Abstract
Anaerobic archaeal methanogens are key players in the global carbon cycle due to their role in the final stages of organic matter decomposition in anaerobic environments such as wetland sediments. Here we present the first draft metagenome-assembled genome (MAG) sequence of an unclassified Methanosarcinaceae methanogen phylogenetically placed adjacent to the Methanolobus and Methanomethylovorans genera that appears to be a distinct genus and species. The genome is derived from sediments of a hypersaline (97–148 ppt chloride) unrestored industrial saltern that has been observed to be a significant methane source. The source sediment is more saline than previous sources of Methanolobus and Methanomethylovorans. We propose a new genus name, Methanosalis, to house this genome, which we designate with the strain name SBSPR1A. The MAG was binned with CONCOCT and then improved via scaffold extension and reassembly. The genome contains pathways for methylotrophic methanogenesis from trimethylamine and dimethylamine, as well as genes for the synthesis and transport of compatible solutes. Some genes involved in acetoclastic and hydrogenotrophic methanogenesis are present, but those pathways appear incomplete in the genome. The MAG was more abundant in two former industrial salterns than in a nearby reference wetland and a restored wetland, both of which have much lower salinity levels, as well as significantly lower methane emissions than the salterns.
1. Introduction
Methanogens play a key role in the global carbon cycle by performing the final steps of organic matter decomposition into methane (CH4) and carbon dioxide (CO2) in anaerobic environments. Natural methanogenesis contributes ~217 Tg CH4 yr−1 to the atmosphere, with wetlands as the single largest natural source of methane at an estimated ~149 Tg CH4 yr−1 of emissions [1]. Three major pathways of anaerobic archaeal methanogenesis have been described in the literature—acetoclastic (acetate splitting, with acetate acting as an electron donor and acceptor), hydrogenotrophic (using hydrogen, formate, carbon monoxide or alcohols as electron donors and carbon dioxide as an electron acceptor), and methylotrophic (involving demethylation of methylamine, dimethylamine, trimethylamine, dimethyl sulfide, methanethiol, tetramethylammonium, methanol, or glycine betaine) methanogenesis [2]. The relative contribution of these different pathways depends on several variables including organic matter quality and composition, salinity, temperature, and pH [2].
While all three of these pathways are performed by members of the Euryarchaeota phylum, there are some distinctions at the order, family, and genus levels. Methanomicrobiales, Methanobacteriales, Methanocellales, and Methanosarcinales have been described as hydrogenotrophic orders, while the Methanosarcinaceae and Methanotrichaceae families within the Methanosarcinales order contain acetoclastic taxa [3]. Methanosarcinaceae is recognized as the most metabolically diverse family, containing genera that can perform all three pathways [4]. Members of the genera Methanolobus and Methanomethylovorans (Methanosarcinaceae) are methylotrophic methanogens, as many experiments on pure cultures have shown growth and methane production with methylotrophic substrates but not acetate or hydrogen/carbon dioxide [5,6,7,8,9,10,11,12,13,14,15].
Methanogenesis in saline to hypersaline environments presents an intriguing situation both in terms of extremophilic adaptations for survival and metabolic adaptations to avoid competition. Two families of methanogens, Methanosarcinaceae and Methanocalculaceae, contain halophilic members [16]. Several studies conducted in hypersaline environments, including solar salterns and hypersaline lakes, have identified key halophilic methanogenic genera inhabiting these environments, including Methanohalobium, Methanohalophilus, Methanosalum, Methanocalculus, and Methanosarcina [16,17,18,19]. These genera not only tolerate extremely high salinity (up to 30%) but grow optimally in hypersaline conditions (15%). By contrast, the genera Methanolobus and Methanomethylovorans have not been observed in the most hypersaline environments. Methanomethylovorans have been isolated from freshwater sediments [10,11,20], while cultivated members of Methanolobus appear to be at least halotolerant (growing in up to 10% salinity) [21] but perhaps not true halophiles based on the salt levels where they achieve optimum growth [22]. The methylotrophic pathway likely predominates in hypersaline environments for two reasons: first, sulfate reducers do not compete for methylated compounds as they do for hydrogen or acetate [23,24]; and second, methyl-group containing compounds are available in these environments due to the breakdown of compatible solutes such as glycine betaine and dimethylsulfoniopropionate that are produced by both prokaryotes and algae [25].
Several options exist for archaea in hypersaline environments to achieve the necessary osmoregulation for their survival. These have been broken down into two categories, the “salt in’’ strategy typically used by Haloarchaea, which involves increasing cytoplasm salt concentrations, usually with potassium chloride (KCl), and the “low salt in” strategy (also sometimes referred to as “salt out” strategy), typically used by bacteria but also some archaea, which involves production of compatible solutes to maintain osmotic balance in the cytoplasm without increased salt [26,27]. In particular, other halophilic archaea (i.e., Halobacteriales) have been shown to produce trehalose and betaine, and the genes required for their synthesis and transport have been described [28,29]. Previous studies have also described several other compounds such as glutamate, sucrose, ectoine, and glycosylglycerol as compatible solutes for halotolerance, as well as the major alkali metal-cation transporters that are involved in the “salt in” strategy [27]. However, the presence of genes involved in synthesis and transport of these salts and solutes has not been assessed in the available Methanolobus and Methanomethylovorans genomes.
In this paper we describe a new draft genome for Methanosalis sp. SBSPR1A, gen. nov., sp. nov., with a particular emphasis on methane cycling genes and halotolerance genes. We also compare the genome to closely related Methanolobus and Methanomethylovorans taxa phylogenetically, as well as in terms of shared and unique orthologous gene groups.
2. Materials and Methods
2.1. Metagenome Information
The genome was assembled from metagenomic sample R1_A_D2 (IMG ID 3300026157, NCBI BioSample accession SAMN06266236) as part of the study published by Zhou et al. [30] on South San Francisco Bay salt ponds, who initially classified the MAG as Methanolobus with Bin Annotation Tool (BAT) [31]. The genome name SBSPR1A is based on the sample from which it is derived (South Bay Salt Pond R1 replicate A). This sample is the 5–15 cm deep section of a sediment core taken from a 57.2 ha former industrial salt pond (37.5° N, −122.13° W) that is no longer used for salt production but has not been restored. Industrial salt production occurred in the pond from the 1850s to 2003. The sample was characterized by high salinity (97.2 total ppt Cl−), a temperature of 24.7 °C, low dissolved oxygen content (0.96 mg L−1) and a pH of 7.75 [30]. With salinity levels over twice that of seawater (~35 ppt), the pond can be considered hypersaline [25]. Elevated methane emissions were measured from this core in the field (737 µmol CH4 m−2 d−1) and in a lab incubation using sediments collected from the same pond [30]. Approximately 10 Gb of shotgun sequence data were generated from this sample on the HiSeq 2500 (Illumina, San Diego, CA, USA) platform in a 2 × 150 paired-end run mode. Reads were filtered and trimmed with BBTools (https://sourceforge.net/projects/bbmap/, accessed 16 April 2018) and assembled with SPAdes [32].
2.2. MAG Assembly
Metagenome assembled genomes (MAGs) were binned using MetaBat [33], MaxBin [34], and CONCOCT [35], the best of which were selected using DAS Tool [36]. For this draft genome, we began with a MAG binned by CONCOCT, with a completeness of 98.37%, contamination of 3.27%, and strain heterogeneity of 0, as calculated by CheckM [37], meeting community standards for a “high-quality draft” [38].
Abundance estimates were calculated by mapping the raw metagenomic reads to the MAG, taking the average read depth, and then transforming to counts per million of assembled reads to take into account differences in library sizes across the 24 metagenome samples [30]. The effect of site (4 separate wetlands) on MAG abundance was calculated with a Kruskal-Wallis test (R package stats) followed by Nemenyi posthoc (R package PMCMR [39]), as assumptions for ANOVA and Tukey HSD were not met (Levene Test, p < 0.05, Shapiro-Wilk test, p < 0.05).
Scaffolds were extended manually using Geneious [40] to remap paired-end reads to each scaffold and extend using reads partially overlapping the ends of the scaffolds [41]. For scaffolds that could not be extended using reads from sample R1_A_D2, reads from sample R1_A_D1, the 0–5 cm section of the same core, were used. After each round of scaffold extension, the new scaffold set was assembled de novo with the Geneious assembler to merge any newly overlapping scaffolds, and then reads were remapped to the reassembled scaffolds to resolve any ambiguous bases created during the assembly. Following two rounds of scaffold extension, we attempted to close two gaps (one group of 71 Ns and one group of 10 Ns) with both Sealer [42] and GapCloser from SOAPdenovo2 [43], but neither program was able to close either gap. The genome was then uploaded to the Joint Genome Institute’s Integrated Microbial Genomes and Microbiomes database (IMG/M) [44] for gene annotation, and is publicly available with IMG ID 2929001634 and on GenBank under BioProject ID PRJNA365332.
2.3. Comparative Genomics
For comparative genomics, we downloaded nine Methanolobus reference genomes available from the RefSeq database, representing the species Methanolobus psychrophilus R15 (NC_018876) [15], Methanolobus psychrotolerans YSF-03 (NZ_MBKP01000078) [21], Methanolobus tindarius DSM 2278 (NZ_AZAJ01000001) [45], Methanolobus vulcani B1d (NZ_VIAQ01000006) [46], Methanolobus vulcani PL 12/M (NZ_FNCA01000018) [47], Methanolobus profundi Mob M (NZ_FOUJ01000018) [8,48], Methanolobus sp. SY-01 (NZ_PGGK01000010) [49], Methanolobus zinderi DSM 21339 (NZ_CP058215) [6], and Methanolobus bombayensis DSM 7082 (NZ_JAGGKD010000001) [9,50]. We also downloaded the one Methanomethylovorans genome on RefSeq, Methanomethylovorans hollandica DSM 15978 (NC_019977) [10,45]. We first used the “Insert Set of Genomes Into SpeciesTree” tool (version 2.2.0) in KBase [51] to place these 11 genomes into a phylogenetic tree with 50 other genomes from RefSeq [52] using 49 single copy clusters of orthologous groups (COGs). Next, a more detailed phylogenetic tree was constructed by first identifying and aligning 122 universal single copy archaeal marker genes with GTDB-Tk [53] and then building a consensus tree from the concatenated alignment with RAxML version 8.12 [54] with the PROTGAMMALG model of amino acid substitution, 1000 bootstraps, and Nitrosopumilus adriaticus (RefSeq GCF_000956175.1) as an outgroup. We used ProtTest 3 [55] to select the best model of amino acid substitution (LG). Furthermore, we constructed a phylogenetic tree of full length or near full length 16S rRNA genes, which were acquired from IMG where available or extracted with ContEst16S [56], by aligning with MUSCLE [57] and then using the IQ-Tree webserver to build the tree [58]. Some additional taxa with available 16S rRNA genes on NCBI were added to this analysis—Methanolobus oregonensis (U20152.1), Methanolobus taylorii (U20154.1), Methanolobus chelungpuianus (EU293796.1), Methanomethylovorans uponensis EK1 (KC876048.1), Methanomethylovorans thermophila (AY672821.1), Methanomethylovorans victoriae TM (AJ276437.1), and Methanomethylovorans sp. Z1 (EF174501.1). Pairwise percentage similarity of these 16S rRNA gene sequences was calculated with NCBI BLAST. Average nucleotide identity (ANI) was calculated with FastANI implemented in KBase. The KBase narrative including the ANI analysis and phylogenetic analysis for this project is publicly available under narrative ID 90991.
To ensure consistent gene annotations across genomes, Prodigal [59] was used to predict protein-coding genes from the nucleotide sequences. Mean isoelectric points of the protein-coding sequences were calculated with EMBOSS [60] using the “iep” tool on the Galaxy website platform (https://usegalaxy.org/, accessed 28 July 2021) [61] with a pH step of 0.5. Analysis of orthologous gene groups was conducted with proteinortho [62] and KEGG orthology (KO) profiles [63] and intersections among the genomes were graphed with the ComplexUpset R package [64]. KO profiles were downloaded from IMG where available; for genomes not yet on IMG, KO profiles were created with BLASTKoala [65]. Selected genes from proteinortho were classified with Pfam [66].
The KO tables were queried for KOs involved in methanogenesis and salt tolerance. A master map of methanogenesis processes containing the genes, enzymes, and reactions of all three pathways, as well as synthesis of precursor molecules such as coenzyme B and coenzyme M and the regeneration of these molecules from heterodisulfide after methane production, was built by synthesizing information in the ModelSeed [67], KEGG [68], and BioCyc [69] databases and mapped with Escher in ModelSeed (Figure S1). A list of KOs was created by utilizing existing KO associations with reactions in these databases, although not all of the reactions were annotated with KOs as enzymes have not yet been identified for some reactions. A list of KOs involved in salt tolerance was developed based on salt tolerance related genes and processes described in the literature [22,26,27,28,29,70]. These KOs include proteins involved in compatible solute biosynthesis and transport (e.g., betaine, trehalose, choline, proline), as well as other cation transporters. Presence or absence of these methanogenesis and salt tolerance related KOs were plotted as heatmaps using the pheatmap R package [71]. All R analyses were performed with version 4.0.2 [72].
3. Results
The draft genome of Methanosalis sp. SBSPR1A consists of 2,476,202 bp in 48 scaffolds, with 2656 protein-coding genes and a GC content of 39.6%. GTDB-Tk classified the genome to the genus level as Methanolobus, in agreement with the original MAG classification by Bin Annotation Tool. However, using 49 single copy COGs to place the genomes into a reference tree, Methanosalis sp. SBSPR1A is placed between the Methanomethylovorans and Methanolobus genera (Figure S2). Similarly, using the set of 122 universal archaeal single copy protein-coding genes in GTDB-Tk, the genome is again placed on its own branch between these two genera (Figure 1). This result is corroborated when just considering the full length 16S rRNA gene phylogenetic analysis (Figure S3). The 16S rRNA gene percent similarity between Methanosalis sp. SBSPR1A and the Methanomethylovorans taxa was between 90.40–91.78% and compared to the Methanolobus taxa was between 91.68–92.69% (Figure S4). These values are below 94.5% and are suggestive of a new genus and species [73]. According to ANI, the Methanosalis sp. SBSPR1A genome is most closely related to Ml. bombayensis, but differences in pairwise ANI between this genome and those of the other taxa were small (Figure S5). ANI values between Ms. sp. SBSPR1A and the other genomes ranged between 76.97 and 77.97, again suggesting that it is a unique genus and species (Figure S5). MASH ANI clustering generally agrees with the marker gene clustering with the exception that Methanosalis sp. SBSPR1A groups more closely with Mmv. hollandica and the zinderi-SP01-psychrophilus Methanolobus clade in MASH ANI clustering (Figure S6).
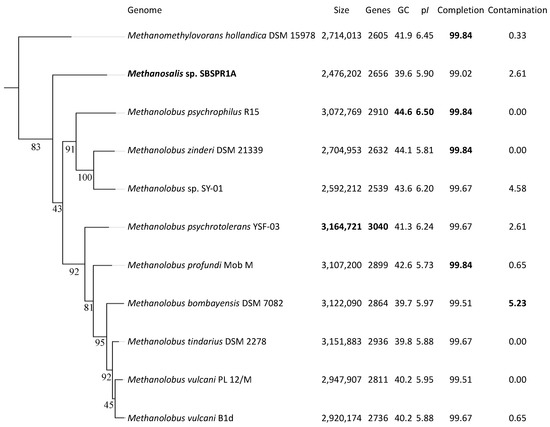
Figure 1.
RAxML phylogenetic tree of the 11 genomes in this study using a concatenated alignment of 122 single copy archaeal genes and the PROTGAMMALG model of amino acid substitution. Branch labels show the bootstrap support, calculated with 1000 bootstraps. Also shown are the genome size, number of protein-coding genes, % G + C content, mean isoelectric point of protein-coding genes (pI), and percent completeness and contamination estimates from CheckM. The greatest values in each column are bolded. The tree is rooted with Nitrosopumilus adriaticus (Thaumarchaeota) as an outgroup (not shown).
There were 3772 orthologous gene groups identified across all the genomes computed by proteinortho; using KEGG Orthology annotations, there were a total of 1439 KOs across the eleven genomes. This is consistent with about 45% of the protein-coding genes in these genomes having a KO annotation. Among the eleven genomes examined here, according to proteinortho there were 1347 orthologous genes shared among all eleven taxa and 61 genes present in the other ten genomes but not the Methanosalis sp. SBSPR1A genome (Figure 2a). The genes missing from our focal genome include a wide variety of functions (Table S1). A small number of genes were found only in Ms. sp. SBSPR1A and one other genome; among these, the largest number (13) was shared with Ml. profundi. For KEGG orthology gene groups, there were 15 KOs present in the other 11 genomes but not that of Ms. sp. SBSPR1A (Figure 2b). Of these, a few notable genes include comDE for biosynthesis of coenzyme M, an important precursor to methane, and mtaBC for methanogenesis from methanol (Table S2). There were 18 KOs present unique to Ms. sp. SBSPR1A. Of these, functions included ABC transporters, sdmt and gsmt betaine biosynthesis genes, ribosomal protein rplT, and the purine metabolism ADK adenosine kinase gene (Table S2).
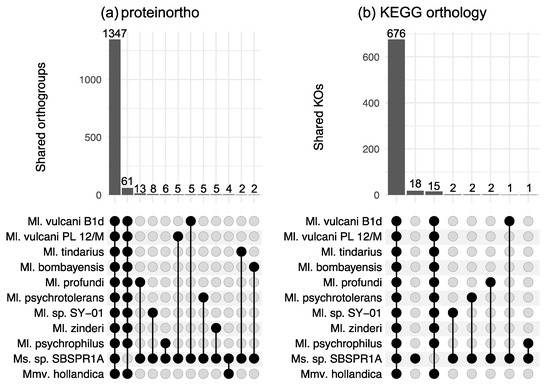
Figure 2.
Shared orthologous gene groups among different combinations of genomes as calculated by (a) proteinortho, and (b) KEGG orthology profiles. Shown here are intersections between all 11 genomes, genes unique to Ms. sp. SBSPR1A (only in panel b), intersections between 10 other genomes but not Methanosalis sp. SBSPR1A, and pairwise comparisons between Ms. sp. SBSPR1A and the other genomes. Numbers above the columns state the number of shared orthogroups or shared KOs. For strain names of all of the species, see the methods section or Figure 1.
In terms of methane cycling, all of the genomes contained the three key mcrABG genes necessary for methyl-CoM reduction to methane, the terminal step in all of the archaeal methanogenesis pathways (Figure 3 and Figure S1). As for the pathways for methyl-CoM formation, at least some genes involved in each of the acetate, hydrogen/CO2, and methylotrophic pathways were present in all of the genomes. However, the acetoclastic and hydrogenotrophic routes appear to be incomplete in the MAG and the Methanolobus and Methanomethylovorans genomes, with all 11 genomes missing ackA, pta, and eutD in the acetate pathway and missing fwdH and hmd in the hydrogenotrophic pathway. Methanosalis sp. SBSPR1A contained all of the genes for demethylation pathways of trimethylamine and dimethylamine, but only some of the genes for methylamine and methanol demethylation, while several of the other taxa contained all of the genes for all four of the methylotrophic pathways with KO annotations (Figure 3). Note that methyl-CoM can also be produced from methanethiol, dimethyl sulfide, tetramethylammonium, and glycine betaine [69], but these pathways did not have KOs assigned to the biochemical reactions as of the time of writing. During methyl-CoM reduction to methane, heterodisulfide is produced as a byproduct, which can then be cycled back into CoM and CoB via several different pathways (Figure S1). Genes involved in these pathways are patchily distributed among the 11 genomes, with the pathway using cofactor F420 being the only consistently complete pathway (hdrA2, hdrB2, hdrC2 genes).
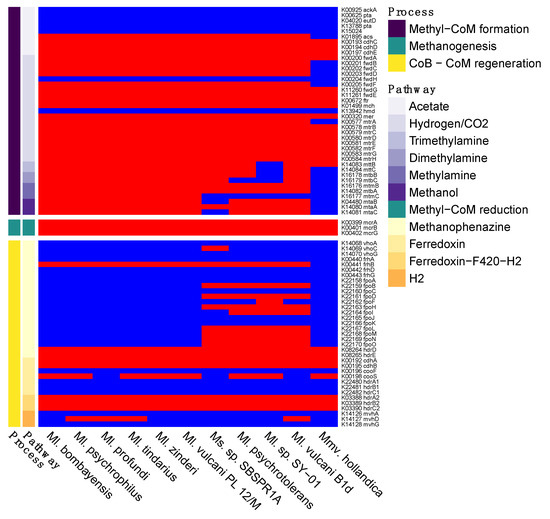
Figure 3.
Presence (red) or absence (blue) of genes involved in methanogenesis. Shown here are six different pathways for methyl-CoM formation (Figure S1), the single methane production step of methyl-CoM reduction, and four pathways for coenzyme B (CoB) and coenzyme M (CoM) regeneration. The names of the genes and their KEGG K number assignments are shown. Columns are clustered by KO presence/absence rather than phylogeny.
Additionally, upstream of the pathways discussed above, two compounds must be synthesized or imported—coenzyme B (CoB) and coenzyme M (CoM) [74], the latter of which can be synthesized from phosphoenol pyruvate (CoM synthesis I) or O-phosphate-L-serine (CoM synthesis II) [69]. Each of these processes involves multiple chemical reactions, only some of which are currently annotated with KOs. Interestingly, even with the limited number of described KOs involved in these pathways, all 11 of the genomes analysed here contain less than half of those described KOs (Figure S7). The KOs present in the genomes are consistent, with the exception of comDE for CoM synthesis which is missing from Ms. sp. SBSPR1A.
Another interesting phenotype of this taxon is its halotolerance, as it was sequenced from a sample with 97 ppt salinity and was also abundant in samples containing up to 232 ppt salinity. Similar to other halotolerant archaea [75], Ms. sp. SBSPR1A had an acidic proteome, with the protein-coding sequences having a mean isoelectric point (pI) of 5.90 ± 0.04 SE. Similarly, the other Methanolobus and Methanomethylovorans genomes analyzed here had mean isoelectric points ranging from 5.73 to 6.50 (Figure 1). Isoelectric point profiles for all 11 genomes were asymmetrically bimodal, with a large peak around pH 4–5 and a second but much smaller peak around pH 9–10 (Figure S8). The profiles of the Methanolobus, Methanomethylovorans and Methanosalis genomes were all similar despite Ms. sp. SBSPR1A being found in a more hypersaline environment than the other ten species; Ms. sp. SBSPR1A also did not have the most acidic proteome. Ms. sp. SBSPR1A was present in all 24 sediment samples we characterized from the South San Francisco Bay but significantly more abundant in the 12 unrestored saltern samples than in the reference wetland and restored saltern which had much lower salinities (Nemenyi posthoc p < 0.05, Figure 4). While other Methanolobus taxa have also been found to be moderately halotolerant, tolerating salinities of up to 117 ppt NaCl, their optimum growth was reported to range from 0 to 35 ppt NaCl, which is the approximate salinity of the ocean [21]. Methanomethylovorans hollandica and other Methanomethylovorans species grow optimally between 0–6 ppt NaCl and only tolerate up to 17.5 ppt NaCl [10,11,12]. Thus, the Methanosalis genome sequenced here comes from a sample with salinity well over both the optimum conditions and upper tolerances reported in Methanolobus and Methanomethylovorans.
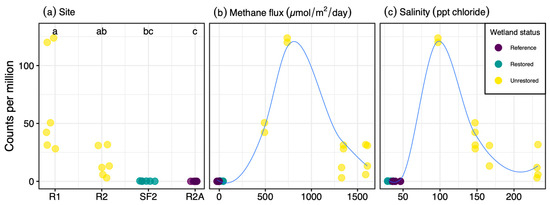
Figure 4.
Abundance of Methanosalis sp. SBSPR1A, expressed as counts per million assembled reads, across 24 metagenomic samples from Zhou et al. (2021) [30], organized by (a) site, showing the four wetland sites sampled, (b) methane flux (µmol/m2/day), and (c) salinity (ppt chloride). Samples were taken from two unrestored salterns, a restored saltern, and a reference wetland. Lines in (b,c) were fitted with loess functions.
Ms. sp. SBSPR1A was the only genome with the sdmt and gsmt genes for betaine biosynthesis (Figure 5). Nine of the genomes contained at least one gene for betaine transport, the exceptions being Ml. psychrophilus and Mmv. hollandica. All 11 genomes contained TrkA potassium uptake uniporter genes, kpdB ATP-driven potassium transport system, lysC and asd for ectoine biosynthesis, and proABC for proline biosynthesis. Some genomes additionally contained genes for sucrose (SPP) and glutamate (gdhA) biosynthesis. Thus, there appear to be some differences among the taxa in compatible solute strategies.
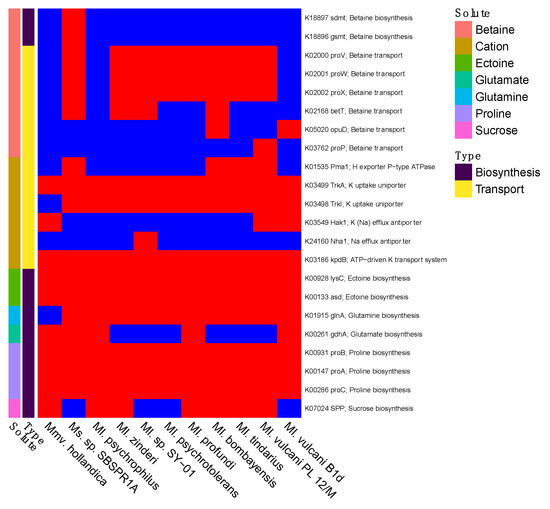
Figure 5.
Presence (red) or absence (blue) of genes involved in compatible solute or salt biosynthesis and transport, for salt tolerance. Compounds include betaine, cations (K+ or Na+), ectoine, glutamate, glutamine, proline, and sucrose. The gene names and their KEGG K number assignments are shown. Columns were clustered according to the phylogeny in Figure 1.
4. Discussion
Methanolobus and Methanomethylovorans taxa have previously been isolated from diverse environmental samples encompassing a range of conditions, but the organism whose genome is presented here originates from a former saltern sediment sample with higher salinity than any samples in which those genera have previously been found [21]. Interestingly, our saline 24.7 °C sample harbored a Methanosarcinaceae species closely related to Methanolobus species isolated from Tibet [15] and Siberia [21], both of which experience below freezing temperatures, demonstrating a broad temperature range for this subgroup of Methanosarcinaceae. At another set of extreme conditions, Ml. zinderi was isolated from deep sea vents [6] and Ml. profundi was isolated from deep subsurface sediments in a natural gas field [8]. Despite these different habitats, Methanosalis sp. SBSPR1A and all of the Methanolobus species, as well as Methanomethylovorans hollandica, possess generally the same architecture for methylotrophic methanogenesis and halotolerance. Ms. sp. SBSPR1A has the highest diversity of halotolerance genes among the genomes examined but is missing some genes for methanol and methylamine demethylation that are present in most of the other genomes.
Methanolobus and Methanomethylovorans taxa have previously been described as methylotrophic methanogens. Methylotrophic methanogenesis involves the demethylation of methylated compounds to produce methyl-CoM, followed by reduction of methyl-CoM to methane (Figure S1). Six of the 11 genomes contained genes for demethylation of trimethylamine, dimethylamine, methylamine, and methanol, which are the four molecules with demethylation reactions currently annotated with KEGG K numbers. These genomic data agree with experimental results for these taxa [5,6,7,8,14,15,76]. Interestingly, the other five genomes, including the new genome described in this paper, were missing some of these methanogenesis KOs. Most notably, the genomic data alone suggest that Methanosalis sp. SBSPR1A might not perform methanogenesis from methanol and methylamine, only from trimethylamine and dimethylamine. This agrees with experimental data from a sediment core from this pond (but note that this was not a pure culture of Methanosalis sp. SBSPR1A) in which methane emissions increased more, and more rapidly, when trimethylamine was added compared to methanol or acetate [30]. Methanol is a product of pectin degradation [77], which proceeds slowly in lake sediments [77]; perhaps for this reason methanol was only a marginal methanogenic precursor in a previous lake sediment study [78] and may not be as important as trimethylamine in the salterns or wetland studied here. It is also possible that these genes are missing from the genome because it is not 100% complete, and this should be confirmed experimentally with pure cultures. Similarly, Methanolobus sp. SY-01, Ml. psychrotolerans, Ml. vulcani B1d, and Mmv. hollandica might not be able to perform methanogenesis from all four methyl-containing compounds examined here. Mmv. hollandica was previously shown to use dimethyl sulfide and methanethiol [10].
Methylotrophic methanogenesis is an important carbon cycling pathway in saline environments [2,25]. While on a global scale it contributes less methane than acetoclastic or hydrogenotrophic methanogenesis, it can be an important local carbon cycling pathway [79,80,81]. Saline environments with high sulfate concentrations suppress acetoclastic and hydrogenotrophic methanogenesis due to the activity of sulfate reducers, which outcompete methanogens for substrates such as acetate and hydrogen; methylotrophic methanogens avoid this competition [2,23,82,83]. Furthermore, the compatible solutes used to tolerate the saline environments can also be degraded to methyl-containing compounds, particularly trimethylamine and dimethyl sulfide, thus providing a source of substrates for methylotrophic methanogenesis [2]. For example, betaine can be fermented to produce trimethylamine [84], while the osmolyte dimethylsulfoniopropionate can be converted to methanethiol and dimethyl sulfide [85]. Thus, both high salinity and high sulfate environments have been shown to differentially select for methylotrophs over acetoclastic and hydrogenotrophic taxa [86,87]. More specifically, in this particular saltern, due to the abundance of betaine, trimethylamine may be more abundant than methanol, thus supporting an organism such as Ms. sp. SBSPR1A that likely performs methanogenesis from trimethylamine but not methanol.
The Ms. sp. SBSPR1A genome contains genes involved in compatible solute biosynthesis and transport, which may explain its ability to adapt to hypersaline former solar salterns. The data presented here do not clearly determine which of the two “salt in’’ or “low salt in” strategies Ms. sp. SBSPR1A uses. In fact, the presence of KOs involved in both compatible solute synthesis and transport, as well as potassium and sodium transporters suggest that both strategies could be at play, as has been discussed for other organisms [75,88]. Increasing salinity levels have been associated with increasing proteome acidity [89,90]. Ms. sp. SBSPR1A has an acidic proteome as has been reported for other halotolerant archaea and bacteria including Haloferax, Halorubrum, Halobacterium, Salinibacter, Halomonas, and Alteromonas [70,75,90]. A mean isoelectric point value of 5.90 (and range of 5.73–5.97 for the other Methanolobus genomes excluding Ml. psychrophilus, Ml. sp. SY-01, and Ml. psychrotolerans) is similar to values reported for Halobacterium NRC-1 (5.03) and Salinibacter ruber (5.92), which are thought to use the “salt in” strategy [75]. However, it has also been shown that organisms such as Halorhodospira halophila, Halomonas elongata, or Chromohalobacter salexigens that produce compatible solutes (i.e., “low salt in” strategy) can also have an acidic proteome and high intracellular KCl concentrations (typical of a “salt in” strategy) [88,91]. Regardless of the strategy used, the mean pI values and pI profile of Ms. sp. SBSPR1A is more similar to moderately and extremely halophilic organisms than to other freshwater, ruminant, or thermophilic methanogens (e.g., mean isoelectric points of 6.32–6.83 [92], symmetrical bimodal profile of Methanococcus jannaschii [93]), perhaps highlighting the adaptation of Ms. sp. SBSPR1A to the conditions of the unrestored salterns compared to the restored saltern or reference wetland.
It is notable that Ms. sp. SBSPR1A is the only genome analyzed here containing the sdmt and gsmt genes which are involved in parallel pathways for betaine biosynthesis (Figure 5 and Figure S9). The sdmt gene produces betaine from sarcosine, while gsmt produces dimethylglycine from glycine, which can then be converted to betaine with bsmB, a gene which was not present in any of the 11 genomes, including Ms. sp. SBSPR1A. Thus, it is more likely that Ms. sp. SBSPR1A produces betaine from sarcosine with sdmt, which bypasses the dimethylglycine intermediate step when synthesizing betaine from glycine [63]. While all of the genomes analyzed except Mmv. hollandica, and Ml. psychrophilus contain at least one gene for betaine transport, this additional ability to synthesize its own betaine could be an adaptation of Ms. sp. SBSPR1A to tolerate more hypersaline conditions than its sister genera have been found in to date. The presence of betaine synthesis and transport genes in these genomes agrees with previous work in solar salterns; on the other hand, one surprising result in these genomes is the lack of trehalose synthesis and transport genes, which were also suggested to be involved in salt tolerance in other salterns [28]. Future work is needed to identify the optimum growth conditions of this taxon. Based on the environmental sequencing data (Figure 4) and previous results from sister taxa [21], we hypothesize that while this organism can tolerate high levels of salt (e.g., ~300 ppt Cl−), its optimum will be at much lower concentrations.
We plan on building on the work presented here by attempting to culture the organism and run growth experiments to confirm methanogenesis substrates, as well as salt tolerance and optimum growth conditions. Such experiments will not only increase our knowledge of this organism, but also increase our understanding of the biogeochemistry and methane cycling in hypersaline ecosystems and inform the restoration of human-made systems such as industrial salt making facilities.
5. Conclusions
Here we have presented the draft genome sequence of Methanosalis sp. SBSPR1A gen. nov., sp. nov., and compared it to closely related taxa in the Methanolobus and Methanomethylovorans genera. Evidence for the discovery of a new genus includes the following: ANI values with other Methanolobus and Methanomethylovorans genomes were less than 80%, marker gene alignments place it between or adjacent to the Methanolobus and Methanomethylovorans taxa with sequenced genomes, and full length 16S rRNA gene percent identities were less than 94.5%. Similar to other Methanolobus and Methanomethylovorans species, Ms. sp. SBSPR1A is a methylotrophic methanogen and likely contributes to the elevated methane emissions observed in former industrial salterns [30]. Ms. sp. SBSPR1A has an acidic proteome similar to other halophilic organisms and is capable of synthesizing betaine, proline, ectoine, glutamate, and glutamine to tolerate environmental salinities between 97 and 232 ppt.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12101609/s1, Figure S1: Map of methanogenesis pathways, Figure S2: The 11 genomes analyzed in this study (highlighted) placed within a reference tree set to display 50 other genomes, Figure S3: Phylogenetic tree of full length 16S rRNA genes extracted from the 11 complete genomes in this study plus seven additional taxa with available 16S rRNA genes on NCBI, Figure S4: Heatmap of pairwise percent similarity of full length 16S rRNA genes, Figure S5: Average nucleotide identity (ANI) between the new Methanosalis sp. SBSPR1A genome, nine other Methanolobus reference genomes, and Methanomethylovorans hollandica, Figure S6: MASH clustering of average nucleotide identities among the 11 genomes in this study, Figure S7: Presence (red) or absence (blue) of genes involved in coenzyme B and coenzyme M synthesis, two important compounds in methanogenesis, Figure S8: Isoelectric point profiles for the 11 genomes analyzed in this study, Figure S9: Betaine synthesis in Methanosalis sp. SBSPR1A, showing the presence of gsmt and sdmt genes suggesting synthesis from sarcosine, and a lack of genes for synthesis from choline, Table S1: Pfam results for 61 genes missing from Methanosalis sp. SBSPR1A according to proteinortho, Table S2: KOs missing from or unique to Methanosalis sp. SBSPR1A.
Author Contributions
Conceptualization, C.P.B.d.M. and S.G.T.; data curation, C.P.B.d.M.; formal analysis, C.P.B.d.M., J.Z. and S.M.T.; funding acquisition, S.G.T.; methodology, C.P.B.d.M., J.Z., S.M.T. and S.G.T.; project administration, S.G.T.; resources, S.G.T.; supervision, S.G.T.; visualization, C.P.B.d.M.; writing—original draft, C.P.B.d.M.; writing—review and editing, C.P.B.d.M. and S.G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the DOE Early Career Research Program, grant no. KP/CH57/1, and includes work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The new genome is publicly available with IMG ID 2929001634 and on GenBank under BioProject ID PRJNA365332 and genome accession JAIPUR000000000. The metagenome from which it was derived is also available with IMG ID 3300026157 and NCBI BioSample accession SAMN06266236. Some aspects of the analysis are publicly available on Kbase narrative ID 90991.
Acknowledgments
We thank the production and sequence analysis teams at JGI for sequencing and bioinformatic support and Frederik Schulz for phylogenomic advice. We thank John Bourgeois, Donna Ball, and John Krause for help accessing the field sites. We thank Kelly Wrighton for permission to use the Ml. profundi MobM and Ml. vulcani PL 12/M genomes and Markus Göker for permission to use the Ml. bombayensis DSM 7082 genome. We acknowledge the German Research Foundation (DFG) SFB/TRR 51 for funding sequencing of Ml. bombayensis DSM 7082.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saunois, M.; Stavert, A.R.; Poulter, B.; Bousquet, P.; Canadell, J.G.; Jackson, R.B.; Raymond, P.A.; Dlugokencky, E.J.; Houweling, S.; Patra, P.K.; et al. The Global Methane Budget 2000–2017. Earth Syst. Sci. Data 2020, 12, 1561–1623. [Google Scholar] [CrossRef]
- Conrad, R. Importance of Hydrogenotrophic, Aceticlastic and Methylotrophic Methanogenesis for Methane Production in Terrestrial, Aquatic and Other Anoxic Environments: A Mini Review. Pedosphere 2020, 30, 25–39. [Google Scholar] [CrossRef]
- Liu, Y.; Whitman, W. Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. The Family Methanosarcinaceae. In The Prokaryotes: Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 259–281. ISBN 978-3-642-38954-2. [Google Scholar]
- König, H.; Stetter, K.O. Isolation and Characterization of Methanolobus Tindarius, Sp. Nov., a Coccoid Methanogen Growing Only on Methanol and Methylamines. Zent. Für Bakteriol. Mikrobiol. Hyg. Abt Orig. C Allg. Angew. Ökol. Mikrobiol. 1982, 3, 478–490. [Google Scholar] [CrossRef]
- Doerfert, S.N.; Reichlen, M.; Iyer, P.; Wang, M.; Ferry, J.G. Methanolobus Zinderi Sp. Nov., a Methylotrophic Methanogen Isolated from a Deep Subsurface Coal Seam. Int. J. Syst. Evol. Microbiol. 2009, 59, 1064–1069. [Google Scholar] [CrossRef]
- Ni, S.; Boone, D.R. Isolation and Characterization of a Dimethyl Sulfide-Degrading Methanogen, Methanolobus Siciliae HI350, from an Oil Well, Characterization of M. Siciliae T4/MT, and Emendation of M. Siciliae. Int. J. Syst. Evol. Microbiol. 1991, 41, 410–416. [Google Scholar] [CrossRef]
- Mochimaru, H.; Tamaki, H.; Hanada, S.; Imachi, H.; Nakamura, K.; Sakata, S.; Kamagata, Y. Methanolobus Profundi Sp. Nov., a Methylotrophic Methanogen Isolated from Deep Subsurface Sediments in a Natural Gas Field. Int. J. Syst. Evol. Microbiol. 2009, 59, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Kadam, P.C.; Ranade, D.R.; Mandelco, L.; Boone, D.R. Isolation and Characterization of Methanolobus Bombayensis Sp. Nov., a Methylotrophic Methanogen That Requires High Concentrations of Divalent Cations. Int. J. Syst. Evol. Microbiol. 1994, 44, 603–607. [Google Scholar] [CrossRef][Green Version]
- Lomans, B.P.; Maas, R.; Luderer, R.; Op den Camp, H.J.M.; Pol, A.; van der Drift, C.; Vogels, G.D. Isolation and Characterization of Methanomethylovorans Hollandica Gen. Nov., Sp. Nov., Isolated from Freshwater Sediment, a Methylotrophic Methanogen Able To Grow on Dimethyl Sulfide and Methanethiol. Appl. Environ. Microbiol. 1999, 65, 3641–3650. [Google Scholar] [CrossRef]
- Cha, I.-T.; Min, U.-G.; Kim, S.-J.; Yim, K.J.; Roh, S.W.; Rhee, S.-K. Methanomethylovorans Uponensis Sp. Nov., a Methylotrophic Methanogen Isolated from Wetland Sediment. Antonie Van Leeuwenhoek 2013, 104, 1005–1012. [Google Scholar] [CrossRef]
- Jiang, B.; Parshina, S.N.; van Doesburg, W.; Lomans, B.P.; Stams, A.J.M.Y. 2005 Methanomethylovorans Thermophila Sp. Nov., a Thermophilic, Methylotrophic Methanogen from an Anaerobic Reactor Fed with Methanol. Int. J. Syst. Evol. Microbiol. 2005, 55, 2465–2470. [Google Scholar] [CrossRef]
- Simankova, M.V.; Kotsyurbenko, O.R.; Lueders, T.; Nozhevnikova, A.N.; Wagner, B.; Conrad, R.; Friedrich, M.W. Isolation and Characterization of New Strains of Methanogens from Cold Terrestrial Habitats. Syst. Appl. Microbiol. 2003, 26, 312–318. [Google Scholar] [CrossRef]
- Oremland, R.S.; Boone, D.R. Methanolobus Taylorii Sp. Nov., a New Methylotrophic, Estuarine Methanogen. Int. J. Syst. Bacteriol. 1994, 44, 573–575. [Google Scholar] [CrossRef]
- Zhang, G.; Jiang, N.; Liu, X.; Dong, X. Methanogenesis from Methanol at Low Temperatures by a Novel Psychrophilic Methanogen, “Methanolobus Psychrophilus” Sp. Nov., Prevalent in Zoige Wetland of the Tibetan Plateau. Appl. Environ. Microbiol. 2008, 74, 6114–6120. [Google Scholar] [CrossRef]
- Oren, A. Taxonomy of Halophilic Archaea: Current Status and Future Challenges. Extremophiles 2014, 18, 825–834. [Google Scholar] [CrossRef]
- Ventosa, A.; Mellado, E.; Sanchez-Porro, C.; Marquez, M.C. Halophilic and Halotolerant Micro-Organisms from Soils. In Microbiology of Extreme Soils; Dion, P., Nautiyal, C.S., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 87–115. ISBN 978-3-540-74231-9. [Google Scholar]
- Vavourakis, C.D.; Andrei, A.-S.; Mehrshad, M.; Ghai, R.; Sorokin, D.Y.; Muyzer, G. A Metagenomics Roadmap to the Uncultured Genome Diversity in Hypersaline Soda Lake Sediments. Microbiome 2018, 6, 168. [Google Scholar] [CrossRef] [PubMed]
- Mirete, S.; Mora-Ruiz, M.R.; Lamprecht-Grandío, M.; de Figueras, C.G.; Rosselló-Móra, R.; González-Pastor, J.E. Salt Resistance Genes Revealed by Functional Metagenomics from Brines and Moderate-Salinity Rhizosphere within a Hypersaline Environment. Front. Microbiol. 2015, 6, 1121. [Google Scholar] [CrossRef] [PubMed]
- Muyodi, F.J. Microbiological analysis of the waters of Lake Victoria in relation to the invasion of the water hyacinth, Eichhornia crassipes (Mart.) Solms. In A Case Study of the Lakeshores of Mwanza Municipality; University of Dar es Salaam: Dar es Salaam, Tanzania, 2000; pp. 158–173. [Google Scholar]
- Chen, S.-C.; Huang, H.-H.; Lai, M.-C.; Weng, C.-Y.; Chiu, H.-H.; Tang, S.-L.; Rogozin, D.Y.; Degermendzhy, A.G. Methanolobus Psychrotolerans Sp. Nov., a Psychrotolerant Methanoarchaeon Isolated from a Saline Meromictic Lake in Siberia. Int. J. Syst. Evol. Microbiol. 2018, 68, 1378–1383. [Google Scholar] [CrossRef] [PubMed]
- Vreeland, R.H. Mechanisms of Halotolerance in Microorganisms. Crit. Rev. Microbiol. 2009, 14, 311–356. [Google Scholar] [CrossRef]
- Oremland, R.S.; Polcin, S. Methanogenesis and Sulfate Reduction: Competitive and Noncompetitive Substrates in Estuarine Sediments. Appl. Environ. Microbiol. 1982, 44, 1270–1276. [Google Scholar] [CrossRef]
- Oremland, R.S.; Marsh, L.M.; Polcin, S. Methane Production and Simultaneous Sulphate Reduction in Anoxic, Salt Marsh Sediments. Nature 1982, 296, 143–145. [Google Scholar] [CrossRef]
- Mcgenity, T.; Sorokin, D. Methanogens and Methanogenesis in Hypersaline Environments. In Biogenesis of Hydrocarbons; Springer International Publishing: New York, NY, USA, 2018; pp. 1–27. ISBN 978-3-319-53114-4. [Google Scholar]
- Matarredona, L.; Camacho, M.; Zafrilla, B.; Bonete, M.-J.; Esclapez, J. The Role of Stress Proteins in Haloarchaea and Their Adaptive Response to Environmental Shifts. Biomolecules 2020, 10, 1390. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of Adaptation of Microorganisms of the Three Domains of Life to High Salt Concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Vera-Gargallo, B.; Ventosa, A. Metagenomic Insights into the Phylogenetic and Metabolic Diversity of the Prokaryotic Community Dwelling in Hypersaline Soils from the Odiel Saltmarshes (SW Spain). Genes 2018, 9, 152. [Google Scholar] [CrossRef]
- Youssef, N.H.; Savage-Ashlock, K.N.; McCully, A.L.; Luedtke, B.; Shaw, E.I.; Hoff, W.D.; Elshahed, M.S. Trehalose/2-Sulfotrehalose Biosynthesis and Glycine-Betaine Uptake Are Widely Spread Mechanisms for Osmoadaptation in the Halobacteriales. ISME J. 2014, 8, 636–649. [Google Scholar] [CrossRef]
- Zhou, J.; Theroux, S.M.; Bueno de Mesquita, C.P.; Hartman, W.H.; Tringe, S.G. Microbial Drivers of Methane Emissions from Unrestored Industrial Salt Ponds. ISME J. 2021. [Google Scholar] [CrossRef] [PubMed]
- von Meijenfeldt, F.A.B.; Arkhipova, K.; Cambuy, D.D.; Coutinho, F.H.; Dutilh, B.E. Robust Taxonomic Classification of Uncharted Microbial Sequences and Bins with CAT and BAT. Genome Biol. 2019, 20, 217. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an Efficient Tool for Accurately Reconstructing Single Genomes from Complex Microbial Communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Tang, Y.-H.; Tringe, S.G.; Simmons, B.A.; Singer, S.W. MaxBin: An Automated Binning Method to Recover Individual Genomes from Metagenomes Using an Expectation-Maximization Algorithm. Microbiome 2014, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-H.; Liao, Y.-C. Accurate Binning of Metagenomic Contigs via Automated Clustering Sequences Using Information of Genomic Signatures and Marker Genes. Sci. Rep. 2016, 6, 24175. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of Genomes from Metagenomes via a Dereplication, Aggregation and Scoring Strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum Information about a Single Amplified Genome (MISAG) and a Metagenome-Assembled Genome (MIMAG) of Bacteria and Archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef]
- Pohlert, T. The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). 2014. Available online: https://CRAN.R-project.org/package=PMCMR (accessed on 28 July 2021).
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Chen, L.-X.; Anantharaman, K.; Shaiber, A.; Eren, A.M.; Banfield, J.F. Accurate and Complete Genomes from Metagenomes. Genome Res. 2020, 30, 315–333. [Google Scholar] [CrossRef]
- Paulino, D.; Warren, R.L.; Vandervalk, B.P.; Raymond, A.; Jackman, S.D.; Birol, I. Sealer: A Scalable Gap-Closing Application for Finishing Draft Genomes. BMC Bioinform. 2015, 16, 230. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. GigaScience 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An Integrated Data Management and Comparative Analysis System for Microbial Genomes and Microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Seshadri, R.; Varghese, N.J.; Eloe-Fadrosh, E.A.; Meier-Kolthoff, J.P.; Göker, M.; Coates, R.C.; Hadjithomas, M.; Pavlopoulos, G.A.; Paez-Espino, D.; et al. 1,003 Reference Genomes of Bacterial and Archaeal Isolates Expand Coverage of the Tree of Life. Nat. Biotechnol. 2017, 35, 676–683. [Google Scholar] [CrossRef]
- Ticak, T.; Hariraju, D.; Arcelay, M.B.; Arivett, B.A.; Fiester, S.E.; Ferguson, D.J. Isolation and Characterization of a Tetramethylammonium-Degrading Methanococcoides Strain and a Novel Glycine Betaine-Utilizing Methanolobus Strain. Arch. Microbiol. 2015, 197, 197–209. [Google Scholar] [CrossRef]
- Kadam, P.C.; Boone, D.R. Physiological Characterization and Emended Description of Methanolobus Vulcani. Int. J. Syst. Bacteriol. 1995, 45, 400–402. [Google Scholar] [CrossRef]
- Roux, S.; Krupovic, M.; Daly, R.A.; Borges, A.L.; Nayfach, S.; Schulz, F.; Sharrar, A.; Matheus Carnevali, P.B.; Cheng, J.-F.; Ivanova, N.N.; et al. Cryptic Inoviruses Revealed as Pervasive in Bacteria and Archaea across Earth’s Biomes. Nat. Microbiol. 2019, 4, 1895–1906. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, S.-C.; Lai, M.-C.; Huang, H.-H.; Chiu, H.-H.; Tang, S.-L.; Rogozin, D.Y.; Degermendzhy, A.G. Methanolobus Halotolerans Sp. Nov., Isolated from the Saline Lake Tus in Siberia. Int. J. Syst. Evol. Microbiol. 2020, 70, 5586–5593. [Google Scholar] [CrossRef]
- Göker, M.; Klenk, H.-P. Phylogeny-Driven Target Selection for Largescale Genome-Sequencing (and Other) Projects. Stand. Genom. Sci. 2013, 8, 360–374. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast Selection of Best-Fit Models of Protein Evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Lee, I.; Chalita, M.; Ha, S.-M.; Na, S.-I.; Yoon, S.-H.; Chun, J. 2017 ContEst16S: An Algorithm That Identifies Contaminated Prokaryotic Genomes Using 16S RNA Gene Sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Blankenberg, D.; Taylor, J.; Schenck, I.; He, J.; Zhang, Y.; Ghent, M.; Veeraraghavan, N.; Albert, I.; Miller, W.; Makova, K.D.; et al. A Framework for Collaborative Analysis of ENCODE Data: Making Large-Scale Analyses Biologist-Friendly. Genome Res. 2007, 17, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)Orthologs in Large-Scale Analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Krassowski, M. Krassowski/complex-upset. GitHub 2020. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Seaver, S.M.D.; Liu, F.; Zhang, Q.; Jeffryes, J.; Faria, J.P.; Edirisinghe, J.N.; Mundy, M.; Chia, N.; Noor, E.; Beber, M.E.; et al. The ModelSEED Biochemistry Database for the Integration of Metabolic Annotations and the Reconstruction, Comparison and Analysis of Metabolic Models for Plants, Fungi and Microbes. Nucleic Acids Res. 2021, 49, D575–D588. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc Collection of Microbial Genomes and Metabolic Pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Becker, E.A.; Seitzer, P.M.; Tritt, A.; Larsen, D.; Krusor, M.; Yao, A.I.; Wu, D.; Madern, D.; Eisen, J.A.; Darling, A.E.; et al. Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-Response. PLoS Genet. 2014, 10, e1004784. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty Heatmaps, R Package Version 1.0.12; 2019. Available online: http://CRAN.R-project.org/package=pheatmap (accessed on 28 July 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the Classification of Cultured and Uncultured Bacteria and Archaea Using 16S rRNA Gene Sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Deppenmeier, U. The unique biochemistry of methanogenesis. In Progress in Nucleic Acid Research and Molecular Biology; Academic Press: Cambridge, MA, USA, 2002; Volume 71, pp. 223–283. [Google Scholar]
- Oren, A. Life at High Salt Concentrations, Intracellular KCl Concentrations, and Acidic Proteomes. Front. Microbiol. 2013, 4, 315. [Google Scholar] [CrossRef] [PubMed]
- Antony, C.P.; Murrell, J.C.; Shouche, Y.S. Molecular Diversity of Methanogens and Identification of Methanolobus Sp. as Active Methylotrophic Archaea in Lonar Crater Lake Sediments. FEMS Microbiol. Ecol. 2012, 81, 43–51. [Google Scholar] [CrossRef][Green Version]
- Schink, B.; Zeikus, J.G.Y. 1982 Microbial Ecology of Pectin Decomposition in Anoxic Lake Sediments. Microbiology 1982, 128, 393–404. [Google Scholar] [CrossRef]
- Lovley, D.R.; Klug, M.J. Methanogenesis from Methanol and Methylamines and Acetogenesis from Hydrogen and Carbon Dioxide in the Sediments of a Eutrophic Lake. Appl. Environ. Microbiol. 1983, 45, 1310–1315. [Google Scholar] [CrossRef]
- Orphan, V.J.; Jahnke, L.L.; Embaye, T.; Turk, K.A.; Pernthaler, A.; Summons, R.E.; Marais, D.J.D. Characterization and Spatial Distribution of Methanogens and Methanogenic Biosignatures in Hypersaline Microbial Mats of Baja California. Geobiology 2008, 6, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, G.-C.; Elling, F.J.; Nigro, L.M.; Samarkin, V.; Joye, S.B.; Teske, A.; Hinrichs, K.-U. Multiple Evidence for Methylotrophic Methanogenesis as the Dominant Methanogenic Pathway in Hypersaline Sediments from the Orca Basin, Gulf of Mexico. Geochim. Cosmochim. Acta 2016, 187, 1–20. [Google Scholar] [CrossRef]
- Zhuang, G.-C.; Heuer, V.B.; Lazar, C.S.; Goldhammer, T.; Wendt, J.; Samarkin, V.A.; Elvert, M.; Teske, A.P.; Joye, S.B.; Hinrichs, K.-U. Relative Importance of Methylotrophic Methanogenesis in Sediments of the Western Mediterranean Sea. Geochim. Cosmochim. Acta 2018, 224, 171–186. [Google Scholar] [CrossRef]
- Weston, N.B.; Dixon, R.E.; Joye, S.B. Ramifications of Increased Salinity in Tidal Freshwater Sediments: Geochemistry and Microbial Pathways of Organic Matter Mineralization. J. Geophys. Res. Biogeosciences 2006, 111. [Google Scholar] [CrossRef]
- Kristjansson, J.K.; Schönheit, P. Why Do Sulfate-Reducing Bacteria Outcompete Methanogenic Bacteria for Substrates? Oecologia 1983, 60, 264–266. [Google Scholar] [CrossRef]
- Oren, A. Formation and Breakdown of Glycine Betaine and Trimethylamine in Hypersaline Environments. Antonie Van Leeuwenhoek 1990, 58, 291–298. [Google Scholar] [CrossRef]
- Kiene, R.P.; Visscher, P.T. Production and Fate of Methylated Sulfur Compounds from Methionine and Dimethylsulfoniopropionate in Anoxic Salt Marsh Sediments. Appl. Environ. Microbiol. 1987, 53, 2426–2434. [Google Scholar] [CrossRef]
- Liu, Y.; Priscu, J.C.; Xiong, J.; Conrad, R.; Vick-Majors, T.; Chu, H.; Hou, J. Salinity Drives Archaeal Distribution Patterns in High Altitude Lake Sediments on the Tibetan Plateau. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef]
- Smith, J.M.; Green, S.J.; Kelley, C.A.; Prufert-Bebout, L.; Bebout, B.M. Shifts in Methanogen Community Structure and Function Associated with Long-Term Manipulation of Sulfate and Salinity in a Hypersaline Microbial Mat. Environ. Microbiol. 2008, 10, 386–394. [Google Scholar] [CrossRef]
- Deole, R.; Challacombe, J.; Raiford, D.W.; Hoff, W.D. An Extremely Halophilic Proteobacterium Combines a Highly Acidic Proteome with a Low Cytoplasmic Potassium Content. J. Biol. Chem. 2013, 288, 581–588. [Google Scholar] [CrossRef]
- Rhodes, M.E.; Fitz-Gibbon, S.T.; Oren, A.; House, C.H. Amino Acid Signatures of Salinity on an Environmental Scale with a Focus on the Dead Sea. Environ. Microbiol. 2010, 12, 2613–2623. [Google Scholar] [CrossRef]
- Paul, S.; Bag, S.K.; Das, S.; Harvill, E.T.; Dutta, C. Molecular Signature of Hypersaline Adaptation: Insights from Genome and Proteome Composition of Halophilic Prokaryotes. Genome Biol. 2008, 9, R70. [Google Scholar] [CrossRef] [PubMed]
- Elevi Bardavid, R.; Oren, A. Acid-Shifted Isoelectric Point Profiles of the Proteins in a Hypersaline Microbial Mat: An Adaptation to Life at High Salt Concentrations? Extremophiles 2012, 16, 787–792. [Google Scholar] [CrossRef]
- Sur, S.; Bothra, A.K.; Sen, A. Proteome Analysis Reveals the Influence of Isoelectric Point and Amino Acid Usages on the Lifestyle of Nitrogen Fixing Microorganisms. Indian J. Biotechnol. 2013, 12, 88–97. [Google Scholar]
- Schwartz, R.; Ting, C.S.; King, J. Whole Proteome PI Values Correlate with Subcellular Localizations of Proteins for Organisms within the Three Domains of Life. Genome Res. 2001, 11, 703–709. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).