery yet, but it is believed to relate with some nucleoside- or nucleoside-like-molecule-involved prebiotic chemistry that occurred in the primordial soup on the early earth, catalyzed by metal ions, nucleic acids, amino acids, or other coenzymes [8].
In this article, we are not trying to seek the certainty that the homochirality of natural nucleic acids agrees with or diverges from the classic RNA world hypothesis in evolution [9]. We are interested in another actuality: L-DNAs/RNAs, as the completely orthogonal biomolecules of native nucleic acids, are featured due to their resistance to nuclease degradation, nontoxicity, and nonimmunogenicity [10]. Those are the properties an ideal nucleic acid drug is expected to possess. Therefore, there has been continuous active research in this field. Importantly, L-DNAs/RNAs are incapable of base pairing with native nucleic acids in vivo or interacting with native nucleic-acid-processing enzymes. This precludes the possibility of using L-nucleic acids such as antisense therapy [11] or a siRNA strand to induce Dicer-involved RNA interference [12]. The current application of L-nucleic acids as therapeutic agents in clinical trials mainly focuses on the L-aptamer development, and it utilizes the versatile secondary/tertiary structures of L-nucleic acids to recognize the disease relevant target with high specificity and affinity. The functional L-aptamer is discovered from the combinatorial library through in vitro SELEX (systematic evolution of ligands by exponential enrichment) [13,14]. Other successful applications of L-nucleic acid drugs include the construction of an entire L-DNA nanoparticle to deliver therapeutic ligands, and the usage of an L-nucleoside with a history of 60 years [15]. This review will mainly discuss three aspects of L-nucleic acids, including L-nucleoside, L-aptamer, and L-nanoparticle, and introduce the instances closely related to disease treatment. Other biotechnological applications of L-nucleic acids, such as biosensors [16], are beyond our emphases in this article. Those related contents have been summarized in other review articles [17,18].
3. L-Nuclei Acid Aptamers to Target Disease-Related Elements
3.1. L-Aptamers Bind to Small Molecules
In 1996, a 58-mer L-RNA aptamer had been isolated from a combinatorial pool to selectively bind to naturally occurring D-adenosine [86]. Although the binding affinity was not optimal (Kd 1.7 μM), the L-RNA aptamer displayed an extraordinary stability in human serum, and it shed light on the potential application of elaborated L-RNA ligands as stable monoclonal antibody analogues.
Malachite green (MG) is a popular organic material, frequently used as a dye and an antimicrobial in aquaculture and the food industry [87]. The wide range of toxicological effects of MG have been identified, including carcinogenesis, mutagenesis, chromosomal fractures, teratogenicity, and respiratory toxicity [88]. Therefore, the sensitive and rapid detection as well as the clearance of MG are critical. The Huang lab has developed an L-RNA aptamer to selectively bind MG [89]. Since malachite green as the selection target is a planar molecule without characteristic chirality, both L-RNA and D-RNA aptamers can bind to it with the identical affinities. However, the L-aptamer exhibited excellent stability and remained durably intact in the standard buffer. This result suggested the promising potential of this L-aptamer as a sensitive and reliable analysis tool in environment monitoring and animal tissue examination.
Clearly, when detecting small molecules without chirality, L-RNA and D-RNA aptamers sharing the same sequence should have the same binding affinities. Malachite green is just one example. Following this principle, several labs have directly replaced the reported D-aptamers with L-aptamers to target nonchiral targets, including Hemin, cationic porphyrin [90], and ethanolamine [91]. As expected, L-aptamers displayed greatly improved biostability and identical binding affinity as D-aptamers. Interestingly, the chiral molecule of natural ATP was also bound to an L-aptamer in the same manner as a D-aptamer, since the intermolecular interactions are independent of chiral atoms in ATP [90].
3.2. L-Aptamers Bind to RNA Motifs
When targeting small RNA motifs, L-aptamers were commonly selected as binding ligands to selectively target disease-relevant noncoding RNAs. SELEX is the main approach of aptamer identification. In the practical isolation, a wild-type D-aptamer is first identified against the synthetic mirror-image of the intended target from the random DNA/RNA libraries. Secondly, the isolated aptamers are converted to L-oligonucleotides binding to the wild-type targets. For instance, different L-RNA aptamers have been obtained by in vitro selection against the precursor microRNAs, which are oncogenic precursor of mature microRNAs. The successful examples include pre-miR-155 aptamers [92,93] and pre-miR-19a aptamers [94]. These L-DNA or L-RNA aptamers were observed to have exceptional affinities and intrinsical stability. They can bind to pre-microRNA targets through unique tertiary structural features and block the classic Dicer-mediated processing of pre-microRNAs to generate mature microRNAs. Given their particularities, these L-aptamers might be further explored and applied as future miRNA inhibitors and cancer therapies.
In addition, L-RNA aptamer has been developed successfully as potential anti-HIV agent. A 43mer L-RNA aptamer was selected to bind the natural HIV-1 trans-activation responsive (TAR) RNA with a Kd of 100 nM [95]. The cross-chiral intermolecular bridging highly depends on the tertiary interactions and the characteristic six-nucleotide distal loop of the TAR element. The L-aptamer has the therapeutic promise to interfere with the TAR-tat protein complex formation in HIV, thereby interrupting the normal TAR function as well as the viral RNA genome expression.
The Kwok lab reported two different L-RNA aptamers, which can target native RNA G-quadruplexes (G4s) with the Kd of 75 and 99 nM, respectively [96,97]. G4s is one featured RNA structural motifs, containing multiple G-quartet planes that can be stabilized by monovalent ions [98]. The structure is important for its interaction with G4-binding proteins, which modulate telomerase activity and regulate gene expression [99]. Therefore, the development of these L-RNA aptamers is of great significance in cancer diagnosis and treatment.
Recently, a novel L-DNA aptamer was successfully identified as targeting the stem-loop II-like motif (s2m), which is a highly conserved structure at the 3′-UTR region of the RNA genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [100]. The 41-nucleotide s2m element is discovered in coronavirus, astrovirus, and equine rhinovirus genomes, and it plays a critical role in viral replication and packaging [101]. Therefore, s2m structure is an effective and popular target in anti-viral drug design. With the optimal binding affinity and selectivity, as well as the outstanding stability against nucleases, this L-DNA aptamer provides the opportunity to offer a new therapeutic molecule to end this unprecedented pandemic.
3.3. L-Aptamers Bind to Amino Acids and Proteins
In 1996, Nolte et al. selected a stable 38-mer L-RNA oligoribonucleotide to bind to L-arginine and a short peptide containing the basic region of HIV-1 Tat-protein [102]. L-RNA was observed to own the exceptional stability against human serum. By binding to HIV Tat-protein and blocking the regulatory function of Tat, this L-aptamer had the potential to interfere with the transcription of the HIV genome as an innovative anti-HIV drug.
The Peyrin lab has applied the mirror-image strategy to design the L-RNA aptamer used in chiral stationary phase (CSP) against different natural amino acid residues [103,104,105,106]. The successful targets included L-arginine, L-tyrosine, and L-histidine. The selected 40–60 nucleotides aptamers displayed prominent resistance to the enzymatic degradation and resolved the different racemates well. This L-RNA aptamer-based CSP is an effective approach for the chromatographic separation of the enantiomers of amino acid residues.
As the mirror-image-structured oligonucleotides, considerable efforts have been delivered to develop L-aptamers to bind and antagonize the disease-relevant target. The most famous instance is the registered trademark of Spiegelmer, belonging to NOXXON Pharma AG. Selection of L-aptamers against proteins follows the similar procedure as those against microRNAs. With the non-natural chirality, Spiegelmers has a abiding survival time in biological fluids [107]. Different Spiegelmer aptamers have been isolated to target various pharmacological molecules, and several of them are in different preclinical animal models and clinical phases.
Complement factor C5a is a potent proinflammatory mediator that contributes to the pathogenesis of a variety of diseases, including autoimmune diseases, acute inflammatory responses, and systemic inflammatory response syndrome caused by ischemia/reperfusion injuries, trauma, or systemic infection in sepsis [108,109]. It has been discovered that neutralizing C5a with binding ligands significantly prevents multiorgan failure and improves survival as well as prevents the exhaustion and dysfunction of neutrophils [110]. Accordingly, effective therapeutic candidates of Spiegelmers, NOX-D20 [111], and NOX-D21 [112] have been screened out. These PEGylated biostable mirror-image mixed (L-)RNA/DNA aptamers bind to mouse and human C5a with pM affinities and demonstrated promising effects on attenuating inflammation and organ damage, preventing the breakdown of the vascular endothelial barrier, and improving survival. Moreover, the crystal structure of this therapeutic Spiegelmer complexed with mouse C5a has been determined, illustrating the molecular insights into the L-aptamer binding basis [113].
Another example of Spiegelmer, NOX-A12, has been identified to inhibit Stromal Cell Derived Factor 1 (SDF-1), which binds to CXCR4 chemokine receptor and mediates hematopoietic cell migration to bone marrow stroma. NOX-A12 could inhibit SDF-1 in vitro and in vivo, against BCR-ABL- and FLT3-ITD-dependent leukemia cells. NOX-A12 itself, or in synergistic application with the Tyrosine Kinase Inhibitor drug, significantly suppressed the SDF-1 induced cell migration and reduced the leukemia burden in mice to a greater extent [114]. Besides, NOX-A12, together with another Spiegelmer mNOX-E36, was evaluated in a syngeneic mouse model of intraportal islet transplantation and in a diabetes induction model containing multiple low doses of streptozotocin (MLD-STZ) [115,116]. Both L-aptamers significantly improved islet engraftment, decreased the recruitment of inflammatory monocytes, and attenuated diabetes progression. The crystal structure of aptamer mNOX-E36 bound to pro-inflammatory chemokine CLL2 was also determined to be 2.05 Å, and it unveiled the structural rationale of L-aptamer targeting certain chemokines [117].
The sphingolipid S1P (sphingosine 1-phosphate) is involved in a number of pathophysiological conditions [118]. It is well understood that S1P plays a vital role by defining the components of the plasma membrane, as well as in various significant cell signaling pathways and physiological processes such as cell migration, survival, and proliferation, cellular architecture, cell-cell contacts and adhesions, inflammation and immunity, and tumorogenesis and metastasis [119]. It is obvious that regulating the S1P synthesis and degradation will be critical to govern a variety of cellular and physiological processes. Another Spiegelmer, NOX-S93, has been employed in vitro to selectively bind to S1P with a Kd of 4.3 nM [120]. The results provided evidence that NOX-S93 inhibited S1P-mediated β-arrestin recruitment and intracellular calcium release in a Chinese hamster ovary cell model. Furthermore, the pro-angiogenic activity of S1P was also dramatically blocked by NOX-S93, in a cellular angiogenesis assay employing primary human endothelial cells. It suggested the therapeutic potential of NOX-S93 when neutralizing pharmacological S1P as a promising approach related to angiogenesis.
Calcitonin gene-related peptide (CGRP) plays a major role in the pathogenesis of migraine and other primary headaches [121]. CGRP receptor inhibitors and anti-CGRP antibodies have been demonstrated to be therapeutically effective in migraines. Fischer et al. have selected an L-aptamer NOX-C89 to target CGRP, and single neuron cell activity in mouse model has been assayed when infusing ascending doses of NOX-C89 [122]. The abolishment of heat-induced neuronal activity could be observed after infusing certain amount of NOX-C89, which indicated the potential of this L-aptamer to control spinal trigeminal activity.
High-mobility group A1 (HMGA1) proteins are usually overexpressed in human malignancies and promote anchorage-independent growth and epithelial-mesenchymal transition [123]. Therefore, HMGA1 is considered a potential therapeutic target in pancreatic cancer treatment. Spiegelmers NOX-A50 and NOX-f33 have been successfully selected in vitro as HMGA1 binders [124]. The in vivo assay using a xenograft mouse model with a pancreatic cancer cell line showed the effectiveness of NOX-A50 to significantly reduce tumor volumes.
In order to select the ligand to inhibit and study the RNase function in biological systems, the Joyce lab has identified an L-RNA aptamer targeting the intact barnase enzyme [125]. The aptamer could specifically bind to the barnase with an affinity of ∼100 nM and function as a competitive inhibitor of the enzymatic cleavage of D-RNA substrates. It provided notable implication for application in molecular diagnostics and therapeutics.
4. L-Nucleic Acid Nanoparticles as Drug Delivery Tools
As the enantiomer of natural DNAs/RNAs, L-type nucleic acids composed of nucleobases, phosphate linkages, and L-(deoxy)ribose sugars (Figure 14A,B) retain the identical structural and physical properties, including aqueous solubility, Watson–Crick base pairing, secondary structural conformation, and duplex thermal stability. Due to the chiral inversion, L-DNAs/RNAs are orthogonal to the stereospecific environment of biology and are unable to interact with native nucleases, leading to complete resistance to nuclease degradation and, substantially, avoiding immune response and nonspecific therapeutic toxicity [126].
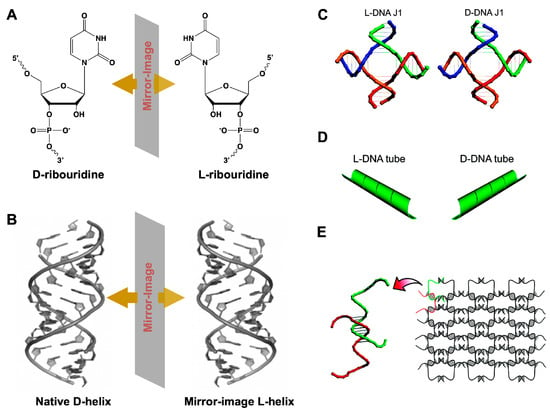
Figure 14.
(A) Chemical structures of D-ribouridine, L-ribouridine, and (B) duplex of native D-DNA and L-DNA. (C) Structural models of L- and D-DNA J1 molecules. (D) The opposite chirality of the L-(left-column) and D-(right column) DNA nanotubes revealed. (E) Schematic drawings of two-dimensional nanoarrays self-assembled from L-DNA. (C–E) reprinted with permission from [126]. Copyright 2009 American Chemical Society.
Since the geometry and periodicity of L-DNA/RNA structures were the same as that of their D-counterparts, the well-established design principles and protocols on nucleic acid nanostructures can be directly employed to L-DNAs/RNAs without further modification. Besides, as above mentioned, L-DNA/RNA molecules have the essential features of selectively binding to molecular targets, after elaborative design and manipulation [127,128]. These observations bring out appealing opportunities for in vivo medical applications of L-DNA/RNA nanostructures.
Early in 2009, Lin et al. constructed a series of short L-DNA architectures that could readily self-assemble into various well-defined 1D and 2D nanostructures with opposite chirality, including a 1D finite four arm junction (J1) (Figure 14C), 2D infinite extended L-DNA nanotubes (Figure 14D), and large infinite two-dimensional nanoarrays (Figure 14E) [129]. These supramolecular L-DNA self-assemblies enriched the library of unnatural nucleic acids, presenting identical physical properties and exceptional nuclease resistance to survive in a biological environment.
A wireframe L-DNA tetrahedron (L-Td) has also been demonstrated by Kim et al. in 2016 [130]. This work was based on the native D-DNA tetrahedra (D-Td), designed with a tetrahedral shape formed by the precise organization of complementary base-pairs [131]. These D-DNA tetrahedra possessed cell-penetrating properties, which enabled the nanoconstructs to effectively carry various bioactive molecules as an intracellular delivery machine in the absence of traditional transfection agents [132,133]. The enantiomeric L-DNA-assembled tetrahedra have shown enhanced cellular uptake, biostability and pharmacokinetics in vivo compared to natural DNA tetrahedra. Due to the extraordinary properties of L-Td, it has exhibited a highly effective tumor-specific delivery of doxorubicin (DOX). Additional application of L-Td includes the streptavidin-Td (STV-Td) hybrid developed as a powerful carrier for intracellular delivery of various enzymes, such as the anti-proliferative proteins/enzymes, leading to significant suppression of tumor growth [134]. Therefore, the STV-Td hybrid potentially enlarged the inventory pool of protein therapeutics for drugging undruggable molecular cancer-relevant targets [135]. However, the restriction on length and size of L-DNAs/RNAs due to the inability of syntheses by traditional enzymatic polymerization largely limits the potential applications of L-nanodevices.
Recent work in the Sczepanski lab has reported a novel methodology taking advantage of peptide nucleic acids (PNAs), which have no chirality but can hybridize to DNA and RNA, thus enabling two toehold-mediated strand displacement [136,137]. The strand displacement utilized a DNA:PNA heteroduplex for the sequence-specific interfacing of the two enantiomers of DNAs. This work notably lays the foundation for an interfacing heterochiral circuit employed as diagnostic and/or therapeutic nanodevices and has great potential in constructing biorthogonal nanoparticles comprised of diverse nucleic acids in living cells and organisms.
5. Conclusions and Outlook
Nucleic acid therapeutics is a rapidly emerging field of biotechnology for disease treatment, especially after the unprecedented and prompt development of mRNA vaccines during the pandemic [138]. As the nucleic acid substitutes containing unusual chirality, L-nucleic acids are featured with unique properties, which can help to address some long-lasting bottlenecks in this area. The rapid growth of L-nucleic acids and L-nucleoside therapeutics can solve the conventional problems of stability in vivo, delivery efficiency and, possibly, immunogenicity. These innovative molecules have created a versatile and powerful platform, which has unlimited potential to meet future clinical needs and care for many diseases.
Whereas, there is room to further improve L-nucleoside and L-nucleic acids therapeutics. The discovery and development of L-nucleoside molecules with broad-spectrum antiviral activities will be a primary target in the coming decades. Moreover, unlike the biotechnology of using native D-antisense, siRNA, microRNA, and aptamer drugs in clinical treatment, there are not many chemically modified L-nucleic acids available. Most of the L-nucleic acid aptamers and nanoparticles under development are based on unmodified L-nucleotides. Though the primary intention of modifying native DNA/RNA drugs was to enhance their stability against serum, this concern is negligible for L-nucleic acids; however, versatile modifications on nucleobases and backbones can also offer unique chemical and structural features [139]. For example, the bulky derivatizations on (deoxy)riboses can increase the thermostabilities, and some hydrophobic substitutions would boost the binding capacities of the aptamers. If L-nucleic acids are equipped with diverse chemical moieties, it will certainly offer more expanded functions to benefit their application. There have been a few examples that chemically derivatized L-DNAs/RNAs have more optimal properties [94,140,141].
Secondly, the preparation of L-oligonucleotides is restricted by the efficiency of solid-phase synthesis. Phosphoramidite chemistry is the main approach to synthesize L-oligonucleotides, and it is inefficient and costly when longer strands are needed, especially in the case of RNA. After billions of years of evolution, there is a lack of competent polymerases to accurately recognize and readily amplify L-nucleic acids. The efforts are now delivered to engineer non-natural polymerases, either via the chemical syntheses of entirely modified D-polymerases [142,143,144,145] or through searching for the ancient polymerase, which can interact with both enantiomeric nucleic acids [146]. It is worth noting, some labs are also focused on developing artificial ribosomes, which contain mutated 23S ribosomal RNA [147,148,149]. The purpose is to engender the ribosome to carry on the D-peptide elongation when the tRNA substrate is aminoacylated with D-amino acid residue. This evolved ribosome would be applied in the translation process to efficiently produce mirror-image D-enzymes, such as polymerases, ligases, helicases, and other L-nucleic acid-contacting enzymes, to eventually realize the high-throughput syntheses of L-nucleic acids. If accomplished, it could generate great potential in the application of mirror-image biological molecules, from the bench of basic science to the bedside of clinical disease treatment.
Author Contributions
All authors (Y.D., Y.Z. and W.Z.) contributed to defining the form and content of this review article and writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We acknowledge the current and past members of Zhang lab for the critical reading of the manuscript and helpful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.A.; Doudna, J.A. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 457–475. [Google Scholar] [CrossRef] [PubMed]
- Bonner, W.A. Chirality and life. Orig. Life Evol. Biosph. 1995, 25, 175–190. [Google Scholar] [CrossRef]
- Ohide, H.; Miyoshi, Y.; Maruyama, R.; Hamase, K.; Konno, R. d-Amino acid metabolism in mammals: Biosynthesis, degradation and analytical aspects of the metabolic study. J. Chromatogr. B 2011, 879, 3162–3168. [Google Scholar] [CrossRef]
- Volkmann, R.; Heck, S. Biosynthesis of d-amino acid-containing peptides: Exploring the role of peptide isomerases. EXS 1998, 85, 87–105. [Google Scholar]
- Chen, Y.; Ma, W. The origin of biological homochirality along with the origin of life. PLoS Comput. Biol. 2020, 16, e1007592. [Google Scholar] [CrossRef]
- Garay, J. Active centrum hypothesis: The origin of chiral homogeneity and the RNA-world. Biosystems 2011, 103, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. The RNA worlds in context. Cold Spring Harb. Perspect. Biol. 2012, 4, a006742. [Google Scholar] [CrossRef] [Green Version]
- Hauser, N.C.; Martinez, R.; Jacob, A.; Rupp, S.; Hoheisel, J.D.; Matysiak, S. Utilising the left-helical conformation of l-DNA for analysing different marker types on a single universal microarray platform. Nucleic Acids Res. 2006, 34, 5101–5111. [Google Scholar] [CrossRef] [Green Version]
- Gleave, M.E.; Monia, B.P. Antisense therapy for cancer. Nat. Rev. Cancer 2005, 5, 468–479. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Acton, E.M.; Ryan, K.J.; Goodman, L. Synthesis of l-ribofuranose and l-adenosine. J. Am. Chem. Soc. 1964, 86, 5352–5354. [Google Scholar] [CrossRef]
- Ke, G.; Wang, C.; Ge, Y.; Zheng, N.; Zhu, Z.; Yang, C.J. l-DNA molecular beacon: A safe, stable, and accurate intracellular nano-thermometer for temperature sensing in living cells. J. Am. Chem. Soc. 2012, 134, 18908–18911. [Google Scholar] [CrossRef]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: The evolution of Spiegelmer® therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Young, B.E.; Kundu, N.; Sczepanski, J.T. Mirror-image oligonucleotides: History and emerging applications. Chemistry 2019, 25, 7981. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. Strategies in the design of antiviral drugs. Nat. Rev. Drug Discov. 2002, 1, 13–25. [Google Scholar] [CrossRef]
- De Clercq, E. Antivirals and antiviral strategies. Nat. Rev. Microbiol. 2004, 2, 704–720. [Google Scholar] [CrossRef] [PubMed]
- Périgaud, C.; Gosselin, G.; Imbach, J. Nucleoside analogues as chemotherapeutic agents: A review. Nucleosides Nucleotides Nucleic Acids 1992, 11, 903–945. [Google Scholar] [CrossRef]
- Clercq, E.D. Antiviral activity spectrum and target of action of different classes of nucleoside analogues. Nucleosides Nucleotides Nucleic Acids 1994, 13, 1271–1295. [Google Scholar] [CrossRef]
- Chien, M.; Anderson, T.K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.Y. Addressing the selectivity and toxicity of antiviral nucleosides. Antivir. Chem. Chemother. 2018, 26, 2040206618758524. [Google Scholar] [CrossRef]
- De Clercq, E.; Cools, M.; Balzarini, J.; Marquez, V.E.; Borcherding, D.R.; Borchardt, R.T.; Drach, J.C.; Kitaoka, S.; Konno, T. Broad-spectrum antiviral activities of neplanocin A, 3-deazaneplanocin A, and their 5′-nor derivatives. Antimicrob. Agents Chemother. 1989, 33, 1291–1297. [Google Scholar] [CrossRef] [Green Version]
- Qi Chen, A.D. Synthesis, conformational study and antiviral activity of l-like neplanocin derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 4436–4439. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Q.; Schneller, S.W. Enantiomeric 3-deaza-1′,6′-isoneplanocin and its 3-bromo analogue: Synthesis by the Ullmann reaction and their antiviral properties. Bioorg. Med. Chem. Lett. 2016, 26, 928–930. [Google Scholar] [CrossRef]
- Kicska, G.A.; Long, L.; Hörig, H.; Fairchild, C.; Tyler, P.C.; Furneaux, R.H.; Schramm, V.L.; Kaufman, H.L. Immucillin H, a powerful transition-state analog inhibitor of purine nucleoside phosphorylase, selectively inhibits human T lymphocytes. Proc. Natl. Acad. Sci. USA 2001, 98, 4593–4598. [Google Scholar] [CrossRef] [Green Version]
- Evans, G.B.; Furneaux, R.H.; Lewandowicz, A.; Schramm, V.L.; Tyler, P.C. Exploring structure− activity relationships of transition state analogues of human purine nucleoside phosphorylase. J. Med. Chem. 2003, 46, 3412–3423. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Ting, L.-M.; Kicska, G.A.; Lewandowicz, A.; Tyler, P.C.; Evans, G.B.; Furneaux, R.H.; Kim, K.; Almo, S.C.; Schramm, V.L. Plasmodium falciparum purine nucleoside phosphorylase: Crystal structures, immucillin inhibitors, and dual catalytic function. J. Biol. Chem. 2004, 279, 18103–18106. [Google Scholar] [CrossRef] [Green Version]
- Ho, M.-C.; Shi, W.; Rinaldo-Matthis, A.; Tyler, P.C.; Evans, G.B.; Clinch, K.; Almo, S.C.; Schramm, V.L. Four generations of transition-state analogues for human purine nucleoside phosphorylase. Proc. Natl. Acad. Sci. USA 2010, 107, 4805–4812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinch, K.; Evans, G.B.; Fleet, G.W.; Furneaux, R.H.; Johnson, S.W.; Lenz, D.H.; Mee, S.P.; Rands, P.R.; Schramm, V.L.; Ringia, E.A.T.; et al. Syntheses and bio-activities of the l-enantiomers of two potent transition state analogue inhibitors of purine nucleoside phosphorylases. Org. Biomol. Chem. 2006, 4, 1131–1139. [Google Scholar] [CrossRef]
- Evans, G.B.; Furneaux, R.H.; Tyler, P.C.; Schramm, V.L. Synthesis of a transition state analogue inhibitor of purine nucleoside phosphorylase via the Mannich reaction. Org. Lett. 2003, 5, 3639–3640. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Med. Res. Rev. 2000, 20, 323–349. [Google Scholar] [CrossRef]
- Götte, M.; Rausch, J.W.; Marchand, B.; Sarafianos, S.; Le Grice, S.F. Reverse transcriptase in motion: Conformational dynamics of enzyme–substrate interactions. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1202–1212. [Google Scholar] [CrossRef] [Green Version]
- D’Andrea, G.; Brisdelli, F.; Bozzi, A. AZT: An old drug with new perspectives. Curr. Clin. Pharmacol. 2008, 3, 20–37. [Google Scholar] [PubMed]
- Perry, C.M.; Noble, S. Didanosine. Drugs 1999, 58, 1099–1135. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Pauwels, R.; Herdewijn, P.; De Clercq, E.; Desmyter, J.; Vandeputte, M. Both 2′,3′-dideoxythymidine and its 2′,3′-unsaturated derivative (2′,3′-dideoxythymidinene) are potent and selective inhibitors of human immunodeficiency virus replication in vitro. Biochem. Biophys. Res. Commun. 1987, 142, 128–134. [Google Scholar] [CrossRef]
- Hitchcock, M. 2′,3′-Didehydro-2′,3′-dideoxythymidine (D4T), an anti-HIV agent. Antivir. Chem. Chemother. 1991, 2, 125–132. [Google Scholar] [CrossRef]
- Gavriliu, D.; Fossey, C.; Fontaine, G.; Benzaria, S.; Ciurea, A.; Delbederi, Z.; Lelong, B.; Laduree, D.; Aubertin, A.; Kirn, A. Synthesis and antiviral activity of C-5 substituted analogues of d4T bearing methylamino-or methyldiamino-linker arms. Nucleosides Nucleotides Nucleic Acids 2000, 19, 1017–1031. [Google Scholar] [CrossRef]
- Park, A.-Y.; Kim, W.H.; Kang, J.-A.; Lee, H.J.; Lee, C.-K.; Moon, H.R. Synthesis of enantiomerically pure d-and l-bicyclo[3.1.0] hexenyl carbanucleosides and their antiviral evaluation. Bioorg. Med. Chem. 2011, 19, 3945–3955. [Google Scholar] [CrossRef]
- Park, A.-Y.; Moon, H.R.; Kim, K.R.; Chun, M.W.; Jeong, L.S. Synthesis of novel l-N-MCd4T as a potent anti-HIV agent. Org. Biomol. Chem. 2006, 4, 4065–4067. [Google Scholar] [CrossRef]
- Gosselin, G.; Schinazi, R.F.; Sommadossi, J.-P.; Mathé, C.; Bergogne, M.-C.; Aubertin, A.-M.; Kirn, A.; Imbach, J.-L. Anti-human immunodeficiency virus activities of the β-L enantiomer of 2′,3′-dideoxycytidine and its 5-fluoro derivative in vitro. Antimicrob. Agents Chemother. 1994, 38, 1292–1297. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; McAtee, J.J.; Schlueter Wirtz, S.; Tharnish, P.; Juodawlkis, A.; Liotta, D.C.; Schinazi, R.F. Synthesis and biological evaluation of 2′,3′-didehydro-2′,3′-dideoxy-5-fluorocytidine (D4FC) analogues: Discovery of carbocyclic nucleoside triphosphates with potent inhibitory activity against HIV-1 reverse transcriptase. J. Med. Chem. 1999, 42, 859–867. [Google Scholar] [CrossRef]
- Lin, T.-S.; Luo, M.-Z.; Liu, M.-C.; Zhu, Y.-L.; Gullen, E.; Dutschman, G.E.; Cheng, Y.-C. Design and synthesis of 2′,3′-dideoxy-2′,3′-didehydro-β-l-cytidine (β-l-d4C) and 2′,3′-dideoxy-2′,3′-didehydro-β-l-5-fluorocytidine (β-l-Fd4C), two exceptionally potent inhibitors of human hepatitis B virus (HBV) and potent inhibitors of human immunodeficiency virus (HIV) in vitro. J. Med. Chem. 1996, 39, 1757–1759. [Google Scholar]
- Schinazi, R.F.; McMillan, A.; Cannon, D.; Mathis, R.; Lloyd, R.M.; Peck, A.; Sommadossi, J.-P.; St Clair, M.; Wilson, J.; Furman, P. Selective inhibition of human immunodeficiency viruses by racemates and enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1, 3-oxathiolan-5-yl] cytosine. Antimicrob. Agents Chemother. 1992, 36, 2423–2431. [Google Scholar] [CrossRef] [Green Version]
- Soudeyns, H.; Yao, X.; Gao, Q.; Belleau, B.; Kraus, J.-L.; Nguyen-Ba, N.; Spira, B.; Wainberg, M.A. Anti-human immunodeficiency virus type 1 activity and in vitro toxicity of 2′-deoxy-3′-thiacytidine (BCH-189), a novel heterocyclic nucleoside analog. Antimicrob. Agents Chemother. 1991, 35, 1386–1390. [Google Scholar] [CrossRef] [Green Version]
- Coates, J.; Cammack, N.; Jenkinson, H.; Mutton, I.; Pearson, B.; Storer, R.; Cameron, J.; Penn, C. The separated enantiomers of 2′-deoxy-3′-thiacytidine (BCH 189) both inhibit human immunodeficiency virus replication in vitro. Antimicrob. Agents Chemother. 1992, 36, 202–205. [Google Scholar] [CrossRef] [Green Version]
- Krooth, R.S.; Wen-Luan, H.; Lam, G.F. Effect of 6-azauracil, and of certain structurally similar compounds, on three pyridoxal-phosphate requiring enzymes involved in neurotransmitter metabolism. Biochem. Pharmacol. 1979, 28, 1071–1076. [Google Scholar] [CrossRef]
- Karon, M.; Sieger, L.; Leimbrock, S.; Finklestein, J.Z.; Nesbit, M.E.; Swaney, J.J. 5-Azacytidine: A new active agent for the treatment of acute leukemia. Blood 1973, 42, 359–365. [Google Scholar] [CrossRef]
- Luo, M.-Z.; Liu, M.-C.; Mozdziesz, D.E.; Lin, T.-S.; Dutschman, G.E.; Gullen, E.A.; Cheng, Y.-C.; Sartorelli, A.C. Synthesis and biological evaluation of l-and d-configuration 1, 3-dioxolane 5-azacytosine and 6-azathymine nucleosides. Bioorg. Med. Chem. Lett. 2000, 10, 2145–2148. [Google Scholar] [CrossRef]
- Pejanović, V.; Stokić, Z.; Stojanović, B.; Piperski, V.; Popsavin, M.; Popsavin, V. Synthesis and biological evaluation of some novel 4′-Thio-l-ribonucleosides with modified nucleobase moieties. Bioorg. Med. Chem. Lett. 2003, 13, 1849–1852. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef]
- Capizzi, R.; White, J.C.; Powell, B.; Perrino, F. Effect of dose on the pharmacokinetic and pharmacodynamic effects of cytarabine. Semin. Hematol. 1991, 28, 54–69. [Google Scholar]
- Galmarini, C.M.; Thomas, X.; Calvo, F.; Rousselot, P.; El Jafaari, A.; Cros, E.; Dumontet, C. Potential mechanisms of resistance to cytarabine in AML patients. Leuk. Res. 2002, 26, 621–629. [Google Scholar] [CrossRef]
- Bankar, A.; Siriwardena, T.P.; Rizoska, B.; Rydergård, C.; Kylefjord, H.; Rraklli, V.; Eneroth, A.; Pinho, P.; Norin, S.; Bylund, J. Novel l-nucleoside analog, 5-fluorotroxacitabine, displays potent efficacy against acute myeloid leukemia. Haematologica 2021, 106, 574. [Google Scholar] [CrossRef] [Green Version]
- Dienstag, J.; Easley, C.; Kirkpatrick, P. Telbivudine. Nat. Rev. Drug Discov. 2007, 6, 267–268. [Google Scholar] [CrossRef]
- Férir, G.; Kaptein, S.; Neyts, J.; De Clercq, E. Antiviral treatment of chronic hepatitis B virus infections: The past, the present and the future. Rev. Med. Virol. 2008, 18, 19–34. [Google Scholar] [CrossRef]
- Venhoff, N.; Setzer, B.; Melkaoui, K.; Walker, U.A. Mitochondrial toxicity of tenofovir, emtricitabine and abacavir alone and in combination with additional nucleoside reverse transcriptase inhibitors. Antivir. Ther. 2007, 12, 1075. [Google Scholar]
- Zhang, H.-W.; Detorio, M.; Herman, B.D.; Solomon, S.; Bassit, L.; Nettles, J.H.; Obikhod, A.; Tao, S.-J.; Mellors, J.W.; Sluis-Cremer, N. Synthesis, antiviral activity, cytotoxicity and cellular pharmacology of l-3′-azido-2′,3′-dideoxypurine nucleosides. Eur. J. Med. Chem. 2011, 46, 3832–3844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, H. The function of the 5′cap of mRNA and nuclear RNA species. Perspect. Biol. Med. 1976, 19, 549–567. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, B.; Shi, P.-Y. Flavivirus methyltransferase: A novel antiviral target. Antiviral Res. 2008, 80, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cools, M.; De Clercq, E. Correlation between the antiviral activity of acyclic and carbocyclic adenosine analogues in murine L929 cells and their inhibitory effect on L929 cell S-adenosylhomocysteine hydrolase. Biochem. Pharmacol. 1989, 38, 1061–1067. [Google Scholar] [CrossRef]
- Wang, J.-F.; Yang, X.-D.; Zhang, L.-R.; Yang, Z.-J.; Zhang, L.-H. Synthesis and biological activities of 5′-ethylenic and acetylenic modified l-nucleosides and isonucleosides. Tetrahedron 2004, 60, 8535–8546. [Google Scholar] [CrossRef]
- Zhu, W.; Gumina, G.; Schinazi, R.F.; Chu, C.K. Synthesis and anti-HIV activity of l-β-3′-C-cyano-2′,3′-unsaturated nucleosides and l-3′-C-cyano-3′-deoxyribonucleosides. Tetrahedron 2003, 59, 6423–6431. [Google Scholar] [CrossRef]
- Hall, C.B.; McBride, J.T.; Walsh, E.E.; Bell, D.M.; Gala, C.L.; Hildreth, S.; Ten Eyck, L.G.; Hall, W.J. Aerosolized ribavirin treatment of infants with respiratory syncytial viral infection: A randomized double-blind study. N. Engl. J. Med. 1983, 308, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Reichard, O.; Norkrans, G.; Frydén, A.; Braconier, J.-H.; Sönnerborg, A.; Weiland, O.; Group, S.S. Randomised, double-blind, placebo-controlled trial of interferon α-2b with and without ribavirin for chronic hepatitis C. Lancet 1998, 351, 83–87. [Google Scholar] [CrossRef]
- Tam, R.C.; Lim, C.; Bard, J.; Pai, B. Contact hypersensitivity responses following ribavirin treatment in vivo are influenced by type 1 cytokine polarization, regulation of IL-10 expression, and costimulatory signaling. J. Immunol. 1999, 163, 3709–3717. [Google Scholar]
- Ramasamy, K.S.; Tam, R.C.; Bard, J.; Averett, D.R. Monocyclic l-nucleosides with type 1 cytokine-inducing activity. J. Med. Chem. 2000, 43, 1019–1028. [Google Scholar] [CrossRef]
- Krauer, F.; Riesen, M.; Reveiz, L.; Oladapo, O.T.; Martínez-Vega, R.; Porgo, T.V.; Haefliger, A.; Broutet, N.J.; Low, N.; Group, W.Z.C.W. Zika virus infection as a cause of congenital brain abnormalities and Guillain–Barré syndrome: Systematic review. PLoS Med. 2017, 14, e1002203. [Google Scholar] [CrossRef] [Green Version]
- Bassetto, M.; Cima, C.M.; Basso, M.; Salerno, M.; Schwarze, F.; Friese, D.; Bugert, J.J.; Brancale, A. Novel Nucleoside Analogues as Effective Antiviral Agents for Zika Virus Infections. Molecules 2020, 25, 4813. [Google Scholar] [CrossRef] [PubMed]
- Miralles-Llumà, R.; Figueras, A.; Busqué, F.; Alvarez-Larena, A.; Balzarini, J.; Figueredo, M.; Font, J.; Alibés, R.; Maréchal, J.D. Synthesis, antiviral evaluation, and computational studies of cyclobutane and cyclobutene l-nucleoside analogues. Eur. J. Org. Chem. 2013, 2013, 7761–7775. [Google Scholar] [CrossRef]
- D’Alonzo, D.; Guaragna, A.; Van Aerschot, A.; Herdewijn, P.; Palumbo, G. Toward l-homo-DNA: Stereoselective de novo synthesis of β-L-erythro-hexopyranosyl nucleosides. J. Org. Chem. 2010, 75, 6402–6410. [Google Scholar] [CrossRef]
- Mathé, C.; Gosselin, G. Synthesis and antiviral evaluation of the β-L-enantiomers of some thymine 3′-deoxypentofuranonucleoside derivatives. Nucleosides Nucleotides Nucleic Acids 2000, 19, 1517–1530. [Google Scholar] [CrossRef]
- Kitano, K.; Kohgo, S.; Yamada, K.; Sakata, S.; Ashida, N.; Hayakawa, H.; Nameki, D.; Kodama, E.; Matsuoka, M.; Mitsuya, H. Attempt to reduce cytotoxicity by synthesizing the l-enantiomer of 4′-C-ethynyl-2′-deoxypurine nucleosides as antiviral agents against HIV and HBV. Antivir. Chem. Chemother. 2004, 15, 161–167. [Google Scholar] [CrossRef]
- Sørensen, M.D.; Kværnø, L.; Bryld, T.; Håkansson, A.E.; Verbeure, B.; Gaubert, G.; Herdewijn, P.; Wengel, J. α-L-ribo-configured locked nucleic acid (α-L-LNA): Synthesis and properties. J. Am. Chem. Soc. 2002, 124, 2164–2176. [Google Scholar] [CrossRef] [PubMed]
- Babu, B.R.; Sørensen, M.D.; Parmar, V.S.; Harrit, N.H.; Wengel, J. Oligodeoxynucleotides containing α-l-ribo configured LNA-type C-aryl nucleotides. Org. Biomol. Chem. 2004, 2, 80–89. [Google Scholar]
- Seth, P.P.; Allerson, C.A.; Østergaard, M.E.; Swayze, E.E. Synthesis and biophysical evaluation of 3′-Me-α-l-LNA–Substitution in the minor groove of α-l-LNA duplexes. Bioorg. Med. Chem. Lett. 2011, 21, 4690–4694. [Google Scholar] [CrossRef]
- Kumar, T.S.; Madsen, A.S.; Wengel, J.; Hrdlicka, P.J. Synthesis and hybridization studies of 2′-amino-α-L-LNA and tetracyclic “Locked LNA”. J. Org. Chem. 2006, 71, 4188–4201. [Google Scholar] [CrossRef]
- Seela, F.; Lin, W.; Kazimierczuk, Z.; Rosemeyer, H.; Glaçon, V.; Peng, X.; He, Y.; Ming, X.; Andrzejewska, M.; Gorska, A. l-nucleosides containing modified nucleobases. Nucleosides Nucleotides Nucleic Acids 2005, 24, 859–863. [Google Scholar] [CrossRef]
- Jessel, S.; Meier, C. Synthesis of 2′,3′-Modified Carbocyclic l-Nucleoside Analogues. Eur. J. Org. Chem. 2011, 2011, 1702–1713. [Google Scholar] [CrossRef]
- Song, W.-S.; Liu, S.-X.; Chang, C.-C. Synthesis of l-Deoxyribonucleosides from d-Ribose. J. Org. Chem. 2018, 83, 14923–14932. [Google Scholar] [CrossRef]
- Aragonès, S.; Bravo, F.; Díaz, Y.; Matheu, M.I.; Castillón, S. Stereoselective synthesis of l-isonucleosides. Tetrahedron Lett. 2003, 44, 3771–3773. [Google Scholar] [CrossRef]
- Mathé, C.; Gosselin, G. l-nucleoside enantiomers as antivirals drugs: A mini-review. Antivir. Res. 2006, 71, 276–281. [Google Scholar] [CrossRef]
- Mahmoud, S.; Hasabelnaby, S.; Hammad, S.; Sakr, T. Antiviral nucleoside and nucleotide analogs: A review. J. Adv. Pharm. Res. 2018, 2, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Klussmann, S.; Nolte, A.; Bald, R.; Erdmann, V.A.; Fürste, J.P. Mirror-image RNA that binds d-adenosine. Nat. Biotechnol. 1996, 14, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Alderman, D. Malachite green: A review. J. Fish Dis. 1985, 8, 289–298. [Google Scholar] [CrossRef]
- Srivastava, S.; Sinha, R.; Roy, D. Toxicological effects of malachite green. Aquat. Toxicol. 2004, 66, 319–329. [Google Scholar] [CrossRef]
- Luo, X.; Chen, Z.; Li, H.; Li, W.; Cui, L.; Huang, J. Exploiting the application of l-aptamer with excellent stability: An efficient sensing platform for malachite green in fish samples. Analyst 2019, 144, 4204–4209. [Google Scholar] [CrossRef]
- Feng, X.-N.; Cui, Y.-X.; Zhang, J.; Tang, A.-N.; Mao, H.-B.; Kong, D.-M. Chiral interaction is a decisive factor to replace d-DNA with l-DNA aptamers. Anal. Chem. 2020, 92, 6470–6477. [Google Scholar] [CrossRef]
- Chen, H.; Xie, S.; Liang, H.; Wu, C.; Cui, L.; Huan, S.-Y.; Zhang, X. Generation of biostable l-aptamers against achiral targets by chiral inversion of existing d-aptamers. Talanta 2017, 164, 662–667. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Joyce, G.F. Specific inhibition of microRNA processing using l-RNA aptamers. J. Am. Chem. Soc. 2015, 137, 16032–16037. [Google Scholar] [CrossRef] [Green Version]
- Dey, S.; Sczepanski, J.T. In vitro selection of l-DNA aptamers that bind a structured d-RNA molecule. Nucleic Acids Res. 2020, 48, 1669–1680. [Google Scholar] [CrossRef]
- Kabza, A.M.; Sczepanski, J.T. An l-RNA aptamer with expanded chemical functionality that Inhibits microRNA biogenesis. ChemBioChem 2017, 18, 1824–1827. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Joyce, G.F. Binding of a structured d-RNA molecule by an l-RNA aptamer. J. Am. Chem. Soc. 2013, 135, 13290–13293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umar, M.I.; Kwok, C.K. Specific suppression of d-RNA G-quadruplex–protein interaction with an l-RNA aptamer. Nucleic Acids Res. 2020, 48, 10125–10141. [Google Scholar] [CrossRef]
- Chan, C.Y.; Kwok, C.K. Specific binding of a d-RNA G-quadruplex structure with an l-RNA aptamer. Angew. Chem. Int. Ed. 2020, 132, 5331–5335. [Google Scholar] [CrossRef]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef]
- Brázda, V.; Hároníková, L.; Liao, J.C.; Fojta, M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sczepanski, J. Targeting a conserved structural element from the SARS-CoV-2 genome using l-DNA aptamers. RSC Chem. Biol. 2022. [Google Scholar] [CrossRef]
- Gilbert, C.; Tengs, T. No species-level losses of s2m suggests critical role in replication of SARS-related coronaviruses. Sci. Rep. 2021, 11, 16145. [Google Scholar] [CrossRef] [PubMed]
- Nolte, A.; Klußmann, S.; Bald, R.; Erdmann, V.A.; Fürste, J.P. Mirror-design of l-oligonucleotide ligands binding to l-arginine. Nat. Biotechnol. 1996, 14, 1116–1119. [Google Scholar] [CrossRef] [PubMed]
- Ravelet, C.; Boulkedid, R.; Ravel, A.; Grosset, C.; Villet, A.; Fize, J.; Peyrin, E. A l-RNA aptamer chiral stationary phase for the resolution of target and related compounds. J. Chromatogr. A 2005, 1076, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Brumbt, A.; Ravelet, C.; Grosset, C.; Ravel, A.; Villet, A.; Peyrin, E. Chiral stationary phase based on a biostable l-RNA aptamer. Anal. Chem. 2005, 77, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Ruta, J.; Ravelet, C.; Grosset, C.; Fize, J.; Ravel, A.; Villet, A.; Peyrin, E. Enantiomeric separation using an l-RNA aptamer as chiral additive in partial-filling capillary electrophoresis. Anal. Chem. 2006, 78, 3032–3039. [Google Scholar] [CrossRef] [PubMed]
- Ruta, J.; Grosset, C.; Ravelet, C.; Fize, J.; Villet, A.; Ravel, A.; Peyrin, E. Chiral resolution of histidine using an anti-d-histidine l-RNA aptamer microbore column. J. Chromatogr. B 2007, 845, 186–190. [Google Scholar] [CrossRef]
- Jørgensen, A.S.; Hansen, L.H.; Vester, B.; Wengel, J. Improvement of a streptavidin-binding aptamer by LNA-and α-l-LNA-substitutions. Bioorg. Med. Chem. Lett. 2014, 24, 2273–2277. [Google Scholar] [CrossRef]
- Ricklin, D.; Hajishengallis, G.; Yang, K.; Lambris, J.D. Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 2010, 11, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Bosmann, M.; Ward, P.A. Role of C3, C5 and anaphylatoxin receptors in acute lung injury and in sepsis. In Current Topics in Innate Immunity II; Springer Nature: London, UK, 2012; pp. 147–159. [Google Scholar]
- Czermak, B.J.; Sarma, V.; Pierson, C.L.; Warner, R.L.; Huber-Lang, M.; Bless, N.M.; Schmal, H.; Friedl, H.P.; Ward, P.A. Protective effects of C5a blockade in sepsis. Nat. Med. 1999, 5, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Hoehlig, K.; Maasch, C.; Shushakova, N.; Buchner, K.; Huber-Lang, M.; Purschke, W.G.; Vater, A.; Klussmann, S. A novel C5a-neutralizing mirror-image (l-) aptamer prevents organ failure and improves survival in experimental sepsis. Mol. Ther. 2013, 21, 2236–2246. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Winkelmann, V.E.; Braumüller, S.; Palmer, A.; Schultze, A.; Klohs, B.; Ignatius, A.; Vater, A.; Fauler, M.; Frick, M. Role of the C5a-C5a receptor axis in the inflammatory responses of the lungs after experimental polytrauma and hemorrhagic shock. Sci. Rep. 2021, 11, 2158. [Google Scholar] [CrossRef] [PubMed]
- Yatime, L.; Maasch, C.; Hoehlig, K.; Klussmann, S.; Andersen, G.R.; Vater, A. Structural basis for the targeting of complement anaphylatoxin C5a using a mixed l-RNA/l-DNA aptamer. Nat. Commun. 2015, 6, 6481. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, E.L.; Sattler, M.; Azab, A.K.; Eulberg, D.; Kruschinski, A.; Manley, P.W.; Stone, R.; Griffin, J.D. Inhibition of SDF-1-induced migration of oncogene-driven myeloid leukemia by the l-RNA aptamer (Spiegelmer), NOX-A12, and potentiation of tyrosine kinase inhibition. Oncotarget 2017, 8, 109973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citro, A.; Pellegrini, S.; Dugnani, E.; Eulberg, D.; Klussmann, S.; Piemonti, L. CCL 2/MCP-1 and CXCL 12/SDF-1 blockade by l-aptamers improve pancreatic islet engraftment and survival in mouse. Am. J. Transplant. 2019, 19, 3131–3138. [Google Scholar] [CrossRef] [PubMed]
- Eulberg, D.; Purschke, W.; Anders, H.-J.; Selve, N.; Klussmann, S. Spiegelmer NOX-E36 for Renal Diseases. In Therapeutic Oligonucleotides, 1st ed.; Royal Society of Chemistry: Cambridge, UK, 2008; Volume 12, pp. 200–225. [Google Scholar]
- Oberthür, D.; Achenbach, J.; Gabdulkhakov, A.; Buchner, K.; Maasch, C.; Falke, S.; Rehders, D.; Klussmann, S.; Betzel, C. Crystal structure of a mirror-image l-RNA aptamer (Spiegelmer) in complex with the natural l-protein target CCL2. Nat. Commun. 2015, 6, 6923. [Google Scholar] [CrossRef] [Green Version]
- Pyne, N.J.; Pyne, S. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer 2010, 10, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Gandy, K.A.O.; Obeid, L.M. Targeting the sphingosine kinase/sphingosine 1-phosphate pathway in disease: Review of sphingosine kinase inhibitors. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2013, 1831, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Purschke, W.G.; Hoehlig, K.; Buchner, K.; Zboralski, D.; Schwoebel, F.; Vater, A.; Klussmann, S. Identification and characterization of a mirror-image oligonucleotide that binds and neutralizes sphingosine 1-phosphate, a central mediator of angiogenesis. Biochem. J. 2014, 462, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.F. Calcitonin gene-related peptide (CGRP): A new target for migraine. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 533–552. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.J.; Schmidt, J.; Koulchitsky, S.; Klussmann, S.; Vater, A.; Messlinger, K. Effect of a calcitonin gene-related peptide-binding l-RNA aptamer on neuronal activity in the rat spinal trigeminal nucleus. J. Headache Pain 2018, 19, 3. [Google Scholar] [CrossRef] [Green Version]
- Sumter, T.; Xian, L.; Huso, T.; Koo, M.; Chang, Y.-T.; Almasri, T.; Chia, L.; Inglis, C.; Reid, D.; Resar, L. The high mobility group A1 (HMGA1) transcriptome in cancer and development. Curr. Mol. Med. 2016, 16, 353–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maasch, C.; Vater, A.; Buchner, K.; Purschke, W.G.; Eulberg, D.; Vonhoff, S.; Klussmann, S. Polyetheylenimine-polyplexes of Spiegelmer NOX-A50 directed against intracellular high mobility group protein A1 (HMGA1) reduce tumor growth in vivo. J. Med. Chem. 2010, 285, 40012–40018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olea Jr, C.; Weidmann, J.; Dawson, P.E.; Joyce, G.F. An l-RNA aptamer that binds and inhibits RNase. Chem. Biol. 2015, 22, 1437–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wlotzka, B.; Leva, S.; Eschgfäller, B.; Burmeister, J.; Kleinjung, F.; Kaduk, C.; Muhn, P.; Hess-Stumpp, H.; Klussmann, S. In vivo properties of an anti-GnRH Spiegelmer: An example of an oligonucleotide-based therapeutic substance class. Proc. Natl. Acad. Sci. USA 2002, 99, 8898–8902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dose, C.; Ho, D.; Gaub, H.E.; Dervan, P.B.; Albrecht, C.H. Recognition of “Mirror-Image” DNA by Small Molecules. Angew. Chem. Int. Ed. 2007, 119, 8536–8539. [Google Scholar] [CrossRef]
- Williams, K.P.; Liu, X.-H.; Schumacher, T.N.; Lin, H.Y.; Ausiello, D.A.; Kim, P.S.; Bartel, D.P. Bioactive and nuclease-resistant l-DNA ligand of vasopressin. Proc. Natl. Acad. Sci. USA 1997, 94, 11285–11290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; Ke, Y.; Li, Z.; Wang, J.H.; Liu, Y.; Yan, H. Mirror image DNA nanostructures for chiral supramolecular assemblies. Nano Lett. 2009, 9, 433–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.-R.; Kim, H.Y.; Lee, Y.-D.; Ha, J.S.; Kang, J.H.; Jeong, H.; Bang, D.; Ko, Y.T.; Kim, S.; Lee, H. Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs. J. Control. Release 2016, 243, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.S.; Yin, H.; Erben, C.M.; Wood, M.J.; Turberfield, A.J. DNA cage delivery to mammalian cells. ACS Nano 2011, 5, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lytton-Jean, A.K.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Yu, T.; Clifford, C.; Liu, Y.; Yan, H.; Chang, Y. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012, 12, 4254–4259. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.-R.; Hwang, D.; Kim, J.; Lee, C.-Y.; Lee, W.; Yoon, D.S.; Shin, D.; Min, S.-J.; Kwon, I.C.; Chung, H.S. Streptavidin-mirror DNA tetrahedron hybrid as a platform for intracellular and tumor delivery of enzymes. J. Control. Release 2018, 280, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lazo, J.S.; Sharlow, E.R. Drugging undruggable molecular cancer targets. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Kabza, A.M.; Young, B.E.; Sczepanski, J.T. Heterochiral DNA strand-displacement circuits. J. Am. Chem. Soc. 2017, 139, 17715–17718. [Google Scholar] [CrossRef] [PubMed]
- Kabza, A.M.; Young, B.E.; Kundu, N.; Sczepanski, J.T. Heterochiral nucleic acid circuits. Emerg. Top. Life Sci. 2019, 3, 501–506. [Google Scholar]
- Zhang, N.-N.; Li, X.-F.; Deng, Y.-Q.; Zhao, H.; Huang, Y.-J.; Yang, G.; Huang, W.-J.; Gao, P.; Zhou, C.; Zhang, R.-R. A thermostable mRNA vaccine against COVID-19. Cell 2020, 182, 1271–1283.e16. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, L.K.; El-Khoury, R.; Thorpe, J.D.; Damha, M.J.; Hollenstein, M. Recent progress in non-native nucleic acid modifications. Chem. Soc. Rev. 2021, 50, 5126–5164. [Google Scholar] [CrossRef] [PubMed]
- Dantsu, Y.; Zhang, Y.; Zhang, W. Synthesis and Structural Characterization of 2′-Deoxy-2′-fluoro-l-uridine Nucleic Acids. Org. Lett. 2021, 23, 5007–5011. [Google Scholar] [CrossRef]
- Dantsu, Y.; Zhang, Y.; Zhang, W. Synthesis of 2′-deoxy-2′-fluoro-l-cytidine and fluorinated l-nucleic acids for structural studies. ChemistrySelect 2021, 6, 10597–10600. [Google Scholar] [CrossRef]
- Jiang, W.; Zhang, B.; Fan, C.; Wang, M.; Wang, J.; Deng, Q.; Liu, X.; Chen, J.; Zheng, J.; Liu, L. Mirror-image polymerase chain reaction. Cell Discov. 2017, 3, 17037. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Deng, Q.; Zhu, T.F. Bioorthogonal information storage in l-DNA with a high-fidelity mirror-image Pfu DNA polymerase. Nat. Biotechnol. 2021, 39, 1548–1555. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, W.; Liu, X.; Wang, J.; Zhang, B.; Fan, C.; Liu, L.; Pena-Alcantara, G.; Ling, J.-J.; Chen, J. Mirror-image gene transcription and reverse transcription. Chem 2019, 5, 848–857. [Google Scholar] [CrossRef] [Green Version]
- Pech, A.; Achenbach, J.; Jahnz, M.; Schülzchen, S.; Jarosch, F.; Bordusa, F.; Klussmann, S. A thermostable d-polymerase for mirror-image PCR. Nucleic Acids Res. 2017, 45, 3997–4005. [Google Scholar] [CrossRef] [Green Version]
- MenáDuong, T.H.; SukáChung, H. The crystal structure of a natural DNA polymerase complexed with mirror DNA. Chem. Commun. 2020, 56, 2186–2189. [Google Scholar]
- Dedkova, L.M.; Fahmi, N.E.; Golovine, S.Y.; Hecht, S.M. Enhanced d-amino acid incorporation into protein by modified ribosomes. J. Am. Chem. Soc. 2003, 125, 6616–6617. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, J.; Jahnz, M.; Bethge, L.; Paal, K.; Jung, M.; Schuster, M.; Albrecht, R.; Jarosch, F.; Nierhaus, K.H.; Klussmann, S. Outwitting EF-Tu and the ribosome: Translation with d-amino acids. Nucleic Acids Res. 2015, 43, 5687–5698. [Google Scholar] [CrossRef] [Green Version]
- Dedkova, L.M.; Fahmi, N.E.; Golovine, S.Y.; Hecht, S.M. Construction of modified ribosomes for incorporation of d-amino acids into proteins. Biochemistry 2006, 45, 15541–15551. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).