Abstract
Bacterial fruit blotch (BFB) causes losses in melon marketable yield. However, until now, there has been no information about the genetic loci responsible for resistance to the disease or their pattern of inheritance. We determined the inheritance pattern of BFB resistance from a segregating population of 491 F2 individuals raised by crossing BFB-resistant (PI 353814) and susceptible (PI 614596) parental accessions. All F1 plants were resistant to Acidovorax citrulli strain KACC18782, and F2 plants segregated with a 3:1 ratio for resistant and susceptible phenotypes, respectively, in a seedling bioassay experiment, indicating that BFB resistance is controlled by a monogenic dominant gene. In an investigation of 57 putative disease-resistance related genes across the melon genome, only the MELO3C022157 gene (encoding TIR-NBS-LRR domain), showing polymorphism between resistant and susceptible parents, revealed as a good candidate for further investigation. Cloning, sequencing and quantitative RT-PCR expression of the polymorphic gene MELO3C022157 located on chromosome 9 revealed multiple insertion/deletions (InDels) and single nucleotide polymorphisms (SNPs), of which the SNP A2035T in the second exon of the gene caused loss of the LRR domain and truncated protein in the susceptible accession. The InDel marker MB157-2, based on the large (504 bp) insertion in the first intron of the susceptible accession, was able to distinguish resistant and susceptible accessions among 491 F2 and 22 landraces/inbred accessions with 98.17% and 100% detection accuracy, respectively. This novel PCR-based, co-dominant InDel marker represents a practical tool for marker-assisted breeding aimed at developing BFB-resistant melon accessions.
1. Introduction
Melon (Cucumis melo L., 2n = 2× = 24) belonging to the Cucurbitaceae family produce delicious, sweet, fruits rich in vitamins, minerals and health-promoting antioxidants [1]. They are an economically important agricultural crop cultivated and consumed all around the world, with an average production of more than 29 million tons per year throughout the past decade [2]. Melons are predominantly cultivated in temperate and tropical countries with all the major producing regions in the world (China, Turkey, Iran, India, Kazakhstan, USA, Egypt, Spain, Guatemala, Italy, etc.) (http://www.fao.org/faostat/en/#rankings/countries_by_commodity). Production is greatly hampered by several pathogens creating a barrier to attaining higher yields of this important fruit crop.
Bacterial fruit blotch (BFB), caused by the aerobic, Gram-negative, rod-shaped and seed-borne bacterium Acidovorax citrulli (formerly Acidovorax avenae subsp. citrulli) [3], is a destructive disease responsible for significant economic losses of melon crops worldwide. It is very difficult to estimate the exact economic losses caused by BFB; however, it can cause 80% to 100% loss in melon production under favorable environmental situations, especially during rainy seasons and highly fluctuating temperature regimes [4,5,6]. The disease also causes significant economic losses in other cultivated cucurbits, such as citron melon, prickly paddy melon, pumpkin, cucumber, squash, several types of gourds and watermelon, and has been reported all over the world, including the United States, Nicaragua, Costa Rica, Brazil, Australia, Turkey, Japan, Korea, Thailand, Taiwan and China [7,8,9,10,11,12,13]. In South Korea, BFB in melon was first reported in 1990, with several outbreaks observed in the last decade [14]. BFB was initially thought to be a severe threat mainly to watermelon. However, recent worldwide increases in BFB outbreaks on other cucurbits, especially melon, are believed to be due to changes in the population structure and worldwide spread of the pathogen [10,12,15,16,17,18].
Melon seedlings and fruits are highly susceptible to A. citrulli. Typical symptoms include water-soaked lesions on cotyledons, leaves and fruit that are often small and irregular and progress throughout the leaf and rind of the fruit, leading to collapse and death of the plant [19]. The disease constitutes a severe threat, particularly because it affects marketable yield, i.e., the fruits, as well as the plants [20].
As of yet, chemical and other cultural practices have not been fully effective in managing BFB. Melon growers rely on chemical treatments to control BFB, even though these show only partial success [15,21,22,23,24]. Host resistance represents the most cost-effective and environmentally friendly approach for BFB management [20]. There has been little research to screen sources of resistance to BFB in melons, although a screen of 332 Cucumis sp. accessions identified only four C. melo and one Cucumis ficifolius accessions resistant to BFB [25]. Furthermore, Bahar, et al. [26] reported four genotypes, 6401, BLB-B, EAD-B and ADIR, that were resistant in seedling transmission assays. The genetic control of BFB resistance in melons has not been extensively investigated and no molecular markers linked to resistance to BFB that can be used for breeding BFB-resistant melon cultivars via marker-assisted selection.
Plant resistance to disease is mainly manifested by R (resistance)-genes [27,28]. R-gene-mediated recognition of pathogen effectors activates a series of defense signaling cascades and stimulates pathogenesis-related gene expression, creating global, durable and broad-spectrum systemic acquired resistance in plants [29]. R-genes have been reported in several plant species, including Arabidopsis, cucumber and rice [30,31,32], and nucleotide binding site (NBS)-encoding genes throughout the genome have recently been identified in melons [33]. The largest class of known R-genes comprises those containing NBS and leucine-rich repeat domains (NBS-LRR proteins). The NBS-LRR (NL)-type R-genes are fundamental for plant responses to various pathogens, including bacteria, fungi, viruses and nematodes [30,32]. TIR-NBS-LRR (TNL) proteins containing a Toll-like domain, and CC-NBS-LRR (CNL) proteins characterized by a coiled-coil domain in the N-terminal portion represent a subset of NBS-LRR proteins that often function as R-genes in plant genomes [34].
Several NBS-LRR-type genes are linked to disease resistance in melons. For example, TNL genes are found in the vicinity of genetic loci controlling resistance to Fusarium wilt and papaya ringspot virus [35]. No such quantitative trait locus (QTL) or R-gene has been linked with BFB so far. The lack of confirmed molecular markers associated with BFB resistance hampers the transfer of resistance loci to commercial melon cultivars. In this study, we determined the mode of inheritance of BFB resistance and developed a molecular marker linked to BFB resistance in melons by identifying polymorphism in NBS-coding genes across the genome.
2. Materials and Methods
2.1. Plant Materials and Population Development
An F2 segregating population consisting of 491 plants was developed using melon genotypes PI 614596 and PI 353814. The parental accessions PI 614596 and PI 353814, reported to be susceptible and resistant, respectively, to BFB [18,25], were obtained from the U.S. National Plant Germplasm System (https://npgsweb.ars-grin.gov/gringlobal/search.aspx), U.S. Department of Agriculture, USA. The resistance and susceptibility of these accessions were confirmed using a controlled inoculated bioassay [18]. In addition, 22 melon accessions (Supplementary Table S1) were used for validation of the marker. All plants were raised in 32-cell trays containing artificial soil mix in a controlled plant growth chamber with 25 ± 2 °C, 16-h-day lengths and 440 μmol/m2/s light intensity at bench level for 3 weeks and then transferred to a greenhouse of the Horticulture Laboratory of Sunchon National University, South Korea.
2.2. Bacterial Culture and Inoculum Preparation
A. citrulli strain KACC18782 was obtained from the Korean Agricultural Culture Collection (KACC), South Korea. Bacteria were grown in King’s B (KB) media [3] supplemented with 100 μg mL−1 ampicillin for 36–48 h at 28 °C until formation of bacterial colonies. For all inoculations, a bacterial suspension was prepared by inundating culture plates with 5 mL of sterile, double-distilled water and gently scraping the surface of the KB media using a sterile, L-shaped rubber spreader to an optical density of 1.0 at 600 nm, as measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Bacterial suspension was diluted to a final concentration of ~1 × 106 colony forming units (mL−1).
2.3. Inoculation, Sample Preparation and Assessment of BFB Resistance
Plants with 3–5 true leaves (4–5 weeks old) were inoculated by spraying with a suspension of A. citrulli KACC18782 using a hand-pump spray bottle until runoff in a greenhouse with a temperature range of 20–25 °C. Inoculated plants were covered with a plastic cover to maintain high relative humidity (96%). Plants were re-inoculated 3 days after the first inoculation to ensure that no plants had escaped inoculation and to eliminate false positives. Samples BFB-resistant and susceptible plants were collected from the fourth and fifth true leaves stage of the plants at the time points of 12 h, 1 day, 3 days and 6 days after inoculation and from control plants at the same time points. The samples were immediately frozen in liquid nitrogen (N2). The disease severity of three individual leaves from each inoculated plant was scored 12 days after inoculation using a scale of 1 to 6 (Supplementary Figure S1) for ≥10%, 11–20%, 21–50%, 51–75%, 76–90% and >90% diseased/infected area per leaf, respectively [18]. Disease ratings were scored as per the methods described by Robin et al. [36] and Hassan et al. [37] with little modification. The percentage infected area (PIA) was measured as the ratio of infected area to total leaf area, multiplied by 100. Individuals with a PIA of ≤20 and >21 were considered as resistant and susceptible, respectively [18,37]. Supplementary Table S2 and the resulting ratio was tested for goodness-of-fit using χ2 analysis.
2.4. Selection of Putative R-Genes and Primer Design
A total of 57 NBS, LRR, TIR and CC domains containing putative disease resistance-related genes covering all 12 chromosomes of melons with particular emphasis on chromosomes that harbor disease and insect resistance-related loci were used for detection of length polymorphism between contrastingly resistant melon genotypes (Table 1). DNA coding sequences were retrieved from the melon (DHL92) genome v3.6.1 from the Cucurbit Genomics database (http://cucurbitgenomics.org/organism/3).

Table 1.
Details of 57 putative disease resistance-related genes and corresponding primers used for detecting length polymorphism between resistant and susceptible melon genotypes.
Moreover, for the quantitative RT-PCR expression analysis of polymorphic (MELO3C022157) gene-specific primers were designed F: CGAATTTGAGTGCTGTTCCA and R: CGAAGTTCTACTGTTGGGCG on the fourth exon. Primers (Table 1), one pair for shorter genes and multiple pairs for longer genes, were designed to cover the entire length of the chromosomes using the Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) web tool. Primer sequences were checked for potential self-dimer and hairpin formation using an oligo calculator (http://biotools.nubic.northwestern.edu/OligoCalc.html).
2.5. Extraction of Genomic DNA and Detection of DNA Polymorphism
Genomic DNA (gDNA) was extracted from fresh 3-week-old leaf samples of the two parental accessions (20 from each parent), 20 F1 plants, 491 F2 plants and 22 melon accessions using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The concentration of extracted gDNA was determined using a NanoDrop Spectrophotometer ND-100 (NanoDrop Technologies, Wilmington, DE, USA) and stored in a refrigerator at −20 °C for further use. PCR was performed in 20 μL reaction mixtures containing 1 μL gDNA (152 ng/μL); 1.0 µl (10 pmol) of each forward and reverse primer; 8 μL Prime Taq premix (2×) (GENETBIO Inc., Gwangmyaong, Korea) and 9 μL ultra-pure H2O. PCR was carried out in a thermo-cycler with initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s and elongation at 72 °C for 45 s and final elongation at 72 °C for 7 min. Electrophoresis of amplified PCR products was performed in 1.2% agarose gel stained with HiQ blue mango (20,000×) (bioD, Gwangmyaong, Korea) and visualized using ultraviolet light in a ENDURO™ GDS gel documentation system (New York, NY, USA).
2.6. Cloning and Sequencing of the Polymorphic Gene
The polymorphic gene MELO3C022157 was PCR-amplified from resistant (PI 353814) and susceptible (PI 614596) accession using Phusion® High-Fidelity DNA Polymerase (New England Biolabs, EVRY Cedex, France). Amplified DNA fragments were purified using a Promega DNA Purification kit (Promega, Madison, WI, USA). Cloning was performed using a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The universal primers M13F and M13RpUC were used to sequence cloned amplicons using an ABI3730XL sequencer (Macrogen Co., Seoul, Korea). Each forward and reverse sequence of resistant and susceptible melon accession was repeated three times to remove all uncertainties. Gene sequences of the resistant and susceptible accessions were aligned using CLUSTALW software (https://www.genome.jp/tools-bin/clustalw) to detect sequence variation.
2.7. Total RNA Extraction and cDNA Synthesis
Control and infected melon leaves were crushed to a powder in liquid nitrogen (N2), and 100 mg of each sample was subjected to total RNA extraction using an RNeasy Mini kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA with a SuperScript III First-Strand Synthesis System kit (Invitrogen, Gaithersburg, MD, USA).
2.8. Quantitative RT-PCR (qRT-PCR) Expression of Polymorphic Gene MELO3C022157
The expression patterns of the polymorphic-gene were analyzed by qRT-PCR in a LightCycler® instrument (Roche, Mannheim, Germany) following the manufacturer’s instructions. The reactions were performed in a 10-µL volume consisting of 5 µL of 2x qPCRBIO SyGreen Mix Lo-ROX (PCR Biosystems, London, UK); 5 pmol of primers and cDNA templates diluted to the appropriate concentrations. The PCR conditions were as follows: 5 min at 95 °C, followed by 3-step amplifications at 95 °C for 15 s, 56 °C for 15 s and 72 °C for 20 s, for 45 cycles. The mean expression level of relevant genes was calculated by the 2−ΔΔ Ct method [38], where Actin was used as an internal control [39].
2.9. Statistical Analysis
To determine significant changes in gene expression levels among the different time points for each treatment, a chi-square (χ2) test for goodness-of-fit was performed to determine deviations of observed data from expected segregation ratios using Minitab18 statistical software (State College, PA, USA). A value of p < 0.05 was considered statistically significant. Turkey’s pairwise comparison test was conducted for mean separation.
3. Results
3.1. Dominant Inheritance of BFB Resistance in Melon
To determine the inheritance pattern of BFB resistance in melon, we crossed a BFB-resistant (PI 353814) and a BFB-susceptible (PI 614596) parental accession for making F1. The resistant parental accessions PI 353814 had less than 20% BFB-infected leaf area, while more than 70% of the leaves of susceptible parental accessions PI 614596 were infected by A. citrulli (KACC18782) (Figure 1). Symptoms of the leaves of F1 hybrids resembled those of the resistant parent, indicating the dominant nature of the inheritance of this trait in melons. We next evaluated the inheritance of BFB resistance in the F2 population consisting of 491 plants.
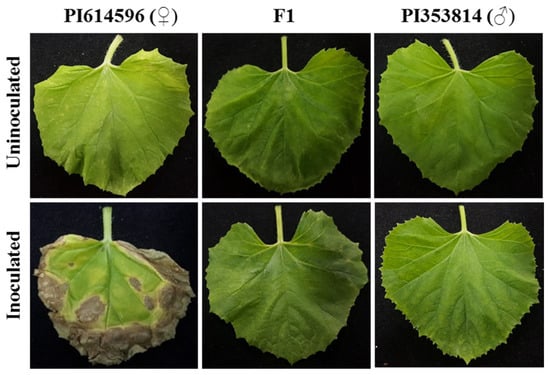
Figure 1.
Bacterial fruit blotch phenotypes on leaves of the two parental melon accessions, PI 353814 (resistant) and PI 614596 (susceptible), and their F1 hybrid (resistant) 12 days after inoculation with A. citrulli strain KACC18782, as compared to uninoculated controls. All leaves were detached just before the photographs were taken.
Bioassay results indicated that 360 F2 plants were resistant, and 131 were susceptible. A chi-square (χ2) test revealed that BFB resistance segregated with a 3:1 ratio (resistant: susceptible), indicating monogenic dominant control of the trait (Table 2 and Supplementary Figures S2 and S3).

Table 2.
Inheritance pattern of bacterial fruit blotch (BFB) resistance in melons (Cucumis melo).
3.2. Microsynteny Analysis of 57 R-Genes of Melon Compared with Watermelon and Cucumber
We performed comparative analysis to identify the homologous R-genes among melon, compared with watermelon and cucumber. Some R-genes from melons share homologous relationships with those of watermelon and cucumber. However, only four chromosome genes of melons (chr.01, chr.03, chr.05 and chr.09) share homologous relationships with those of watermelon, while remaining chromosome genes are not homologous. Moreover, genes of melon on chr.06, chr.07 and chr.12 lack homologues in watermelon, whereas other chromosome genes are homologous (Figure 2).
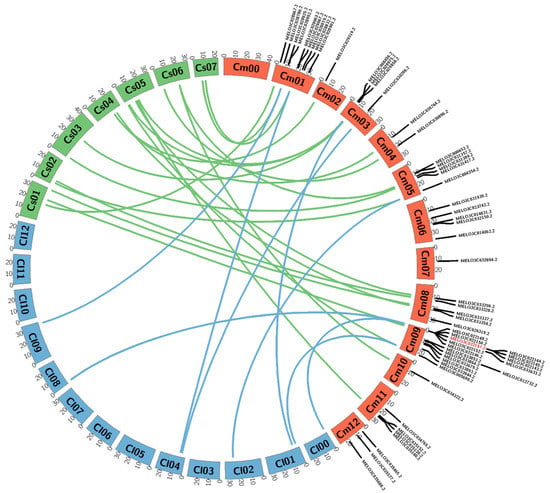
Figure 2.
Microsynteny analysis of melon R-genes compared with watermelon and cucumber. Brown orange, blue and green indicate melon, watermelon and cucumber chromosomes, respectively. Red marking indicates genes showing polymorphism between the resistant (PI 353814) and susceptible (PI 614596) parents of the melon. Microsynteny analysis of genes on the melon chromosomes were drawn using the web-based tool Circos (http://circos.ca/software/download/) circos-0.69-9.tgz.
3.3. Identification of Length Polymorphisms in Putative R-genes
It is essential to identify BFB-resistant genotypes to develop BFB-resistant melon cultivars. Since no definitive R-gene is known to be associated with BFB resistance in melon, we identified polymorphisms in putative R-genes with NBS, LRR, TIR and CC domains throughout the genome based on a functional annotation of sequences in the Cucurbit Genomics database (http://cucurbitgenomics.org/search/genome/3).
We selected 57 NBS-encoding genes covering all chromosomes of melon, with a maximum 17 and 9 genes on chromosomes 9 and 1, respectively (Table 1). Among these 57 genes, 44 were NBS-LRR (NL) type, while 17 and 2 were TIR-NBS-LRR (TNL) and TIR-NBS (TN) types, respectively. Only one gene each was selected from the NBS and CC-NBS-LRR (CNL) types. We designed gene-specific primers (one set for shorter genes and multiple sets for longer genes) covering the entire length of these genes for PCR amplification and detection of length polymorphisms between resistant and susceptible accessions. Among these genes, conspicuous length polymorphism was detected only for the disease resistance-related TNL-type gene MELO3C022157 (Supplementary Figure S4).
3.4. Cloning and Sequencing of the Polymorphic Gene
We divided the polymorphic gene MELO3C022157 into six consecutive fragments amplified by six sets of primers (Table 3) to narrow down the specific polymorphic region. The polymorphic region was identified in the first intron of the gene ( Figure 3; Figure 4b). We cloned and sequenced the six MELO3C022157 gene fragments from both resistant and susceptible accessions, revealing insertion of a 504 bp fragment in the first intron of the susceptible parent (Figure 4c).

Table 3.
List of primers designed along the entire length of gene MELO3C022157 to narrow down the position of length polymorphism and to clone the gene.
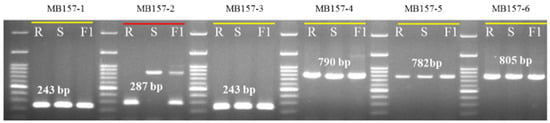
Figure 3.
Banding patterns of six pairs of primers designed against the gene MELO3C022157. Polymorphic primer MB157-2 is marked with red underline. R—resistant parent PI 353814, S—susceptible parent PI 614596 and F1—their F1 hybrid. Primer details are given in Table 3.
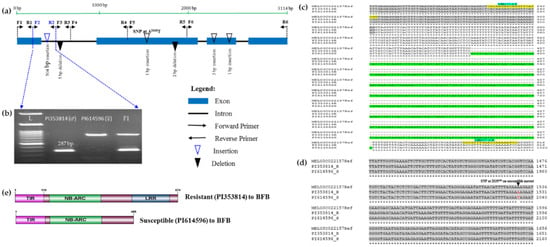
Figure 4.
Polymorphism in the TIR-NBS-LRR gene MELO3022157 linked with bacterial fruit blotch (BFB) resistance in melon. (a) Idiogram of gene MELO3022157 showing the positions of the six sets of primers used (Table 3) and of insertion/deletion (InDel) and SNP polymorphisms between the resistant (PI 353814) and susceptible (PI 614596) parental accessions. (b) Polymorphic PCR amplicons generated by the MB157-2 primer pair in resistant and susceptible parents and their F1 progeny. (c) Sequence alignment showing a long insertion (green highlighted region) in the susceptible parent. Grey shaded regions indicate exons and yellow highlighted segments indicate MB157-2-F and MB157-2-R primer sequences with primer names marked by blue highlights. (d) SNP at the 2035th bp causing a frameshift mutation marker is indicated in red (Supplementary Figure S6b). (e) Loss of LRR domain in the susceptible accession. The complete alignment is shown in Supplementary Figure S6b.
In addition, we detected a 5 bp deletion in the 1st. intron of the resistant accession; a 4 bp deletion in the 2nd. exon of the susceptible accession; three insertions: 1 bp in the 2nd. exon and 3 and 1 bp in the 3rd. exon of the susceptible accession and several single nucleotide polymorphisms (SNPs: “A/G, A/T, A/G and A/T), all ”A” in the resistant parent at positions 1355, 1526, 2454 and 2893 in the 2nd., 2nd., 3rd. and 4th. exons, respectively, and ”G”,”T”, ”G” and ”T” in the susceptible parent at positions 1864, 2035, 2959 and 3398 in the 2nd., 2nd., 3rd. and 4th. exons, respectively, and ”G”,”T” and ”G” in the susceptible parent at positions 1864, 2035 and 2959 in the 2nd., 2nd. and 3rd. exons, respectively (Figure 4a and Supplementary Figure S6b). The SNP ”A/T” (”A” and ”T” in resistant and susceptible parents at positions 1526 and 2035, respectively) generates a premature stop codon in the susceptible parent that produces a truncated protein (Figure 4d, Supplementary Figure S6b,c). In silico domain analysis of the translated protein sequences of the cloned gene of resistant and susceptible accessions using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence-search) revealed that this SNP causes loss of the LRR domain in the susceptible accessions PI 614596, whereas this is present in the resistant accessions PI 353814 (Figure 4e).
3.5. qRT-PCR Expression Analysis of Polymorphic Gene MELO3C022157
The expression of the polymorphic gene via qRT-PCR gradually increased in the leaf tissue of the resistant (PI 353814), as compared to the susceptible (PI 614596) parent at different time points. The gene was induced within 12 h of infection by A. citrulli in both the resistant and susceptible parents. In general, the transcript level in the resistant parent was higher at 12 h, 1 d, 3 d and 6 d after inoculation followed by a decrease in expression (Figure 5) and, after 6d, expression level was very low in the susceptible parent. The gene expression levels revealed a 5-fold peak 6d after inoculation in the resistant parent, whereas transcript levels were lowest in the susceptible parent at this time point.
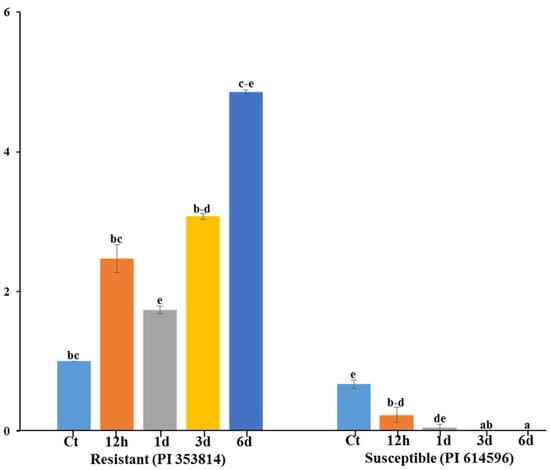
Figure 5.
Relative expression levels of putative candidate R-gene (MELO3022157) in A. citrulli-resistant and susceptible melon accession. Error bars represent (± SE) of three individual observations. Different letters above the bars indicate significant differences. Ct—control, h—hour and d—day.
3.6. Development and Validation of InDel Marker Linked to BFB Resistance in Melon
Among the polymorphisms, we targeted the long insertion/deletion (InDel) mutation between resistant and susceptible accessions and designed a set of primers (MB157-2-F/R) around this mutation (Table 3) by producing a 287 bp amplicon for the resistant accessions, a 791 bp amplicon for the susceptible accessions and both bands for the heterozygous F1 hybrid (Figure 4b). To validate the efficiency of the marker, we genotyped 491 F2 individuals generated from the resistant and susceptible accessions. The genotypes of 482 individuals matched those from the bioassay results, indicating a detection accuracy of about 98.17% (Figure 6 and Supplementary Table S2). We then used the marker to genotype 22 landraces/inbred melon accessions collected from various sources (Supplementary Table S1), of which we detected two accessions, PI 140471 and PI 420145, as resistant (Figure S5b). A bioassay test of these 22 landraces/inbred accessions indicated that these two accessions were indeed resistant to BFB (Figure S5a). These results suggest that the InDel MB157-2 marker can be effectively used for detecting resistant and susceptible melon genotypes using a PCR-based assay.
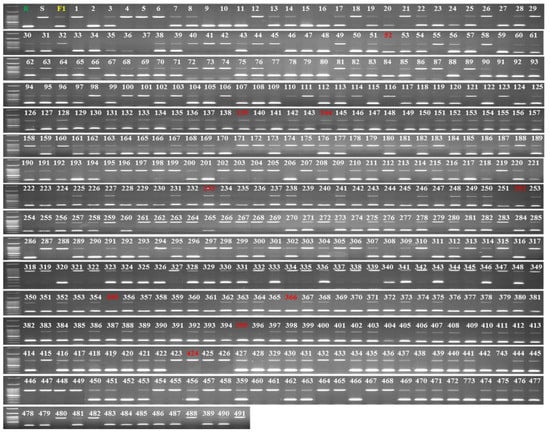
Figure 6.
Validation of the InDel marker MB175-2 in 491 plants of an F2 population raised from resistant and susceptible parental accessions PI 353814 and PI 614596, respectively. Red numbers indicate accessions with a mismatch between phenotypic and genotypic results.
4. Discussion
Developing BFB-resistant melon accessions is essential, since chemical control measures and other cultural practices are only partially effective, increase the cost of production, are not sustainable and are hazardous to the environment [40]. Even though BFB causes extensive damage, there has been no genetic information about any causal loci or genes controlling the resistants or their pattern of inheritance. Here, we determined the inheritance of BFB disease in melon and developed an InDel marker for distinguishing resistant and susceptible melon genotypes through a PCR assay.
Field evaluation after inoculation with A. citrulli strain KACC18782 indicated that seedlings of all F1 plants from the cross PI 614596 × PI 353814 were resistant, demonstrating the completely dominant nature of the trait in melon (Figure 1). Similar evaluations of 491 F2 individuals showed a Mendelian segregation phenotypic ratio of 3:1 (resistant:susceptible) and the genotypic ratio of 1:2:1 (homozygous (resistant):heterozygous (resistant):homozygous (susceptible)), indicating monogenic control of resistance to the disease (Table 2 and Supplementary Figures S2 and S3).
We then focused on developing molecular markers to detect resistant vs. susceptible genotypes by a simple PCR assay. However, since no QTL or functional gene has been reported for BFB resistance in melon, we examined putative R-genes genome-wide.
Functional analysis has revealed that plant disease resistance genes (R-genes) mostly encode proteins containing NBS, LRR, TIR, CC and RLK domains [27,28,41], which play roles in defense against various phyto-pathogens, such as bacteria, viruses and fungi [34,42]. For example, NBS-LRR genes control resistance to Fusarium oxysporum races 0 and 2, papaya ringspot virus [43] and powdery mildew [44]; a CNL gene controls Aphis gossypii-mediated virus resistance [45] and a TNL gene controls resistance against gummy stem blight disease [37] in melon. The NBS domain acts as a molecular switch of the plant defense mechanism, LRR domains are responsible for pathogen recognition and TIR and kinase domains play roles in defense signaling [46,47,48]. Such R-genes have been identified throughout the genomes of various crop species. such as rice, Arabidopsis, potato, soybean, maize, rapeseed and cabbage [49,50,51,52,53], including different cucurbits, such as cucumber, bottle gourd, luffa, watermelon and squash [30,32,54]. Several studies have identified NBS-encoding R-genes genome-wide in melon [33,55,56].
We investigated length polymorphism in 57 putative disease resistance genes encoding proteins containing NBS, LRR, TIR and CC domains and covering all 12 chromosomes of melon, with particular emphasis on chromosomes and regions harboring disease and insect resistance-related loci. Conspicuous length polymorphism between resistant and susceptible melon accessions was only observed for the TIR-NBS-LRR gene MELO3C022157 (Table 1 and Supplementary Figure S4). Besides, the presence of homologues of this R-gene in watermelon and cucumber (Figure 3) indicates that this gene is likely to play a similar role in melon [35,37]. After cloning and sequencing of the entire polymorphic gene (resistant and susceptible parent) revealed a long insertion (504 bp) in the first intron in the susceptible parent and several other small InDels and SNPs between the resistant and susceptible parents (Figure 4c and Supplementary Figure S6b). Of these, the SNP A2035T in the second exon caused loss of the LRR domain, resulting in a truncated protein in the susceptible accession (Figure 4e and Supplementary Figure S6c). So, the polymorphic gene belongs to the LRR domain; qRT-PCR transcription level is higher at 6d after inoculation in resistants, whilst loss of LRR and transcription level is little at 6d in the susceptible (Figure 4e and Figure 5), indicating that 6d after inoculation in the susceptible parent fails to develop a hypersensitive response, i.e., causing BFB disease, suggesting that the LRR domain may be responsible for the susceptible reaction of the mutant accession. LRR domains are involved in resistance against a yellow strain of cucumber mosaic virus (CMV[Y]) in Arabidopsis [57,58], which strongly support our results. LRR domains also implicate LepR3 loci mediate resistance against blackleg in Brassica napus [59] and against Fusarium wilt (Fom2) in melon [57,58]. Since LRR domains provide specificity in pathogen recognition [48,60], the NBS-LRR class of plant innate immune receptors uses its LRR domain to accomplish many other roles [61].
In developing a molecular marker, we targeted the 504 bp InDel region in the first intron of MELO3C022157 and designed primer pair MB157-2-F/R, which identified resistant and susceptible accessions among 491 F2 individuals with 98.17% detection accuracy and those representing 22 landraces/inbred accessions with 100% detection accuracy (Table 3, Supplementary Tables S1 and S2 and Figure 6 and Supplementary Figure S5b). Introns play diverse roles in processes such as transcription coupling [62], gene expression regulation, formation of noncoding RNA [63,64] and exon shuffling and alternative splicing [65]. In addition, intronic polymorphism is a valuable source for developing genetic markers with high interspecies transferability [66,67], since introns are under much lower selective pressure than other functional elements such as exons [68]. Several markers have been developed using intronic polymorphism for a range of traits in different crops, including cabbage, melon, maize and sunflower [37,66,69,70]. Notably, a pair of InDel markers located in the same first intron of MELO3C022157 are reportedly linked to gummy stem blight resistance in melon [37].
5. Conclusions
We determined that BFB resistance in melon is controlled by a single dominant gene, allowing the development of the PCR-based co-dominant InDel marker MB157-2 for identifying resistant and susceptible accessions. The co-dominant InDel marker MB157-2 developed in this study will have practical implications for marker-assisted breeding for improving BFB resistance in melons. This is the first report, to our knowledge, of a molecular marker linked to BFB resistance in melons. Work is underway to map the resistant loci using partial genome sequence-based approaches.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/2/220/s1; Table S1: Details of 22 melon accessions used and their disease response, as determined by bioassay and PCR-based assay using polymorphic InDel marker MB157-2. Table S2: Comparison of Acidovorax citrulli bioassay results and InDel marker BM157-2 bands for the putative R-gene MELO3C022157 in a population of 491 F2 melon individuals raised from resistant and susceptible parental accessions PI 353814 and PI 614596, respectively. ”+”, ”ᴎ” and ”-“ indicate resistant, heterozygous and susceptible bands for the InDel marker BM157-2. Figure S1: Disease scores used to assess the severity of bacterial fruit blotch (BFB) caused by A. citrulli at 12 days after inoculation. Scores range from 1 to 6, with 1 representing leaves with no damage and 6 representing maximum damage. All leaves were detached just before photographs were taken. Figure S2: Disease symptoms on resistant (R) and susceptible (S) melon parental accessions PI 353814 and PI 614596, respectively, their F1 hybrid and 491 individuals of the F2 population at 12 days after inoculation with A. citrulli. All leaves were detached just before photographs were taken. Figure S3: Frequency distribution of BFB scores in melon P1, P2, F1 and F2 populations. Figure S4: Detection of length polymorphism in 57 putative disease resistance genes containing NBS, LRR, CC and TIR domains by PCR-based assay. Details of the genes and corresponding primer specifications are presented in Table 1. Genes 40-2 and 57 were not amplified. Gene 41 (red underline) showed conspicuous length polymorphism. R—resistant parent PI 353814, S—susceptible parent PI 614596 and F1—hybrid (PI 614596 × PI 353814). Figure S5: (a) Bacterial fruit blotch (BFB) disease symptoms on 22 melon accessions at 12 days after inoculation with A. citrulli. Details of these 22 melon accessions are shown in Supplementary Table S1. All leaves were detached just before photographs were taken. (b) Validation of resistance status to BFB using InDel marker MB157-2 in the 22 melon accessions. R—resistant (PI 353814) and S—susceptible (PI 614596) used as control. Figure S6: Sequences and alignments of cloned and sequenced TIR-NBS-LRR gene MELO3C022157 from resistant (PI353814) and susceptible (PI614596) melon accessions. (a) Genomic sequences of reference, resistant and susceptible accessions. (b) Their alignment with reference sequences retrieved from the Cucurbit Genomics database (http://cucurbitgenomics.org) considering DHL92 as the reference genome. In the sequence alignment, asterisks (*) indicate sequence similarity and absence of asterisks indicates sequence dissimilarity; em-dash (—) and green color designate insertion/deletion of nucleotides. In the gDNA sequences, grey highlights illustrate the position of exons, yellow highlights indicate InDel primer (MB157-2-F and MB157-2-R) sequences and light blue indicates the primer name and InDel position. (c) Translated protein sequences of reference gene, resistant and susceptible parent sequences determined using the “Translate Tool-ExPASy” (https://web.expasy.org/translate/) web-based database and showing amino acid alterations and truncated protein in the susceptible parent.
Author Contributions
M.R.I. designed the study, conducted the entire experiment and DNA extraction, performed wet lab experiments, analyzed the data, interpreted the results and wrote the first draft of the manuscript. D.M.I.J. helped with in silico analysis. H.-J.J. assisted in bioassays and PCR assays. M.R.H. comprehensively revised and finalized the manuscript. I.-S.N., H.-T.K. and J.-I.P. conceived and supervised the project. All authors read and approved the final draft of the manuscript.
Funding
This study was supported by the Golden Seed Project (grant no. 213007-05-4-CG100) of the Ministry of Agriculture, Food and Rural Affairs (MAFRA), Republic of Korea.
Acknowledgments
We thank the U.S. National Plant Germplasm System, USA and Korean Agricultural Culture Collection, Korea, as well as the National Institute of Horticultural and Herbal Science, Korea, for providing melon seeds and bacterial strains.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Lester, G.E. Antioxidant, Sugar, Mineral, and Phytonutrient Concentrations across Edible Fruit Tissues of Orange-Fleshed Honeydew Melon (Cucumis melo L.). J. Agric. Food Chem. 2008, 56, 3694–3698. [Google Scholar] [CrossRef] [PubMed]
- FAOstat. Agriculture Organization of the United Nations Statistics Division. Economic and Social Development Department, Rome, Italy. Available online: http://faostat3.fao.org/home/E (accessed on 24 December 2019).
- Schaad, N.W.; Postnikova, E.; Sechler, A.; Claflin, L.E.; Vidaver, A.K.; Jones, J.B.; Agarkova, I.; Ignatov, A.; Dickstein, E.; Ramundo, B.A. Reclassification of subspecies of Acidovorax avenae as A. Avenae (Manns 1905) emend., A. cattleyae (Pavarino, 1911) comb. nov., A. citrulli Schaad et al., 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 2008, 31, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Sales Júnior, R.; Menezes, J. Mapeamento das doenças fúngicas, bacterianas e viróticas do cultivo do melão no Estado do RN. Mossoró. Escola Superior de Agricultura de Mossoró 2001. [Google Scholar]
- Conceição, C.S.; Felix, K.C.S.; Mariano, R.L.; Medeiros, E.V.; Souza, E.B. Combined effect of yeast and silicon on the control of bacterial fruit blotch in melon. Sci. Hortic. 2014, 174, 164–170. [Google Scholar] [CrossRef]
- De Melo, E.A.; Rosa de Lima, R.M.; Laranjeira, D.; dos Santos, L.A.; de Omena Gusmão, L.; de Souza, E.B. Efficacy of yeast in the biocontrol of bacterial fruit blotch in melon plants. Trop. Plant Pathol. 2015, 40, 56–64. [Google Scholar] [CrossRef]
- Isakeit, T.; Black, M.; Barnes, L.; Jones, J. First report of infection of honeydew with Acidovorax avenae subsp. citrulli. Plant Dis. 1997, 81, 694. [Google Scholar] [CrossRef]
- Isakeit, T.; Black, M.; Jones, J. Natural infection of citronmelon with Acidovorax avenae subsp. citrulli. Plant Dis. 1998, 82, 351. [Google Scholar] [CrossRef]
- Langston, D., Jr.; Walcott, R.; Gitaitis, R.; Sanders, F., Jr. First report of a fruit rot of pumpkin caused by Acidivorax avenae subsp. citrulli in Georgia. Plant Dis. 1999, 83, 199. [Google Scholar] [CrossRef]
- O’brien, R.; Martin, H. Bacterial blotch of melons caused by strains of Acidovorax avenae subsp. citrulli. Aust. J. Exp. Agric. 1999, 39, 479–485. [Google Scholar]
- Schaad, N.; Postnikova, E.; Randhawa, P. Emergence of Acidovorax avenae Subsp. citrulli as a Crop Threatening Disease of Watermelon and Melon. In Pseudomonas Syringae and Related Pathogens; Springer: Berlin/Heidelberg, Germany, 2003; pp. 573–581. [Google Scholar]
- Walcott, R.; Fessehaie, A.; Castro, A. Differences in pathogenicity between two genetically distinct groups of Acidovorax avenae subsp. citrulli on cucurbit hosts. J. Phytopathol. 2004, 152, 277–285. [Google Scholar] [CrossRef]
- Martin, G.B. Functional analysis of plant disease resistance genes and their downstream effectors. Curr. Opin. Plant. Biol. 1999, 2, 273–279. [Google Scholar] [CrossRef]
- Noh, J.; Kim, J.-H.; Lim, J.H.; Kim, T.B.; Seong, M.H.; Jung, G.T.; Kim, J.M.; Cheong, S.-S.; Oh, N.K.; Lee, W.-H. Occurrence of diseases and case of clinical diagnosis on watermelon in South Korea, 2008–2012. Res. Plant Dis. 2014, 20, 8–14. [Google Scholar] [CrossRef][Green Version]
- Schaad, N.W.; Sowell, G., Jr.; Goth, R.; Colwell, R.; Webb, R. Pseudomonas pseudoalcaligenes subsp. citrulli subsp.nov. Int. J. Syst. Evol. Microbiol. 1978, 28, 117–125. [Google Scholar]
- Somodi, G.C.; Jones, J.; Hopkins, D.; Stall, R.; Kucharek, T.; Hodge, N.; Watterson, J. Occurrence of a bacterial watermelon fruit blotch in Florida. Plant Dis. 1991, 75, 1053–1056. [Google Scholar] [CrossRef]
- Walcott, R.; Gitaitis, R. Detection of Acidovorax avenae subsp. citrulli in watermelon seed using immunomagnetic separation and the polymerase chain reaction. Plant Dis. 2000, 84, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Hossain, M.R.; Kim, H.-T.; Nath, U.K.; Abuyusuf, M.; Jung, H.-J.; Park, J.-I.; Nou, I.-S. Molecular characterization of Acidovorax citrulli strain NIHHS15-280 causing bacterial fruit blotch disease in Korea and screening of resistance sources in melon. Hortic. Environ. Biotechnol. 2019, 62, 115–126. [Google Scholar] [CrossRef]
- Carvalho, F.C.; Santos, L.A.; Dias, R.C.; Mariano, R.L.; Souza, E.B. Selection of watermelon genotypes for resistance to bacterial fruit blotch. Euphytica 2013, 190, 169–180. [Google Scholar] [CrossRef]
- Burdman, S.; Walcott, R. Acidovorax citrulli: Generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 2012, 13, 805–815. [Google Scholar] [CrossRef]
- Hopkins, D. The Hypothetical Exam Question Becomes Reality. Plant Dis. 1995, 79, 761–765. [Google Scholar]
- Hopkins, D.L.; Thompson, C.M.; Hilgren, J.; Lovic, B. Wet seed treatment with peroxyacetic acid for the control of bacterial fruit blotch and other seedborne diseases of watermelon. Plant Dis. 2003, 87, 1495–1499. [Google Scholar] [CrossRef]
- Stephens, D.; Schneider, R.; Walcott, R.; Johnson, C. A Procedure, Based on Exposure to Chlorine Gas, for Disinfesting Watermelon Seeds. Phytopathology 2018, S150–S151. [Google Scholar]
- Feng, J.; Li, J.; Randhawa, P.; Bonde, M.; Schaad, N.W. Evaluation of seed treatments for the eradication of Acidovorax avenae subsp. citrulli from melon and watermelon seeds. Can J. Plant Pathol 2009, 31, 180–185. [Google Scholar] [CrossRef]
- Wechter, W.P.; Levi, A.; Ling, K.-S.; Kousik, C.; Block, C.C. Identification of resistance to Acidovorax avenae subsp. citrulli among melon (Cucumis spp.) plant introductions. HortScience 2011, 46, 207–212. [Google Scholar] [CrossRef]
- Bahar, O.; Kritzman, G.; Burdman, S. Bacterial fruit blotch of melon: Screens for disease tolerance and role of seed transmission in pathogenicity. Eur. J. Plant Pathol. 2009, 123, 71–83. [Google Scholar] [CrossRef]
- Ellis, J.; Dodds, P.; Pryor, T. Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 2000, 3, 278–284. [Google Scholar] [CrossRef]
- Harris, C.J.; Slootweg, E.J.; Goverse, A.; Baulcombe, D.C. Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. USA 2013, 110, 21189–21194. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Monosi, B.; Wisser, R.; Pennill, L.; Hulbert, S. Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet 2004, 109, 1434–1447. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Bo, K.; Shen, J.; Pang, X.; Chen, J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013, 14, 109. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.; Laidò, G.; De Leonardis, A.; Mastrangelo, A. Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. Int. J. Mol. Sci. 2013, 14, 7302–7326. [Google Scholar] [CrossRef] [PubMed]
- Morata, J.; Puigdomènech, P. Variability among Cucurbitaceae species (melon, cucumber and watermelon) in a genomic region containing a cluster of NBS-LRR genes. BMC Genom. 2017, 18, 138. [Google Scholar] [CrossRef] [PubMed]
- Robin, A.H.K.; Larkan, N.J.; Laila, R.; Park, J.-I.; Ahmed, N.U.; Borhan, H.; Parkin, I.A.; Nou, I.-S. Korean Brassica oleracea germplasm offers a novel source of qualitative resistance to blackleg disease. Eur. J. Plant Pathol. 2017, 149, 611–623. [Google Scholar] [CrossRef]
- Hassan, M.; Rahim, M.; Natarajan, S.; Robin, A.; Kim, H.-T.; Park, J.-I.; Nou, I.-S. Gummy Stem Blight Resistance in Melon: Inheritance Pattern and Development of Molecular Markers. Int. J. Mol. Sci. 2018, 19, 2914. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Gao, L.; Cao, L.; Liu, Y.; Saba, H.; Huang, Y.; Bie, Z. Assessment of suitable reference genes for quantitative gene expression studies in melon fruits. Front. Plant Sci. 2016, 7, 1178. [Google Scholar] [CrossRef]
- Wolukau, J.N.; Zhou, X.; Chen, J. Identification of amplified fragment length polymorphism markers linked to gummy stem blight (Didymella bryoniae) resistance in melon (Cucumis melo L.) PI 420145. HortScience 2009, 44, 32–34. [Google Scholar] [CrossRef]
- Kourelis, J.; van der Hoorn, R.A. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Upadhyaya, C.P.; Nookaraju, A.; Pandey, S.K.; Park, S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012, 78, 51–65. [Google Scholar] [CrossRef]
- Brotman, Y.; Normantovich, M.; Goldenberg, Z.; Zvirin, Z.; Kovalski, I.; Stovbun, N.; Doniger, T.; Bolger, A.M.; Troadec, C.; Bendahmane, A. Dual resistance of melon to Fusarium oxysporum races 0 and 2 and to Papaya ring-spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol. Plant 2013, 6, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Yuste-Lisbona, F.J.; Capel, C.; Gómez-Guillamón, M.L.; Capel, J.; López-Sesé, A.I.; Lozano, R. Codominant PCR-based markers and candidate genes for powdery mildew resistance in melon (Cucumis melo L.). Theor. Appl. Genet 2011, 122, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Dogimont, C.; Chovelon, V.; Pauquet, J.; Boualem, A.; Bendahmane, A. The V at locus encodes for a CC--NBS--LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii—mediated virus resistance. Plant J. 2014, 80, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef]
- Van Ooijen, G.; Mayr, G.; Kasiem, M.M.; Albrecht, M.; Cornelissen, B.J.; Takken, F.L. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef]
- Eitas, T.K.; Dangl, J.L. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010, 13, 472–477. [Google Scholar] [CrossRef]
- Martin, G.B.; Bogdanove, A.J.; Sessa, G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 2003, 54, 23–61. [Google Scholar] [CrossRef]
- Zhou, T.; Wang, Y.; Chen, J.-Q.; Araki, H.; Jing, Z.; Jiang, K.; Shen, J.; Tian, D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genom. 2004, 271, 402–415. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, X.; Jiang, H.; Ma, W.; Miao, W.; Yamada, T.; Zhang, M. Systematic analysis and comparison of nucleotide--binding site disease resistance genes in maize. FEBS J. 2012, 279, 2431–2443. [Google Scholar] [CrossRef]
- Kang, Y.J.; Kim, K.H.; Shim, S.; Yoon, M.Y.; Sun, S.; Kim, M.Y.; Van, K.; Lee, S.-H. Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 2012, 12, 139. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, Y.; Mason, A.S.; Lin, B.; Zhang, D.; Yu, H.; Fu, D. NBS-Encoding Genes in Brassica napus Evolved Rapidly After Allopolyploidization and Co-localize With Known Disease Resistance Loci. Front. Plant Sci. 2019, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Brotman, Y.; Silberstein, L.; Kovalski, I.; Perin, C.; Dogimont, C.; Pitrat, M.; Klingler, J.; Thompson, G.; Perl-Treves, R. Resistance gene homologues in melon are linked to genetic loci conferring disease and pest resistance. Theor. Appl. Genet. 2002, 104, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, H.; Garcia-Mas, J.; Coca, M.; Puigdoménech, P.; Monfort, A. Analysis of the melon genome in regions encompassing TIR-NBS-LRR resistance genes. Mol. Genet. Genom. 2005, 273, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Ruggieri, V.; Alexiou, K.G.; Morata, J.; Argyris, J.; Pujol, M.; Yano, R.; Nonaka, S.; Ezura, H.; Latrasse, D.; Boualem, A. An improved assembly and annotation of the melon (Cucumis melo L.) reference genome. Sci. Rep. 2018, 8, 8088. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Choi, W.; Thomas, C.E.; Dean, R.A. Cloning of disease-resistance homologues in end sequences of BAC clones linked to Fom-2, a gene conferring resistance to Fusarium wilt in melon (Cucumis melo L.). Genome 2002, 45, 473–480. [Google Scholar] [CrossRef]
- Takahashi, H.; Shoji, H.; Ando, S.; Kanayama, Y.; Kusano, T.; Takeshita, M.; Suzuki, M.; Masuta, C. RCY1-mediated resistance to Cucumber mosaic virus is regulated by LRR domain-mediated interaction with CMV (Y) following degradation of RCY1. Mol. Plant Microbe Interact. 2012, 25, 1171–1185. [Google Scholar] [CrossRef]
- Larkan, N.; Lydiate, D.; Parkin, I.; Nelson, M.; Epp, D.; Cowling, W.; Rimmer, S.; Borhan, M. The Brassica napus blackleg resistance gene LepR3 encodes a receptor--like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013, 197, 595–605. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243. [Google Scholar] [CrossRef]
- Padmanabhan, M.; Cournoyer, P.; Dinesh--Kumar, S. The leucine--rich repeat domain in plant innate immunity: A wealth of possibilities. Cell. Microbiol. 2009, 11, 191–198. [Google Scholar] [CrossRef]
- Maniatis, T.; Reed, R. An extensive network of coupling among gene expression machines. Nature 2002, 416, 499. [Google Scholar] [CrossRef]
- Rose, A.B. The effect of intron location on intron--mediated enhancement of gene expression in Arabidopsis. Plant J. 2004, 40, 744–751. [Google Scholar] [CrossRef]
- Chorev, M.; Carmel, L. The function of introns. Front. Genet. 2012, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-F.; Zhu, T.; Niu, D.-K. Association of intron loss with high mutation rate in Arabidopsis: Implications for genome size evolution. Genome Biol. Evolut. 2013, 5, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lin, Y.; Chen, G.; Shen, Y.; Liu, J.; Zhang, S. Genome-scale identification of resistance gene analogs and the development of their intron length polymorphism markers in maize. Mol. Breed. 2012, 29, 437–447. [Google Scholar] [CrossRef]
- Poczai, P.; Varga, I.; Laos, M.; Cseh, A.; Bell, N.; Valkonen, J.P.; Hyvönen, J. Advances in plant gene-targeted and functional markers: A review. Plant Methods 2013, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, W.; Jin, G.; Zhu, J. Genome-wide identification of R genes and exploitation of candidate RGA markers in rice. Chin. Sci. Bull. 2005, 50, 1120. [Google Scholar] [CrossRef]
- Slabaugh, M.B.; Yu, J.K.; Tang, S.; Heesacker, A.; Hu, X.; Lu, G.; Bidney, D.; Han, F.; Knapp, S.J. Haplotyping and mapping a large cluster of downy mildew resistance gene candidates in sunflower using multilocus intron fragment length polymorphisms. Plant Biotechnol. J. 2003, 1, 167–185. [Google Scholar] [CrossRef]
- Abuyusuf, M.; Nath, U.K.; Kim, H.-T.; Biswas, M.K.; Park, J.-I.; Nou, I.-S. Intronic Sequence Variations in a Gene with Peroxidase Domain Alter Bolting Time in Cabbage (Brassica oleracea var. capitata). Plant Mol. Biol. Rep. 2018, 36, 725–737. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).