Abstract
Staphylococcus aureus is a bacterium that mainly colonizes the nasal cavity and skin. To colonize the host, it is necessary for S. aureus to resist many antibacterial factors derived from human and commensal bacteria. Among them are the bacteria-derived antimicrobial peptides (AMPs) called bacteriocins. It was reported that some two-component systems (TCSs), which are signal transduction systems specific to bacteria, are involved in the resistance to several bacteriocins in S. aureus. However, the TCS-mediated resistance is limited to relatively low concentrations of bacteriocins, while high concentrations of bacteriocins still exhibit antibacterial activity against S. aureus. To determine whether we could obtain highly bacteriocin-resistant mutants, we tried to isolate highly nisin A-resistant mutants by exposing the cells to sub-minimum inhibitory concentrations (MICs) of nisin A. Nisin A is one of the bacteriocins produced by Lactococcus lactis and is utilized as a food preservative worldwide. Finally, we obtained highly nisin A-resistant mutants with mutations in one TCS, BraRS, and in PmtR, which is involved in the expression of pmtABCD. Notably, some highly resistant strains also showed increased pathogenicity. Based on our findings, this review provides up-to-date information on the role of TCSs in the susceptibility to antibacterial peptides. Additionally, the mechanism for high antimicrobial peptides resistance and its association with pathogenicity in S. aureus is elucidated.
1. Introduction
Commensal bacteria inhabit the parts of the human body that come in contact with the external environment (oral cavity, digestive organs, vagina, anus, skin, etc.). Commensal bacteria compete and cooperate with each other in the environment. In organs where microorganisms are originally resident, the number of bacteria is controlled by the immune system to prevent infectious diseases. However, in the compromised hosts such as elderly people and patients with systemic diseases, the immune activity in these individuals is considered to be weakened. Under such conditions, the proportion of each bacterial species in some sites of the human body is altered, causing dysbiosis; in some cases, infectious diseases occur. Antibiotics (currently called “antibacterial chemotherapeutic agents”) are used to treat bacterial infections. However, depending on the dose and frequency of antibiotic administration, drug-resistant bacteria sometimes emerge.
Staphylococcus aureus is known as a commensal bacterium in humans; it generally localizes in the nasal cavity, skin, and intestine. S. aureus is a highly adaptable bacterium causing opportunistic infections, such as suppurative diseases, pneumonia, and sepsis [1,2,3]. Additionally, S. aureus causes food poisoning because it produces several heat-stable enterotoxins [2]. S. aureus is a pathogenic bacterium with a wide variety of virulence factors, and antibiotic resistance is likely to occur with long-term exposure to antibacterial chemotherapeutic agents [4]. According to the 2013 Centers for Disease Control and Prevention (CDC) report, 80,000 people were affected by methicillin-resistant Staphylococcus aureus (MRSA) in the USA in that year. In addition, the O’Neill Report [5] estimated that the number of deaths from drug-resistant bacteria would exceed that from cancer in 2050. Among the infectious disease-causing microorganisms listed in this report are various drug-resistant bacteria, such as MRSA, penicillin-resistant Streptococcus pneumoniae (PRSP), and carbapenem-resistant enterobacteriaceae (CRE). In response to these reports, counterplans against drug-resistant bacteria, known as antimicrobial resistance (AMR) action plans, are advocated worldwide.
Antibacterial chemotherapeutic agents are generally administered to cure S. aureus infections. However, the emergence of MRSA has become a growing challenge of this treatment approach. Additionally, disinfectants are also widely used for the prevention of nosocomial infection. It is reported that the qac genes, which encodes a multidrug efflux pump that expels toxic molecules, contributes to the development of resistance to quaternary ammonium compounds such as benzalkonium chloride [6,7]. In the genus Staphylococcus, the qac genes are encoded in a plasmid, and six types of Qac efflux pumps are reported. Among the Qac proteins, QacA and QacB are highly conserved among Staphylococcus species, while QacC, QacG, QacH, and QacJ, which belong to the small multidrug resistance (SMR) family, are known to have amino acid sequence diversity among the Staphylococcus species. Therefore, S. aureus shows resistance not only to several antibacterial chemotherapeutic agents but also to the other antibacterial agents such as disinfectants. In recent years, antimicrobial peptides (AMPs) have attracted attention as antibacterial chemotherapeutic agents. These AMPs are derived from various living organisms, such as humans, plants, and bacteria [8,9,10,11]. Bacterial AMPs are also called bacteriocins. Some of these antibacterial peptides and bacteriocins were also shown to be effective against MRSA [12,13,14] and have potential applications in the clinic [15,16]. Therefore, these peptides are attracting attention as candidates for next-generation antibacterial chemotherapeutic agents because of their high stability and the establishment of purification methods in recent years. In this review, we provide up-to-date information for understanding the role of the potentially present strains that found by applying high concentrations of antimicrobial chemotherapeutic agents, focusing on the genetic characteristics and high resistance mechanisms of isolated strains. Then, we explain the pathogenicity of isolated endogenous highly nisin A-resistant strains and the underlying mechanism. This information reveals the existence of these endogenous antibiotic-resistant strains, which may be an “outbreak reserve force”, and it is thought that these results will help suppress the potential emergence of highly resistant strains of S. aureus.
2. Bacteriocins
Bacteriocins are ribosomally synthesized peptides or proteins that exhibit antibacterial activity against bacterial species that are closely related to bacteriocin producers [17,18]. Bacteriocins are mainly classified into classes I and II [19]. Class I bacteriocins (peptides <5 kDa) are called “lantibiotics” and contain a ring bridged by lanthionine and 3-methyllanthionine residues [20], whereas class II bacteriocins comprise unmodified amino acids [20]. Lantibiotics are subdivided into types A and B [21]. Type A lantibiotics bind to lipid II, which is involved in peptidoglycan synthesis, and then inhibit cell wall biosynthesis and disturb the bacterial membrane [17,20], while type B lantibiotics are globular peptides that inhibit cell wall biosynthetic steps such as transglycosylation [22]. Type A lantibiotics are further classified into two subtypes: type A(I), while lactin 481 and nukacin ISK-1 are classified as subtypes of type A(II) [19]. Class II bacteriocins are classified into the following three subclasses: IIa, IIb, and IIc [23].
Nisin A is a bacteriocin produced by L. lactis [24]. Nisin A is a lantibiotic that contains unusual amino acids such as lanthionine, β-methyllanthionine, and dehydrated amino acids [20]. Nisin A binds to lipid II, resulting in membrane disturbance. Recently, it was reported that nisin A is associated with DNA condensation by interfering with chromosome replication or segregation in S. aureus [25]. Nisin A has broad-spectrum antimicrobial activity, mainly against gram-positive bacteria [26,27,28,29,30,31]. Due to its broad-spectrum activity, nisin A is widely used as a food additive worldwide for the prevention of food poisoning [26,32,33]. In addition, Alves DCB et al. reported the potential use of nisin combined with oxacillin for methicillin-resistant S. aureus [34]. Bacteriocins, including nisin A, were also investigated as potential antibacterial chemotherapeutic agents for clinical application [26,29,35].
3. Two-Component Systems and Their Association with AMP Resistance
Recently, two-component systems (TCSs) were reported to be associated with the resistance to several types of antibacterial agents, such as bacitracin, vancomycin, human β defensins (hBDs), LL37, and bacteriocins [36,37,38,39,40,41]. TCSs are predominantly found in prokaryotes. TCSs comprise a sensory histidine kinase (HK) and a cognate response regulator (RR) [42,43]. The sensor is a transmembrane protein that senses changes in the external environment, resulting in autophosphorylation of histidine residues (HKs) in the sensor and transfer the phosphate to aspartate residues of the cognate response regulator (RR) [43,44]. The phosphorylated RR then binds to target DNA elements with strong affinity, activating or repressing the transcription of target genes. Thus, bacteria are able to quickly adapt to the external environment by regulating the expression of the respective genes.
It was revealed that S. aureus has 16 sets of TCSs. The function of each TCS is shown in Table 1. A well-studied TCS is the Agr system, which is known to be widely involved in the regulation of virulence factor expression. Agr has a central role in the quorum-sensing system, which senses cell density via autoinducer peptides (AIPs) [45]. Agr is involved in the expression of many factors, including virulence factors mediated by RNAIII, a gene product of hld (delta-hemolysin). RNAIII was demonstrated to directly upregulate hla (α-haemolysin) expression [45] and downregulate the expression of spa (protein A) [46] and the transcription factor rot gene, which is responsible for the repression of toxins [47]. RNAIII binds to the target mRNA directly, resulting in the up- or down-regulation of gene expression. Phenol soluble modulins (PSMs) were demonstrated to be regulated by AgrA directly and have versatile virulence activities such as epithelial colonization, cytotoxic activity, biofilm formation, and antimicrobial activity [48,49,50]. However, the precise mechanism of the expression of other virulence factors mediated by the Agr system is still unknown. SaeRS, one of the TCSs in S. aureus, is also known as a global regulator of virulence factors and is important for the regulation of coagulase, α-toxin, β-haemolysin, γ-haemolysin, staphylococcal immunoglobulin-binding protein, nuclease, leucocidin, toxic shock syndrome toxin-1 (TSST-1), epidermis deprivation toxin, etc. SaeRS promotes the expression of coagulase [51,52,53].

Table 1.
Function of two-component system in Staphylococcus aureus MW2 strain.
TCSs were reported to be involved in controlling the susceptibility to human-derived AMPs. AMPs are innate immune factors and are produced in various tissues and organs, such as the skin, lungs, and intestines [8,9,10,11,54]. The most well-known AMPs are defensins. Defensins are classified into two types: α-defensins from neutrophils and Paneth cells and β-defensins (hBDs) from mainly epithelial cells [8,10,55]. Another major peptide is human cathelicidin (LL37), which is found in various cells, including neutrophils and epithelial cells [10]. The Aps system is related to resistance against human β-defensin-3 (hBD3), LL37, and bacteriocins (nisin A, nukacin ISK-1), which possess a strong positive charge. ApsR regulates dlt and mprF (fmtC) expression, causing an increase in cell surface charge [36,41,56]. Dlt is involved in alanine addition to teichoic acids in cell walls, while MprF is involved in lysine addition to phosphatidylglycerol in cell membranes [57,58]. Alanylation of teichoic acids and lysyl-phosphatidylglycerol contribute a shift to a weak negative charge on the cell surfaces (Figure 1). Since apsRS expression is negatively controlled by Agr, this resistance system is mainly observed in the early stage of bacterial growth with low expression of Agr, while in the stationary phase, Agr expression is increased, leading to suppression of the expression of ApsRS. Therefore, the charge on the surface of the bacterial cells is altered during growth. As a result, the susceptibility to antibacterial peptides changes during growth, with low susceptibility observed in the exponential phase and high susceptibility in the stationary phase [37].
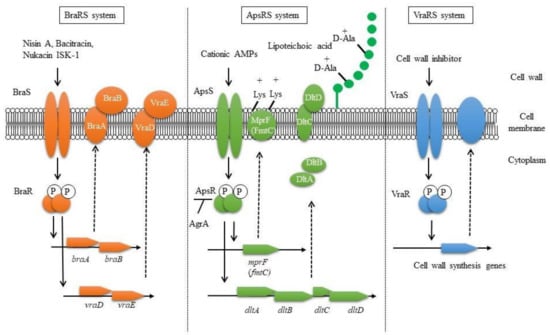
Figure 1.
Proposed bacteriocin resistance mechanism mediated by TCSs in S. aureus.
VraSR regulates many factors involved in cell wall biosynthesis and is associated with the susceptibility to cell wall synthesis inhibitors such as β-lactams, vancomycin, cycloserine, teicoplanin, and bacitracin [38,39,59]. Upon the addition of cell wall synthesis inhibitors, VraSR is activated, resulting in the upregulation of several cell wall synthesis genes, including the transpeptidase pbp2 and the transglycosylase sgtB [38].
BraRS, which is involved in the acquisition of bacteriocin resistance, was first discovered to be involved in the resistance to bacitracin, one of the bacteriocins produced by Bacillus subtilis [60]. The resistance mechanism involves the sensing of low concentrations of bacitracin by the complex of the BraRS TCS and the upstream ABC transporter BraDE. As a result, the regulator BraR promotes the expression of the ABC transporter VraDE, an intrinsic resistance factor for bacitracin [61]. The BraRS-VraDE system is considered to be a TCS system that widely supports the sensing of several bacteriocins because it is also involved in the resistance to nukacin ISK-1 produced by Staphylococcus warneri and nisin A produced by L. lactis [40,41]. In addition, it was reported that the ABC transporter BraDE, encoded with BraRS regulon, was also associated with nisin resistance by directly interacting with BraS [62].
In conclusion, regarding bacteriocin resistance, it was clarified that BraRS regulates the expression of ABC transporters to promote resistance against several bacteriocins. ApsRS and VraRS also participate in bacteriocin resistance by changing the charge and increasing the expression of cell wall synthesis genes, respectively (Figure 1) [41]. In this way, it is expected that S. aureus performs precise TCS-mediated control to survive even in the presence of many bacteriocins produced by some other bacteria colocalized in the bacterial flora.
Three TCSs are known to be involved in bacteriocin resistance: BraRS, ApsRS, and VraRS. BraRS is a TCS that senses various bacteriocins and induces the expression of the ABC transporter vraDE. BraAB is required for BraS to sense bacteriocins, and braAB expression is also induced via BraRS (left). ApsRS is involved in resistance to positively charged antimicrobial peptides (AMPs). Aps controls the cell surface charge by regulating the expression of dlt and mprF. Aps is negatively controlled by Agr, the quorum sensing system (middle). VraRS is involved in resistance to cell wall synthesis inhibitors and regulates several genes in the cell wall synthesis system such as pbp2, sgtB, and murZ (right).
Amino acid sequences of ApsRS and BraRS from S. aureus are compared with those from the other staphylococci (Figure 2 and Table 2). Although ApsS in S. aureus does not have a high similarity with that of the other staphylococci, the response regulator ApsR in S. aureus shows relatively high similarity (above 79% identity) with that of other staphylococcal species except for S. pseudintermedius. It is speculated that this system, which changes the surface charge, is widely conserved among staphylococci. On the contrary, BraRS in S. aureus shows low similarity with that of the other staphylococci. Therefore, BraRS, which senses nisin A, bacitracin, and Nukacin ISK-1, may be specific to S. aureus.

Figure 2.
Comparison of two-component systems showing homology with ApsRS and BraRS of S. aureus among staphylococci. Alignment of ApsRS (A) of BraRS (B) among 8 staphylococcal species. 1. Staphylococcus aureus (Saur), 2. Staphylococcus epidermidis (Sepi), 3. Staphylococcus haemolyticus (Shae), 4. Staphylococcus lugdunensis (Slug), 5. Staphylococcus pseudintermedius (Spse), 6. Staphylococcus saprophyticus (Ssap), 7. Staphylococcus warneri (Swar), 8. Staphylococcus carnosus (Scar).

Table 2.
% amino acid sequence identity of ApsRS and BraRS in S. aureus compared to eight Staphylococcal species.
4. Isolation of Highly Nisin-Resistant Strains with Point Mutations by Exposure to Sub-MICs of Nisin
As explained above, S. aureus has several systems for resistance against AMPs, including some bacteriocins. Since these systems are effective in low concentrations of AMPs, AMPs are sometimes utilized at a high concentration for a treatment or a food preservative. Consequently, S. aureus cells may have a chance to be exposed to high concentrations of bacteriocins, leading to the emergence of highly resistant strains. To clarify our hypothesis, we tried to investigate whether S. aureus can acquire high resistance by exposing the cells to bacteriocins.
In a previous study, high nisin A-resistant strains were isolated by exposing S. aureus to sub-MICs of nisin A and designated them S. aureus nisin-resistant (SAN) strains [63]. Some SAN strains showed resistance to not only nisin A but also to the other antibacterial agents such as bacitracin and human AMPs. Our findings suggest that the acquisition of bacteriocin resistance may result in cross-resistance to the other antibacterial agents. S. aureus can adapt to not only antibacterial chemotherapeutic agents but also preservatives like nisin A.
Some of the SAN strains showed constitutively high expression of vraDE, the expression of which is normally induced by nisin A. In contrast, no induction of vraDE expression was observed in the other SAN strains. By analyzing the sequences of vraDE and braRS, we found point mutations in the BraRS region of three strains with high VraDE expression (Figure 3A). These three strains, designated SAN1, SAN8, and SAN87, showed mutations in different genes in the braRS region [63].
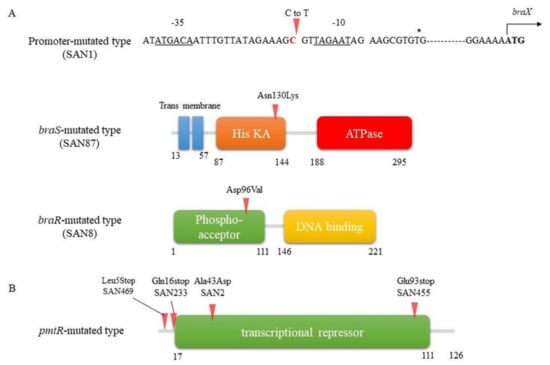
Figure 3.
Mutation sites in the high nisin A-resistant strain. (A) Mutation sites in the braXRS region. The mutation sites in the braXRS region in the isolated mutants are indicated by red arrows. The nucleotide sequence upstream of braXRS in the MW2 strain is shown, with the –35 and –10 regions indicated by the underline. The transcription initiation start sites are labelled with an asterisk, and the ATG translation initiation codons are indicated in bold. Mutation sites in SAN1, SAN87, and SAN8 are found in the braXRS promoter region (upper), His KA of braS region (middle), and phospho-acceptor domain of braR region (lower), respectively. His KA, dimerisation and phospho-acceptor domain of histidine kinases. (B) Mutation sites in the pmtR. The pmtR in MW2 strain is shown. In the isolated mutants from MW2 (SAN2 and SAN469), COL (SAN233) and TY34 (SAN455) strains, all mutations were found within the pmtR (red arrows).
On the other hand, no mutation in the braRS-vraDE region was observed in the high nisin-resistant strain (designated SAN2), in which vraDE expression was not induced upon the addition of nisin A. DNA microarray analysis showed high expression of the pmtRABCD gene. By determining the DNA sequence of this region, a point mutation was found in the gene encoding PmtR (Figure 3B) [64].
4.1. Mechanism Underlying High Nisin Resistance in the BraXRS Mutant
In the SAN1 type strain, only one point mutation was observed between the –35 and –10 boxes in the braXRS promoter region (Figure 3A upper). The strains with mutations in the promoter region showed increased promoter activity, resulting in the high expression of braRS. Based on these results, we speculated that a high amount of BraRS in the SAN1 strain conferred increased levels of phosphorylated BraR upon the addition of nisin A, which resulted in increased expression of VraDE compared to that in the wild-type strain (Figure 4). One point mutation occurred in braS (SAN87) and braR (SAN8), resulting in one amino acid substitution. Due to this mutation, SAN8 and SAN87 showed constitutively high VraDE expression, even in the absence of nisin A. Therefore, the unphosphorylated form of the mutant BraR protein and mutated BraS are capable of inducing VraDE expression (Figure 4).
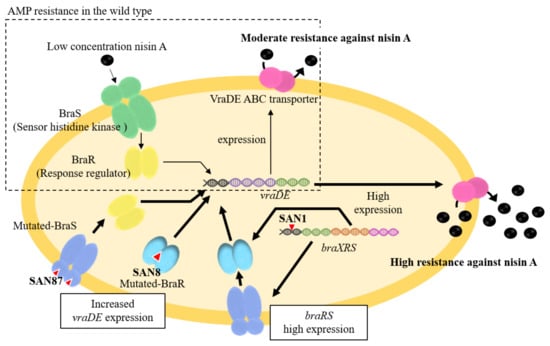
Figure 4.
Mechanism underlying the high nisin A resistance of the BraRS mutant. In the MW2 strain, the BraRS-VraDE system responds to low concentrations of nisin A (upper side). In highly nisin A-resistant strains, mutation (red triangle) occurs in the braXRS region, resulting in high expression of vraDE and resistance to nisin A (lower side).
There were only two reports of increased bacteriocin resistance as a result of mutations in the braXRS region. The mutation sites in the braXRS region are the promoter region of braXRS (one report), the braS sensor region (two reports), and the braR regulator region (one report). All of the mutant strains showed a single nucleotide point mutation. Although it is known that vraDE is regulated by BraRS and its expression is induced in a nisin concentration-dependent manner [41,61], this response is only observed under low concentrations of nisin. Mutations in the braXRS region of the TCS may ultimately trigger the development of high nisin resistance.
According to the structural analysis of NsrR, which shares homology with BraR, the mutation site observed in SAN8 type is an aspartic acid residue at the 96th position (Asp to Val) (Figure 3A lower), adjacent to a conserved phenylalanine residue that is a switch residue [65]. In general, in RRs, phosphorylation of aspartic acid causes conformational changes in two amino acids of the RR called switch residues, resulting in dimer formation [41,66]. Due to the different properties of aspartic acid (hydrophilic and acidic) and valine (hydrophilic and nonpolar), the dimerization interface region undergoes a structural change, leading to BraR forming a dimer and binding upstream of vraDE even in the nonphosphorylated state.
In the SAN87 type strain, a mutation was found at position 130 (asparagine to lysine) of the sensor protein BraS (Figure 3A middle). The sensor protein is composed of three regions: the sensing region (which includes the transmembrane region), the histidine kinase (His KA) region, and the ATPase region. The mutation site in the sensor protein in SAN87 is in the histidine kinase region, next to the sensing region. Previous reports have isolated nisin-resistant S. aureus strains with mutations in the histidine kinase and ATPase regions [67]. Since BraS is necessary to activate the BraR regulator, mutations in these regions may result in the activation of the BraR by increased autophosphorylation, which in turn enhances the induction of target factor gene expression.
4.2. Mechanism Underlying High Nisin Resistance in the PmtR Mutant
BraRS-VraDE-independent nisin A high resistant S. aureus was also isolated (SAN2, SAN233, SAN455, and SAN469) [64]. This mutant has a mutation in pmtR, which encodes a transcriptional regulator that controls the expression of the pmtR and pmtABCDD (pmtA-D) operon (Figure 3B). As a result, these mutants exhibited increased expression of PmtA-D, a transporter responsible for the export of PSMs. High nisin A-resistant mutants were also isolated from not only the MW2 strain but also the MRSA COL strain and TY34 strain. All these mutants had a point mutation in the pmtR gene, yielding a mutant PmtR with an amino acid replacement of alanine to aspartic acid (SAN2 from MW2) or a truncated PmtR (SAN469 from MW2, SAN233 from COL and SAN455 from TY34) (Figure 3B).
Previous studies have shown that PmtR is a negative transcriptional regulator of the pmtRABCD (pmtR-D) operon (Figure 5) [68]. The EMSA results showed that the mutated PmtR from the SAN2 (point mutation type) and the other three SAN (truncation type) strains lost their ability to bind to the DNA region upstream of pmtR-D. The mutated PmtR lost their ability to bind to the DNA region upstream of pmtR-D, resulting in increased expression of pmtR-D. This result suggests two possibilities. One possibility is that the mutation site is important for DNA binding. The other is that mutations alter the structure of PmtR, resulting in the loss of DNA binding. In conclusion, the increased expression of PmtA-D is involved in high nisin A resistance. Since PmtA-D is responsible for the secretion of PSMs, it is speculated that PmtA-D excretes some antibacterial agents, including PSMs and human-derived AMPs externally.
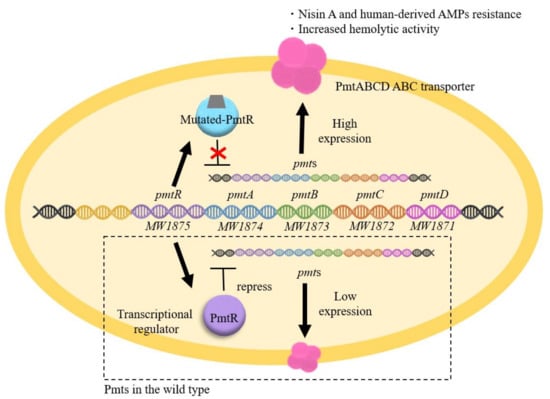
Figure 5.
Mechanism underlying the high nisin A resistance of the PmtR mutant. In the MW2 strain, PmtRABCD expression was low (lower side) because PmtR negatively regulates pmtRABCD expression. In the PmtR mutation strains, mutations in PmtR result in high expression of pmtRABCD (upper side). As a result, high nisin A resistance and an increase in haemolysis are observed.
5. Nisin Resistance Affects Virulence
In addition to having high nisin A resistance, the mutants with BraRS mutations were also found to be highly resistant to bacitracin and gallidermin derived from Bacillus subtilis and Streptococcus agalactiae, respectively. Since the BraRS-VraDE system was originally involved in the acquisition of resistance to low concentration of these bacteriocins, this acquisition of high resistance was considered to be due to the high expression of VraDE.
Unlike the BraRS mutation, the PmtR mutation in SAN2 caused increased pathogenicity. The SAN2 strain showed reduced susceptibility to the innate immune factors hBD3 and LL37 and high haemolytic activity [64]. In addition, in a mouse infection model, the SAN2 strain showed a higher survival rate than the wild-type MW2 strain [64].
In the wild-type MW2 strain, the PmtA-D proteins form an ABC transporter consisting of two membrane proteins (PmtA and C) and two ATPases (PmtB and D) [69]. This transporter is involved in the transport of PSMs and delta-hemolysin from the cytoplasm to the extracellular space [69,70]. PSM is involved in a wide range of pathogenic activities, such as epithelial colonization, biofilm formation, proinflammatory activity, cytolytic activity, and surface diffusion activity, leading to antibacterial effects [48,49,71]. In addition, Pmt transporters were reported to be associated with human-derived AMPs such as hBD3 and LL37 [72]. In the SAN2 strain, mutated PmtR loses its ability to bind to the target DNA region, resulting in a high expression of pmtRABCD, followed by enhanced PmtA-D function [64]. Although there are no clear structural similarities among PSMs, delta-haemolysin, hBD3 and LL37, it is speculated that PmtA-D may be involved in the export of these peptides. From these results, it is clear that the increased expression of pmtA-D affects not only the high resistance to antibacterial peptides but also the pathogenicity of S. aureus.
6. Conclusions
In general, bacteria acquire resistance against antibacterial agents via endogenous mutations and exogenous resistance genes. Several TCSs such as ApsRS, VraRS, and BraRS were demonstrated to be associated with the resistance to antibacterial AMPs, including human-derived AMPs, bacteriocins, and antibacterial chemotherapeutic agents. However, these TCS-mediated resistances are effective to low concentrations of AMPs. We and other laboratories demonstrated that S. aureus could acquire high nisin A resistance via endogenous mutations upon exposure to nisin A [63,64,67]. Since several AMPs including nisin A are used in food preservatives and are considered to be candidates for clinical use, the development of AMP resistance in S. aureus will continue to occur. It is possible that some portion of AMP-resistant strains become highly pathogenic. Therefore, highly pathogenic S. aureus strains are potentially present and may represent an “outbreak reserve”. Antibacterial chemotherapeutic agent abuse and secondary infection followed by unpredictable infectious diseases, such as COVID-19, may potentially enhance the occurrence of S. aureus infections. In such situations, we have to suppress the emergence of highly pathogenic S. aureus strains. Looking ahead, surveillance and research of endogenous resistant mutant strains of S. aureus will help suppress the emergence of potentially highly pathogenic strains.
Author Contributions
Conceptualization, M.K.-M. and H.K.; Methodology, M.K.-M. and H.K.; Validation, M.K.-M., H.K. and M.N.-T.L.; Formal Analysis, M.K.-M., H.K. and M.N.-T.L.; Investigation, M.K.-M.; Resources, Data Curation, Writing—Original Draft Preparation, M.K.-M.; Writing—Review and Editing, H.K. and M.N.-T.L.; Visualization, M.K.-M. and M.N.-T.L.; Supervision and Project Administration, H.K.; Funding Acquisition, M.K.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Grants-in-Aid for Scientific Research (C) (Grant No: 15K11017, 18K09553) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan. The APC were funded by Grants-in-Aid for Scientific Research (C) (Grant No: 21K09858) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
Institutional Review Board Statement
No applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Manders, S.M. Toxin-mediated streptococcal and staphylococcal disease. J. Am. Acad. Dermatol. 1998, 39, 383–400. [Google Scholar] [CrossRef]
- Foster, T.J. The Staphylococcus aureus “superbug”. J. Clin. Investig. 2004, 114, 1693–1696. [Google Scholar] [CrossRef] [PubMed]
- Diep, B.A.; Otto, M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008, 16, 361–369. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a Crisis For The Health and Wealth of Nations. London: Review on Antimicrobial Resistance. 2014. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 21 September 2021).
- McDanel, J.S.; Murphy, C.R.; Diekema, D.J.; Quan, V.; Kim, D.S.; Peterson, E.M.; Evans, K.D.; Tan, G.L.; Hayden, M.K.; Huang, S.S. Chlorhexidine and mupirocin susceptibilities of methicillin-resistant Staphylococcus aureus from colonized nursing home residents. Antimicrob. Agents Chemother. 2013, 57, 552–558. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wassenaar, T.M.; Ussery, D.; Nielsen, L.N.; Ingmer, H. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. 2015, 5, 44–61. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 1985, 76, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Lehrer, R.I. Defensins. Pharmacol. Ther. 1995, 66, 191–205. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Ganz, T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 1999, 11, 23–27. [Google Scholar] [CrossRef]
- Zaiou, M.; Gallo, R.L. Cathelicidins, essential gene-encoded mammalian antibiotics. J. Mol. Med. 2002, 80, 549–561. [Google Scholar] [CrossRef]
- Karczewski, J.; Brown, C.M.; Maezato, Y.; Krasucki, S.P.; Streatfield, S.J. Efficacy of a novel lantibiotic, CMB001, against MRSA. J. Antimicrob. Chemother. 2021, 76, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Walsh, L.; Johnson, C.N.; Hill, C.; Ross, R.P. Efficacy of Phage- and Bacteriocin-Based Therapies in Combatting Nosocomial MRSA Infections. Front. Mol. Biosci. 2021, 8, 654038. [Google Scholar] [CrossRef]
- Ouhara, K.; Komatsuzawa, H.; Kawai, T.; Nishi, H.; Fujiwara, T.; Fujiue, Y.; Kuwabara, M.; Sayama, K.; Hashimoto, K.; Sugai, M. Increased resistance to cationic antimicrobial peptide LL-37 in methicillin-resistant strains of Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 61, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Bendjeddou, K.; Hamma-Faradji, S.; Meddour, A.A.; Belguesmia, Y.; Cudennec, B.; Bendali, F.; Daube, G.; Taminiau, B.; Drider, D. Gut microbiota, body weight and histopathological examinations in experimental infection by methicillin-resistant Staphylococcus aureus: Antibiotic versus bacteriocin. Benef. Microbes 2021, 12, 295–305. [Google Scholar] [CrossRef]
- Mao, Y.; Hoffman, T.; Singh-Varma, A.; Duan-Arnold, Y.; Moorman, M.; Danilkovitch, A.; Kohn, J. Antimicrobial Peptides Secreted From Human Cryopreserved Viable Amniotic Membrane Contribute to its Antibacterial Activity. Sci. Rep. 2017, 7, 13722. [Google Scholar] [CrossRef] [PubMed]
- Nissen-Meyer, J.; Nes, I.F. Ribosomally synthesized antimicrobial peptides: Their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 1997, 167, 67–77. [Google Scholar] [CrossRef]
- Jack, R.W.; Tagg, J.R.; Ray, B. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 1995, 59, 171–200. [Google Scholar] [CrossRef]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef]
- Nagao, J.; Asaduzzaman, S.M.; Aso, Y.; Okuda, K.-I.; Nakayama, J.; Sonomoto, K. Lantibiotics: Insight and foresight for new paradigm. J. Biosci. Bioeng. 2006, 102, 139–149. [Google Scholar] [CrossRef]
- Nes, I.F.; Holo, H. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 2000, 55, 50–61. [Google Scholar] [CrossRef]
- Brötz, H.; Bierbaum, G.; Leopold, K.; Reynolds, P.E.; Sahl, H.G. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 1998, 42, 154–160. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Delves-Broughton, J.; Blackburn, P.; Evans, R.J.; Hugenholtz, J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 1996, 69, 193–202. [Google Scholar] [CrossRef]
- Jensen, C.; Li, H.; Vestergaard, M.; Dalsgaard, A.; Frees, D.; Leisner, J.J. Nisin Damages the Septal Membrane and Triggers DNA Condensation in Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 2020, 11, 1007. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical applications of nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef] [PubMed]
- Le Lay, C.; Dridi, L.; Bergeron, M.G.; Ouellette, M.; Fliss, I.L. Nisin is an effective inhibitor of Clostridium difficile vegetative cells and spore germination. J. Med. Microbiol. 2016, 65, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Udompijitkul, P.; Paredes-Sabja, D.; Sarker, M.R. Inhibitory effects of nisin against Clostridium perfringens food poisoning and nonfood-borne isolates. J. Food Sci. 2012, 77, M51–M56. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Ni, L.; Ling, J. Antibacterial peptide nisin: A potential role in the inhibition of oral pathogenic bacteria. Peptides 2014, 60, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef]
- Field, D.; O’ Connor, R.; Cotter, P.D.; Ross, R.P.; Hill, C. In Vitro Activities of Nisin and Nisin Derivatives Alone and In Combination with Antibiotics against Staphylococcus Biofilms. Front. Microbiol. 2016, 7, 508. [Google Scholar] [CrossRef]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef]
- Juturu, V.; Wu, J.C. Microbial production of bacteriocins: Latest research development and applications. Biotechnol. Adv. 2018, 36, 2187–2200. [Google Scholar] [CrossRef] [PubMed]
- Alves, F.C.B.; Albano, M.; Andrade, B.F.M.T.; Chechi, J.L.; Pereira, A.F.M.; Furlanetto, A.; Rall, V.L.M.; Fernandes, A.A.H.; Dos Santos, L.D.; Barbosa, L.N.; et al. Comparative Proteomics of Methicillin-Resistant Staphylococcus aureus Subjected to Synergistic Effects of the Lantibiotic Nisin and Oxacillin. Microb. Drug Resist. 2020, 26, 179–189. [Google Scholar] [CrossRef]
- Van Harten, R.M.; Willems, R.J.L.; Martin, N.I.; Hendrickx, A.P.A. Multidrug-Resistant Enterococcal Infections: New Compounds, Novel Antimicrobial Therapies? Trends Microbiol. 2017, 25, 467–479. [Google Scholar] [CrossRef]
- Li, M.; Lai, Y.; Villaruz, A.E.; Cha, D.J.; Sturdevant, D.E.; Otto, M. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 2007, 104, 9469–9474. [Google Scholar] [CrossRef]
- Matsuo, M.; Oogai, Y.; Kato, F.; Sugai, M.; Komatsuzawa, H. Growth-phase dependence of susceptibility to antimicrobial peptides in Staphylococcus aureus. Microbiology 2011, 157, 1786–1797. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Kuroda, H.; Oshima, T.; Takeuchi, F.; Mori, H.; Hiramatsu, K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 2003, 49, 807–821. [Google Scholar] [CrossRef]
- Belcheva, A.; Golemi-Kotra, D. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 2008, 283, 12354–12364. [Google Scholar] [CrossRef]
- Kawada-Matsuo, M.; Yoshida, Y.; Nakamura, N.; Komatsuzawa, H. Role of two-component systems in the resistance of Staphylococcus aureus to antibacterial agents. Virulence 2011, 2, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Kawada-Matsuo, M.; Yoshida, Y.; Zendo, T.; Nagao, J.; Oogai, Y.; Nakamura, Y.; Sonomoto, K.; Nakamura, N.; Komatsuzawa, H. Three Distinct Two-Component Systems Are Involved in Resistance to the Class I Bacteriocins, Nukacin ISK-1 and Nisin A, in Staphylococcus aureus. PLoS ONE 2013, 8, e69455. [Google Scholar] [CrossRef]
- Mitrophanov, A.Y.; Groisman, E.A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008, 22, 2601–2611. [Google Scholar] [CrossRef] [PubMed]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000, 3, 165–170. [Google Scholar] [CrossRef]
- Rampersaud, A.; Harlocker, S.L.; Inouye, M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J. Biol. Chem. 1994, 269, 12559–12566. [Google Scholar] [CrossRef]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Boisset, S.; Saveanu, C.; Benito, Y.; Geissmann, T.; Namane, A.; Lina, G.; Etienne, J.; Ehresmann, B.; Ehresmann, C.; et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005, 24, 824–835. [Google Scholar] [CrossRef]
- Geisinger, E.; Adhikari, R.P.; Jin, R.; Ross, H.F.; Novick, R.P. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006, 61, 1038–1048. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Joo, H.-S.; Chatterjee, S.S.; Otto, M. Phenol-soluble modulins--critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.-H.L.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef]
- Giraudo, A.T.; Raspanti, C.G.; Calzolari, A.; Nagel, R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 1994, 40, 677–681. [Google Scholar] [CrossRef]
- Voyich, J.M.; Vuong, C.; DeWald, M.; Nygaard, T.K.; Kocianova, S.; Griffith, S.; Jones, J.; Iverson, C.; Sturdevant, D.E.; Braughton, K.R.; et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 2009, 199, 1698–1706. [Google Scholar] [CrossRef]
- Baroja, M.L.; Herfst, C.A.; Kasper, K.J.; Xu, S.X.; Gillett, D.A.; Li, J.; Reid, G.; McCormick, J.K. The SaeRS Two-Component System Is a Direct and Dominant Transcriptional Activator of Toxic Shock Syndrome Toxin 1 in Staphylococcus aureus. J. Bacteriol. 2016, 198, 2732–2742. [Google Scholar] [CrossRef]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Cunliffe, R.N. Alpha-defensins in the gastrointestinal tract. Mol. Immunol. 2003, 40, 463–467. [Google Scholar] [CrossRef]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 2007, 66, 1136–1147. [Google Scholar] [CrossRef]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Götz, F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef]
- Peschel, A.; Jack, R.W.; Otto, M.; Collins, L.V.; Staubitz, P.; Nicholson, G.; Kalbacher, H.; Nieuwenhuizen, W.F.; Jung, G.; Tarkowski, A.; et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 2001, 193, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- McCallum, N.; Meier, P.S.; Heusser, R.; Berger-Bächi, B. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob. Agents Chemother. 2011, 55, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Matsuo, M.; Oogai, Y.; Kato, F.; Nakamura, N.; Sugai, M.; Komatsuzawa, H. Bacitracin sensing and resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 320, 33–39. [Google Scholar] [CrossRef]
- Hiron, A.; Falord, M.; Valle, J.; Débarbouillé, M.; Msadek, T. Bacitracin and nisin resistance in Staphylococcus aureus: A novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 2011, 81, 602–622. [Google Scholar] [CrossRef]
- Randall, C.P.; Gupta, A.; Utley-Drew, B.; Lee, S.Y.; Morrison-Williams, G.; O’Neill, A.J. Acquired Nisin Resistance in Staphylococcus aureus Involves Constitutive Activation of an Intrinsic Peptide Antibiotic Detoxification Module. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Arii, K.; Kawada-Matsuo, M.; Oogai, Y.; Noguchi, K.; Komatsuzawa, H. Single mutations in BraRS confer high resistance against nisin A in Staphylococcus aureus. Microbiologyopen 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Kawada-Matsuo, M.; Watanabe, A.; Arii, K.; Oogai, Y.; Noguchi, K.; Miyawaki, S.; Hayashi, T.; Komatsuzawa, H. Staphylococcus aureus Virulence Affected by an Alternative Nisin A Resistance Mechanism. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Khosa, S.; AlKhatib, Z.; Smits, S.H.J. NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 2013, 394, 1543–1549. [Google Scholar] [CrossRef]
- Toro-Roman, A.; Mack, T.R.; Stock, A.M. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: A symmetric dimer mediated by the alpha4-beta5-alpha5 face. J. Mol. Biol. 2005, 349, 11–26. [Google Scholar] [CrossRef]
- Blake, K.L.; Randall, C.P.; O’Neill, A.J. In vitro studies indicate a high resistance potential for the lantibiotic nisin in Staphylococcus aureus and define a genetic basis for nisin resistance. Antimicrob. Agents Chemother. 2011, 55, 2362–2368. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-S.; Chatterjee, S.S.; Villaruz, A.E.; Dickey, S.W.; Tan, V.Y.; Chen, Y.; Sturdevant, D.E.; Ricklefs, S.M.; Otto, M. Mechanism of Gene Regulation by a Staphylococcus aureus Toxin. MBio 2016, 7. [Google Scholar] [CrossRef]
- Chatterjee, S.S.; Joo, H.-S.; Duong, A.C.; Dieringer, T.D.; Tan, V.Y.; Song, Y.; Fischer, E.R.; Cheung, G.Y.C.; Li, M.; Otto, M. Essential Staphylococcus aureus toxin export system. Nat. Med. 2013, 19, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Duong, A.C.; Otto, M. Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: Implications for diagnosis and pathogenesis. Microbes Infect. 2012, 14, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Fisher, E.L.; McCausland, J.W.; Choi, J.; Collins, J.W.M.; Dickey, S.W.; Otto, M. Antimicrobial Peptide Resistance Mechanism Contributes to Staphylococcus aureus. Infection. J. Infect. Dis. 2018, 217, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).