Abstract
Neurodevelopmental disorders (NDDs) are a group of highly prevalent, clinically and genetically heterogeneous pediatric disorders comprising, according to the Diagnostic and Statistical Manual of Mental Disorders 5th edition (DSM-V), intellectual disability, developmental delay, autism spectrum disorders, and other neurological and cognitive disorders manifesting in the developmental age. To date, more than 1000 genes have been implicated in the etiopathogenesis of NNDs. Among them, AUTS2 (OMIM # 607270) encodes a protein involved in neural migration and neuritogenesis, and causes NNDs with different molecular mechanisms including copy number variations, single or multiple exonic deletion and single nucleotide variants. We describes a 9-year-old boy with global developmental delay, absent speech, minor craniofacial anomalies, hypoplasia of the cerebellar vermis and thinning of the corpus callosum, resulted carrier of the de novo AUTS2 c.1603_1626del deletion at whole exome sequencing (WES) predicted to cause the loss of eight amino acids [p.(His535_Thr542del)]. Notably, our patient is the first reported so far in medical literature carrying an in-frame deletion and the first in which absent language, hypoplasia of the cerebellar vermis and thinning of the corpus callosum has been observed thus useful to expand the molecular spectrum of AUTS2 pathogenic variants and to broaden our knowledge on the clinical phenotype associated.
1. Introduction
Neurodevelopmental disorders (NDDs) are a group of clinically and genetically heterogeneous disorders, which are diagnosed in childhood and encompass, but are not limited to, intellectual disability (ID), developmental delay (DD), autism spectrum disorders (ASDs), communication and learning disorders, attention deficit/hyperactivity disorders (ADHD) and developmental motor disorders. Emerging evidence is prompting to include epilepsy, developmental regression, sleep disturbance, mood and behavioral disorders, and aggression in the field of NDDs. Overall NNDs have an estimated prevalence of approximately 1–3% in the general population and represent one of the major challenges in medicine being the most frequent cause of disability in children [1]. The molecular milieu underpinning NDDs is increasingly complex with more than 1000 genes identified so far as implicated in their etiopathogenesis. Many of these genes converge on common pathways and protein networks, a fact that mirrors the clinical and molecular variability of NDDs.
Among them, genes involved in neuronal migration, extension, branching of the neurites, synaptic function, transcriptional regulation and construction of neuronal network are strongly represented [2]. AUTS2 (OMIM #607270) belongs to a gene family involved in neural migration and neuritogenesis, pivotal steps to form a functional brain, and regulates these processes both at cytoplasmic and nuclear level. Molecular alterations of AUTS2 causing NDDs currently include copy number variations (CNVs) and intragenic single or multi-exon deletions, as well as a restricted spectrum of single nucleotide variants, while patients carriers of likely pathogenic small in-frame variants involving functional relevant regions of the gene are still missing in medical literature.
In this report, we describe a 9-year-old boy with global developmental delay, absent speech, dysmorphic features, and cerebral anomalies, resulted carrier of a novel, de novo in-frame deletion of AUTS2 (OMIM # 607270) detected at whole exome sequencing (WES).
2. Materials and Methods
2.1. Genomic DNA Extraction and Quantification
This family provided written informed consent to molecular testing and to the full content of this publication. This study was conducted in accordance with the 1984 Declaration of Helsinki and its subsequent revisions. Molecular testing carried out in this report is based on the routine clinical care of our institution. Peripheral blood samples were taken from both the proband and his parents, and genomic DNA was isolated by using Bio Robot EZ1 following manufacturer’s instructions (http://geneious.mx/catalogos/EZ1_AdvanXL_lr2.pdf (accessed on 1 February 2021)) (Quiagen, Solna, Sweden). The quality of DNA was tested on 1% electrophorese agarose gel, and the concentration was quantified by Nanodrop 2000 C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.2. SNP-Array Analysis
High resolution SNP-array analysis on the proband’s DNA was carried out by using the CytoScan HD array (Thermo Fisher Scientific) as previously described [3]. Data analysis was performed using the Chromosome Analysis Suite Software version 4.1 (Thermo Fisher Scientific) following a standardized pipeline [4]. Briefly: (i) the raw data file (.CEL) was normalized using the default options; (ii) an unpaired analysis was performed using 270 HapMap samples as a baseline in order to obtain copy numbers value from .CEL files. The amplified and/or deleted regions were detected using a standard Hidden Markov Model (HMM) method. We retained CNVs ≥ 15 Kb in length and overlapping ≥ 10 consecutive probes to reduce the detection of false-positive calls. The significance of each CNV detected was determined by comparison with all chromosomal alterations identified in the patient to those collected in an internal database of ~4500 patients studied by SNP arrays since 2010, and public databases including Database of Genomic Variants (DGV; available on line at: http://projects.tcag.ca/variation/ (accessed on 1 February 2021)), DECIPHER (available on line at: https://decipher.sanger.ac.uk/ (accessed on 1 February 2021)), and ClinVar (available on line at: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 1 February 2021)). Base pair positions, information about genomic regions and genes affected by CNVs, and known associated diseases have been derived from the University of California Santa Cruz (UCSC) Genome Browser (available online at: http://genome.ucsc.edu/cgi-bin/hgGateway (accessed on 1 February 2021)), build GRCh37 (hg19). The clinical significance of each rearrangement detected was assessed following the American College of Medical Genetics (ACMG) guidelines [5].
2.3. Whole Exome Sequencing (WES)
Proband’s DNA was analyzed by WES by using SureSelect Human All Exome V6 (Agilent Technologies, Santa Clara, CA, USA) following manufacturer instructions as previously described [6]. This is a combined shearing-free transposase-based library prep and target-enrichment solution, which enables comprehensive coverage of the entire exome. This system enables a specific mapping of reads to target deep coverage of protein-coding regions from RefSeq, GENCODE, CCDS, and UCSC Known Genes, with excellent overall exonic coverage and increased coverage of HGMD, OMIM, ClinVar, and ACMG targets. Sequencing was performed on a NextSeq 500 System (Illumina, San Diego, CA, USA) by using the Mid Output flow cells (300 cycles), with a minimum expected coverage depth of 100×. All variants obtained from WES were called by means of the HaplotypeCaller tool of GATK ver. 3.58 [7] and were annotated based on frequency, impact on the encoded protein, conservation, and expression using distinct tools, as appropriate (ANNOVAR, dbSNP, gnomAD, 1000 Genomes, EVS, ExAC, ESP, KAVIAR, and ClinVar) [8,9,10,11,12], and retrieving pre-computed pathogenicity predictions of ad-hoc tools from dbNSFP v 3.0 (e.g., PolyPhen-2, SIFT, Mutation Assessor, FATHMM, LRT and CADD) [13] and evolutionary conservation measures. Variants were discarded if reported as benign or likely-benign in ClinVar and/or if have a minor allele frequency (MAF) > 0.01. Next, variants prioritization was performed as following: (i) nonsense/frameshift variants in genes previously described as disease-causing by haploinsufficiency or loss-of-function; (ii) variants located in a critical or functional domain; (iii) variants affecting canonical splicing sites (i.e., ± 1 or ± 2 positions); (iv) variants absent in allele frequency population databases; (v) variant reported in allele frequency population databases, but with MAF significantly lower than expected for the gene; (vi) variant predicted and/or annotated as (probable) pathogenic in ClinVar and/or LOVD. Variant analysis was carried out considering the ethnicity of the proband.
Candidate variants were confirmed by Sanger sequencing in both the proband and the parents’ DNA. PCR products were sequenced by using BigDye Terminator v1.1 sequencing Kit following manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA) and ABI Prism 3100 Genetic Analyzer (Thermo Fisher Scientific). The clinical significance of the identified putative variants was interpreted according to the American College of Medical Genetics and Genomics (ACMG) [14].
Nucleotide variants nomenclature follows the format indicated in the Human Genome Variation Society (HGVS, http://www.hgvs.org (accessed on 1 February 2021)) recommendations and reported in the Leiden Open Variation Databases (LOVD) (https://databases.lovd.nl/shared/variants/0000659804 (accessed on 1 February 2021)). The data have been deposited in the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/ (accessed on 1 February 2021)) under the accession number E-MTAB-10053.
Putative impact of candidate variants on protein was firstly assessed by searching for known functional/structural annotations in Uniprot [15]. Then, the reference FASTA sequence (Uniprot accession: Q8WXX7-1) was scanned for the presence of Eukaryotic Linear Motifs (ELM), using the ELM web-service (http://elm.eu.org/ (accessed on 1 February 2021)) [16]. Parameters were maintained at default values, while the “Taxonomy Context” was set to “Homo sapiens”. Results were filtered to consider only patterns with elevate conservation scores that overlapped with the mutant protein site.
3. Results
3.1. Clinical Description
This is a 9-year-old boy, second child of healthy non-consanguineous parents of Caucasian origin (southern Italy). No family history of congenital anomalies or ID/NDD was referred. He was born at 37 + 4 weeks of gestation by cesarean section for breech presentation. At birth, his weight was 3560 g (87th percentile), length 50 cm (66th percentile), head circumference was 34 cm (47th percentile), and Apgar scores were 9 and 10 at 1′ and 5′, respectively. The newborn was admitted to intensive care due to jaundice, limb hypertonia and facial dysmorphisms. Standard karyotype performed at birth was 46, XY. Subsequently, psychomotor development was severely delayed as he walked unsupported at 2.5 years and did not develop speech. At 7 years and 5 months, specialistic assessment showed cognitive delay. He did not acquire sphincter control and suffered from chronic constipation. He had difficulty falling asleep with partial improvement by melatonin intake. Parents reported only an episode of febrile seizure at 2 years old. Electroencephalogram at rest was normal. Brain MRI, showed hypoplasia of the cerebellar vermis and thinning of the corpus callosum. Echocardiogram and abdominal ultrasound were normal.
At last clinical evaluation, performed at 9 years old, he showed distinctive facial features including bitemporal narrowing, left posterior plagiocephaly, low anterior hairline, synophrys, strabismus, prominent nasal bridge, underdeveloped nasal alae, malar flattening, narrow palate, and slight anteverted ears (Figure 1). Examination of the oral cavity, limbs, extremities, skin and external genitalia was normal. Language was absent: he vocalized, but without communication purposes.
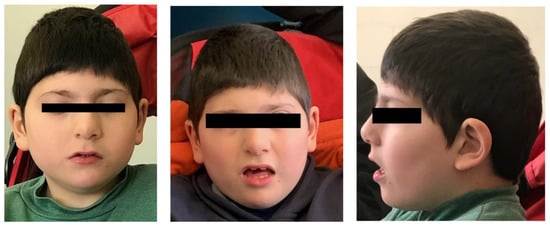
Figure 1.
Craniofacial features observed in the investigated subject.
3.2. Molecular Findings
High-resolution SNP-array analysis did not identify any pathogenic copy number variations (microdeletions or microduplications) in the proband. The molecular karyotype of the patient, according with the International System For Human Cytogenetic Nomenclature (ISCN 2016), is: arr[GRCh37](1-22) × 2,(X,Y) × 1. WES revealed an in-frame deletion at heterozygous state in the exon 9 of the AUTS2 gene (OMIM 607270) (AUTS2:NM_015570) c.1603_1626del resulting in a p.(His535_Thr542del) substitution (GRCh37/hg19). The variant was detected with a depth of coverage greater than 150×, and with elevate quality scores (i.e., Phread quality > 3000 and genotype quality = 99). This variation is not reported in gnomAD and ExAC populations’ database and it is predicted to cause the depletion of 4 out of 9 Histidine of one of the Histidine rich domain of the protein. Complete bioinformatics details are reported in Table 1.

Table 1.
Characteristics of the variant identified in the AUTS2 gene.
The variant was confirmed by Sanger sequencing using specific primers (AUTS2, exon 9, Forward Primer: TGGTCTCGTCGTCTTCATTG; AUTS2, exon 9, Reverse Primer: CGTCAGTCCCCATTCGATCT). Parental DNA analysis showed that it is a de novo event (Figure 2).
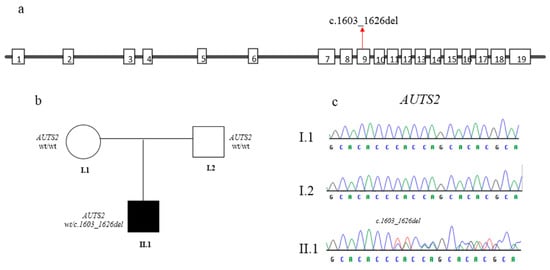
Figure 2.
(a) Schematic representation of the structure of AUTS2 gene. The variant identified here is indicated by red arrow. (b) Pedigree of the family displaying the de novo onset of the variant. Filled and unfilled circles/squares represent affected and unaffected individuals respectively. (c) Electropherograms of the proband (II.1) and his parents (I.1, I.2).
The variant was classified as likely pathogenic according to ACMG guidelines [14]. No further variant classified as pathogenic or likely pathogenic, according to ACMG guidelines in other genes and previously associated with phenotypes compatible with the clinical features observed in the patients, were identified by the bioinformatics analysis.
This variant was predicted to cause the loss of a stretch of eight amino acids, including four histidines, [p.(His535_Thr542del)] in the histidine-rich domain 1 (amino acids 525-548) (Figure 3).
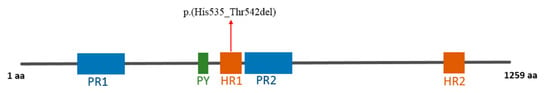
Figure 3.
Schematic representation of the structure of AUTS2 protein [17]. The variant identified here is indicated by a red arrow (PR: proline-rich domains; PY: py domain; HR: histidine-rich domains).
Within this compositionally biased region, the ELM resource predicted the presence of two high-scoring instances of the “MOD_GSK3_1” motif, at positions 531-538 and 535-542, respectively. These “HQHTHQHT” sequences would contain the linear motif “...([ST])...[ST]”, that in turn would represent the GSK3 phosphorylation site (http://elm.eu.org/elms/MOD_GSK3_1.html (accessed on 1 February 2021)). Along the whole AUTS2 sequence, four perfect matches were found for this motif. Motifs that perfectly match an ELM-annotated regular expression exhibit conservation scores of 1. Unfortunately, no crystal structures have been deposited (https://www.rcsb.org/ (accessed on 1 February 2021)) for the whole protein or protein fragments, while data on structural domains are scarce (details on https://www.uniprot.org/uniprot/Q8WXX7 (accessed on 1 February 2021)). However, the predicted linear motif co-locating the deletion would pave the way to functional studies.
4. Discussion
AUTS2 comprises 19 exons, spans about 1.2 Mb of genomic DNA in the proximal 7q11.2 regions. Although the function of the gene has been poorly characterized for a long time, recent research papers provided strong evidence on its role in brain development. Interestingly, the AUTS2 protein has dual physiological roles: cytoplasmic AUTS2 regulates actin cytoskeleton to control neuronal migration and extension, while nuclear AUTS2 is involved in gene expression regulation of various genes [18,19,20,21]. Among the AUTS2 target genes identified, ~35.2% comprise the top 25% highly tran-scribed genes in mouse brain. Among them, PRC1 and SEMA5A. Polycomb-group repressive complex 1 (PRC1), a polycomb-group gene involved in transcriptional repression, physically interacts with AUTS2, implicating a role for AUTS2 in developmental transcriptional regulation [22]. In 2013, the regulatory pathway for SEMA5A (semaphorin 5A), an autism candidate gene, was mapped in silico, using expression quantitative trait locus (eQTL) mapping. The authors found that the SEMA5A regulatory network significantly overlaps with rare CNVs around ASD-associated genes, including AUTS2. Performing eQTL mapping for expression levels of the eQTL-associated genes within the network (eQTLs of the eQTLs of SEMA5A), the authors identified 12 regions associated with the expression of 10 or more primary SEMA5A eQTL genes, including AUTS2. This study suggests that AUTS2 is involved, and may be a master regulator in ASD-related pathways [23]. Sequence analysis of AUTS2 identified two proline-rich domains (amino acids 288-471 and 545-646), two histidine-rich domains (HR1 and HR2) (amino acids 525-548 and 1122-1181) and a predicted PY motif (PPPY) (amino acids 515-519) (domains available at https://www.uniprot.org/uniprot/Q8WXX7#family_and_domains (accessed on 1 February 2021)) [17]. The PY motif is a potential WW-domain-binding region involved in protein-protein interactions. Moreover, it is present in the activation domain of several transcription factors, suggesting the involvement of AUTS2 in transcriptional regulation. Other predicted protein motifs include several cAMP and cGMP-dependent protein kinase phosphorylation sites and putative N-glycosylation sites. Functional evidence and data from the literature demonstrate that the expression of AUTS2 is regulated by a well characterized post-mitotic projection-neuron specific transcription factor, TBR1, which binds the AUTS2 promoter and activates the gene in developing neocortex [24].
AUTS2 was firstly linked to a clinical condition in a paper by Sultana et al., in which the gene was found disrupted due to a balanced translocation in a pair of monozygotic (MZ) twins with ASD. In addition, the authors observed that AUTS2 was strongly expressed in human fetal brain (frontal, parietal and temporal regions, telencephalon, ganglionic eminence, cerebellum anlagen, medulla oblongata, cortical plate and ventricular zone) with high expression in regions associated with higher cognitive brain functions [17]. Since then, more than 50 unrelated patients with neurodevelopmental (ID, ASD, speech and language disorders) and/or neuropsychiatric disorders (schizophrenia, ADHD, dyslexia and depression as well as addiction-related behaviors) have been shown to carry distinct heterozygous alterations of the AUTS2 gene [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42]. For these reasons, the term “AUTS2 syndrome” was coined to describe the wide spectrum of phenotypes, predominantly affected the cognition, observed in individuals with a germline AUTS2 alteration [28]. In addition to a variable NDD, satellite features frequently observed in affected individuals are feeding difficulties, short stature, hypotonia, and cerebral palsy, minor craniofacial anomalies.
From a clinical perspective, our patient shows several clinical manifestations associated to “AUTS2 syndrome” namely growth problems, dysmorphic features, skeletal abnormalities. In addition, he showed absent language at 9 years of age, and hypoplasia of the cerebellar vermis and thinning of the corpus callosum, which have never been reported in this condition. Thus, the description of our patient is useful to better delineate the clinical phenotype associated to the syndrome. Obviously, further reports will help in evaluating the significance of such a provisionally novel association.
From a molecular perspective, to our knowledge, this is the first reported individual with a small in-frame deletion in AUTS2, which involve 4 out of 9 Histidine of the first Histidine-rich domain of the protein. This variant, which results to be de novo, is absent from population databases such as TOPMED and gnomAD, thus useful to expand the mutational spectrum of AUTS2. A putative functional role of the motif involved by this in-frame deletion might be theorized/hypothesized by considering the computational screening of ELM along the AUTS2 amino acid sequence. Short linear motifs are one of the main components of proteins, consisting of functional modules, usually with a length of 3–10 amino acids. Among the functions that they mediate there are: protein-protein interactions, targeting signals, degradation signals, phosphorylation sites or affinity control. Abnormalities of ELMs by genetic alterations or alternative splicing can provide different isoforms of some proteins with different or altered functionality. The “HQHTHQHT” sequences deleted in this subject would contain a linear motif that in turn seems to represent the GSK3 phosphorylation site. GSK3, a serine/threonine protein kinase, comprises two highly related proteins (GSK3-α and GSK3-β) that phosphorylate a wide variety of target proteins with a final inhibitory effect.
In our case, the deletion of part of the HR1 motive could have caused the loss of a phosphorylation site which could be the molecular etiopathogenetic mechanism underlying the clinical phenotype observed in the patient. In supported by additional observations and functional studies, our finding could shed more lights on the molecular pathogenesis of AUTS2 syndrome.
Furthermore, our case suggest to further investigate the biological role of the His-rich motives, in order to elucidate their role in protein function and regulation. Obviously, being this the first patient carrying this kind of genetic variation of AUTS2, further functional studies are needed to confirm the pathogenic mechanism supposed.
5. Conclusions
The subject presented here is the first known individual with NDD and carrying an in-frame deletion of AUTS2. Our findings provisionally expand the AUTS2 syndrome associated clinical spectrum to absent speech, hypoplasia of the cerebellar vermis and hypoplasia of the corpus callosum. Taken together, these findings expand the mutation spectrum of AUTS2 syndrome and pave the way to a deeper understanding if its molecular pathogenesis.
Author Contributions
P.P., O.P., M.C. (Massimo Carella) conceived the study; P.P. and O.P. wrote the draft; O.P. performed SNP-array analysis and interpreted the data; P.P. and E.D.M. performed WES and data analysis; M.B. performed Sanger sequencing; M.A. and M.C.D.G. provided the clinical evaluation of the patient; S.C. and T.M. carried out the bioinformatics analysis; M.C. (Massimo Carella) and M.C. (Marco Castori) supervised the study and reviewed the final draft. All authors contributed to writing and reviewing the manuscript and approved its final version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ricerca Corrente Program from the Italian Ministry of Health to Massimo Carella.
Informed Consent Statement
This family provided written informed consent to molecular testing and to the full content of this publication. This study was conducted in accordance with the 1984 Declaration of Helsinki and its subsequent revisions. Molecular testing carried out in this report is based on the routine clinical care of our institution.
Data Availability Statement
The data presented in this study are openly available in ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-10053.
Acknowledgments
The authors thank the patient’s family for their kind availability.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Mitchell, K.J. The genetics of neurodevelopmental disease. Curr. Opin. Neurobiol. 2011, 21, 197–203. [Google Scholar] [CrossRef] [PubMed]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef]
- Palumbo, O.; Fichera, M.; Palumbo, P.; Rizzo, R.; Mazzolla, E.; Cocuzza, D.M.; Carella, M.; Mattina, T. TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am. J. Med. Genet. Part A 2014, 164, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, O.; Palumbo, P.; Di Muro, E.; Cinque, L.; Petracca, A.; Carella, M.; Castori, M. A Private 16q24.2q24.3 Microduplication in a Boy with Intellectual Disability, Speech Delay and Mild Dysmorphic Features. Genes 2020, 11, 707. [Google Scholar] [CrossRef] [PubMed]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257. [Google Scholar] [CrossRef]
- Genovesi, M.L.; Guadagnolo, D.; Marchionni, E.; Giovannetti, A.; Traversa, A.; Panzironi, N.; Bernardo, S.; Palumbo, P.; Petrizzelli, F.; Carella, M.; et al. GDF5 mutation case report and a systematic review of molecular and clinical spectrum: Expanding current knowledge on genotype-phenotype correlations. Bone 2021, 12, 115803. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- Glusman, G.; Caballero, J.; Mauldin, D.E.; Hood, L.; Roach, J.C. KAVIAR: An accessible system for testing SNV novelty. Bioinformatics 2011, 27, 3216–3217. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Non-synonymous and Splice Site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Kumar, M.; Zeke, A.; Lang, B.; Bely, B.; Chemes, L.B.; Davey, N.E.; Deng, Z.; et al. The eukaryotic linear motif resource—2018 update. Nucleic Acids Res. 2018, 46, D428–D434. [Google Scholar] [CrossRef]
- Sultana, R.; Yu, C.E.; Yu, J.; Munson, J.; Chen, D.; Hua, W.; Estes, A.; Cortes, F.; de la Barra, F.; Yu, D.; et al. Identification of a Novel Gene on Chromosome 7q11.2 Interrupted by a Translocation Breakpoint in a Pair of Autistic Twins. Genomics 2002, 80, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Oksenberg, N.; Haliburton, G.D.; Eckalbar, W.L.; Oren, I.; Nishizaki, S.; Murphy, K.; Pollard, K.S.; Birnbaum, R.Y.; Ahituv, N. Genome-wide distribution of Auts2 binding localizes with active neurodevelopmental genes. Transl. Psychiatry 2014, 4, e431. [Google Scholar] [CrossRef]
- Gao, Z.; Lee, P.; Stafford, J.M.; von Schimmelmann, M.; Schaefer, A.; Reinberg, D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 2014, 516, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Taya, S.; Hashimoto, R.; Hayashi, T.; Abe, M.; Yamazaki, M.; Nakao, K.; et al. Cytoskeletal regulation by AUTS2 in neuronal migration and neuritogenesis. Cell Rep. 2014, 9, 2166–2179. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Hoshino, M. Neuronal Migration and AUTS2 Syndrome. Brain Sci. 2017, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Quinn, J.F.; Weiss, L.A. An eQTL mapping approach reveals that rare variants in the SEMA5A regulatory network impact autism risk. Hum. Mol. Genet. 2013, 22, 2960–2972. [Google Scholar] [CrossRef] [PubMed]
- Bedogni, F.; Hodge, R.D.; Elsen, G.E.; Nelson, B.R.; Daza, R.A.; Beyer, R.P.; Bammler, T.K.; Rubenstein, J.L.; Hevner, R.F. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc. Natl. Acad. Sci. USA 2010, 107, 13129–13134. [Google Scholar] [CrossRef]
- Amarillo, I.E.; Li, W.L.; Li, X.; Vilain, E.; Kantarci, S. De novo single exon deletion of AUTS2 in a patient with speech and language disorder: A review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am. J. Med. Genet. A 2014, 164, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Beunders, G.; Voorhoeve, E.; Golzio, C.; Pardo, L.M.; Rosenfeld, J.A.; Talkowski, M.E.; Simonic, I.; Lionel, A.C.; Vergult, S.; Pyatt, R.E.; et al. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 2013, 92, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Jolley, A.; Corbett, M.; McGregor, L.; Waters, W.; Brown, S.; Nicholl, J.; Yu, S. De novo intragenic deletion of the autism susceptibility candidate 2 (AUTS2) gene in a patient with developmental delay: A case report and literature review. Am. J. Med. Genet. A 2013, 161, 1508–1512. [Google Scholar] [CrossRef] [PubMed]
- Beunders, G.; van de Kamp, J.; Vasudevan, P.; Morton, J.; Smets, K.; Kleefstra, T.; de Munnik, S.A.; Schuurs-Hoeijmakers, J.; Ceulemans, B.; Zollino, M.; et al. A detailed clinical analysis of 13 patients with AUTS2 syndrome further delineates the phenotypic spectrum and underscores the behavioural phenotype. J. Med. Genet. 2016, 53, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xu, Y.H.; Wei, S.G.; Zhang, H.B.; Fu, D.K.; Feng, Z.F.; Guan, F.L.; Zhu, Y.S.; Li, S.B. Association study identifying a new susceptibility gene (AUTS2) for schizophrenia. Int. J. Mol. Sci. 2014, 15, 19406–19416. [Google Scholar] [CrossRef]
- Elia, J.; Gai, X.; Xie, H.M.; Perin, J.C.; Geiger, E.; Glessner, J.T.; D’Arcy, M.; deBerardinis, R.; Frackelton, E.; Kim, C.; et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 2010, 15, 637–646. [Google Scholar] [CrossRef]
- Girirajan, S.; Brkanac, Z.; Coe, B.P.; Baker, C.; Vives, L.; Vu, T.H.; Shafer, N.; Bernier, R.; Ferrero, G.B.; Silengo, M.; et al. Relative burden of large cnvs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011, 7, e1002334. [Google Scholar] [CrossRef]
- Mefford, H.C.; Muhle, H.; Ostertag, P.; von Spiczak, S.; Buysse, K.; Baker, C.; Franke, A.; Malafosse, A.; Genton, P.; Thomas, P.; et al. Genome-wide copy number variation in epilepsy: Novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010, 6, e1000962. [Google Scholar] [CrossRef] [PubMed]
- Myung, W.; Kim, J.; Lim, S.W.; Shim, S.; Won, H.H.; Kim, S.; Kim, S.; Lee, M.S.; Chang, H.S.; Kim, J.W.; et al. A genome-wide association study of antidepressant response in koreans. Transl. Psychiatry 2015, 5, e633. [Google Scholar] [CrossRef]
- Schumann, G.; Coin, L.J.; Lourdusamy, A.; Charoen, P.; Berger, K.H.; Stacey, D.; Desrivieres, S.; Aliev, F.A.; Khan, A.A.; Amin, N.; et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. USA 2011, 108, 7119–7124. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Wang, J.C.; Wetherill, L.; Le, N.; Bertelsen, S.; Hinrichs, A.L.; Budde, J.; Agrawal, A.; Bucholz, K.; Dick, D.; et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum. Genet. 2013, 132, 1141–1151. [Google Scholar] [CrossRef]
- Dang, W.; Zhang, Q.; Zhu, Y.S.; Lu, X.Y. The evidence for the contribution of the autism susceptibility candidate 2 (auts2) gene in heroin dependence susceptibility. J. Mol. Neurosci. 2014, 54, 811–819. [Google Scholar] [CrossRef]
- McCarthy, S.E.; Gillis, J.; Kramer, M.; Lihm, J.; Yoon, S.; Berstein, Y.; Mistry, M.; Pavlidis, P.; Solomon, R.; Ghiban, E.; et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry 2014, 19, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, E.; Granot-Hershkovitz, E.; Monderer-Rothkoff, G.; Lerer, E.; Levi, S.; Yaari, M.; Ebstein, R.P.; Yirmiya, N.; Shifman, S. Identification of a functional rare variant in autism using genome-wide screen for monoallelic expression. Hum. Mol. Genet. 2011, 20, 3632–3641. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.L.; Zou, Y.S.; Maher, T.A.; Newton, S.; Milunsky, J.M. A de novo balanced translocation breakpoint truncating the autism susceptibility candidate 2 (AUTS2) gene in a patient with autism. Am. J. Med. Genet. A 2010, 152A, 2112–2114. [Google Scholar] [CrossRef]
- Pinto, D.; Pagnamenta, A.T.; Klei, L.; Anney, R.; Merico, D.; Regan, R.; Conroy, J.; Magalhaes, T.R.; Correia, C.; Abrahams, B.S.; et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010, 466, 368–372. [Google Scholar] [CrossRef]
- Bakkaloglu, B.; O’Roak, B.J.; Louvi, A.; Gupta, A.R.; Abelson, J.F.; Morgan, T.M.; Chawarska, K.; Klin, A.; Ercan-Sencicek, A.G.; Stillman, A.A.; et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008, 82, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Talkowski, M.E.; Rosenfeld, J.A.; Blumenthal, I.; Pillalamarri, V.; Chiang, C.; Heilbut, A.; Ernst, C.; Hanscom, C.; Rossin, E.; Lindgren, A.M.; et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 2012, 27, 525–537. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).