Abstract
RNA editing is a relatively unexplored process in which transcribed RNA is modified at specific nucleotides before translation, adding another level of regulation of gene expression. Cephalopods use it extensively to increase the regulatory complexity of their nervous systems, and mammals use it too, but less prominently. Nevertheless, little is known about the specifics of RNA editing in most of the other clades and the relevance of RNA editing from an evolutionary perspective remains unknown. Here we analyze a key element of the editing machinery, the ADAR (adenosine deaminase acting on RNA) gene family, in an animal with a key phylogenetic position at the root of chordates: the cephalochordate amphioxus. We show, that as in cephalopods, ADAR genes in amphioxus are predominantly expressed in the nervous system; we identify a number of RNA editing events in amphioxus; and we provide a newly developed method to identify RNA editing events in highly polymorphic genomes using orthology as a guide. Overall, our work lays the foundations for future comparative analysis of RNA-editing events across the metazoan tree.
1. Introduction
Changes in the regulation of gene functions are at the basis of morphological evolution, from the simplest metazoans to the possessors of amazingly complex nervous systems. Contemporary textbook examples of mechanisms that allow exquisite fine-tuning of this regulation are alternative splicing in vertebrates and RNA gene editing in cephalopods [1,2,3]. Both enable an increase of proteome and interactome complexity [4] without affecting genome complexity by addition of extra genes. In this regard, RNA-editing is a post-transcriptional modification that has not received much attention. It adds an extra layer of regulation at the translational level, as well as an increase in proteome diversity without changes in the original DNA sequence. In an RNA-editing event, a specific nucleotide of the RNA transcript undergoes a chemical change. This process transforms the edited nucleotide into a different one, changing the sequence of the messenger RNA molecule and thus, potentially modifying the amino acid sequence [5]. RNA-editing is present in all eukaryotic organisms [6,7,8] in different forms, such as the C-to-U (cytidine to uracil) editing mediated by the Apobec protein family [7] or the most prevalent in metazoans, the A-to-I (adenosine to inosine) editing, mediated by the ADAR (adenosine deaminase acting on RNA) protein family [9].
Although increasing the levels of regulation and the diversity of the proteome has clear benefits, the evolutionary implications of RNA-editing are yet to be discovered. The role of the ADAR family itself is well known in vertebrates and cephalopods [10]. Its removal, for example, is known to be lethal in Mus musculus. Mouse embryos lacking ADAR will die before birth due to stress-induced apoptosis [11] and mice with null ADARB1 will become prone to seizures and die young [12]. In humans, one of its roles is the regulation of the innate immunity via the modulation of the antiviral response. In this case, ADAR would edit the viral dsRNA, which in some cases inactivates the virus. However, this also generates mismatched pairs on the viral dsRNA that dampen the interferon response, making ADAR in some cases a pro-viral agent [13]. Several studies have demonstrated how some level of conservation exists on the edited amino acid in vertebrates [14,15,16]; however, since the mechanisms or the features that restrict editing to specific nucleotides remain unknown, we are not yet able to properly predict RNA-editing sites in silico. This is partially due to the difficulty of detecting bona fide RNA-editing events in enough species, which ideally requires the sequencing of both DNA and RNA from the same individual [17]. This is particularly challenging when dealing with highly polymorphic invertebrate species such as the cephalochordate amphioxus [18]. In this key evolutionary model for the invertebrate/vertebrate transition, and in many other less-established laboratory species, extracting both genomic DNA and RNA in sufficient quantities from the same individual may be difficult to achieve and the sequencing quality required for in-depth RNA-editing studies unaffordable. On the other hand, the study of conservation and changes in RNA-editing in key evolutionary models is crucial to understand if this regulatory mechanism might have had a role in generating morphological novelty.
Here, we present for the first time the characterization of the ADAR family and its expression through the development of the amphioxus Branchiostoma lanceolatum, and propose a method to deal with its highly polymorphic genome when identifying in silico RNA-editing sites, that can be used with any organism with genomic and RNA-seq data available. This novel method, combined with an original pipeline, allows the identification of orthologous editing events between two species. Overall, our work lays the foundations for future comparative analysis of RNA-editing events across the metazoan tree.
2. Materials and Methods
2.1. Phylogenetic Tree of ADAR Family Genes
We selected the following sequences from NCBI: NP_001180424.1, NP_001344887, ALX72798.1, XP_031746878, XP_026690338.1, NP_571671.2, XP_030849200.1, NP_001103.1, NP_001020008, NP_001104544.1, NP_001096190.1, XP_021334694.1, AAF78094.1, NP_443209.2, XP_418563.4, XP_031759801.1, XP_686426.5, XP_781832.1, XP_002128212.1, XP_015888627.1, XP_020412580.1, XP_019848877.1, XP_003387867.1 for alignment with MAFFT (v7.307) [19] alongside the two translated genes from amphioxus BL01761 and BL03778. The aligned sequence was trimmed to select the conserved domain and a maximum likelihood tree was generated using IQ-TREE [20]. The tree is rooted on two plant ADAR-like proteins from Ziziphus jujuba and Prunus persica.
2.2. Third Generation In Situ Hybridization Chain Reaction (HCR) for Detecting Gene Expression
We followed the third generation in situ Hybridization Chain Reaction protocol, which has been recently optimized for amphioxus [21]. We used the following sequences to generate the probes: BL03778 (Adar), BL01761 (Adarb), BL13719 (Prox1), and BL11117 (Elav) from the transcriptome produced by Marletaz et al. [18].
2.3. Basic Single Nucleotide Variants Filter for RNA-Editing Candidates
In the first step of our pipeline, we used bowtie2 (v2.3.5.1) [22] to align, for each available tissue and developmental stage, the amphioxus RNA-seq reads (accessions SRX7351856 to SRX7351867) to the reference B. lanceolatum genome [18] and then the SAMtools library [23] to perform the Single Nucleotide Variants (SNV) analysis. We filtered the SNVs to get only the changes from A to G if the transcript was on the forward strand and T to C if the transcript was on the reverse strand, adding a depth cut-off of 10 and only considering the positions within coding regions. The resulting list of SNVs is the basic candidate list of RNA-editing events.
2.4. Using Genomic Reads to Filter out Confirmed SNPs
We repeated the previous steps with the DNA reads (accession PRJEB13665) instead of the RNA-seq reads in order to get the confirmed SNPs. In this step we did not use the depth cut-off. We then filtered out the candidate list of SNVs that appeared as confirmed SNPs in order to get the putative RNA-editing events. We then made an aggregative analysis of all the tissue and stage putative events.
2.5. GO Analysis
The analyzed sequences were blasted against the RefSeq protein database and run through InterProScan. Then, the blasted sequences were mapped and annotated using Blast2GO [24] and merged with the InterProScan GO annotations.
2.6. Orthologous RNA-Editing with Human Editing
From the candidate list and a human RNA-editing database (REDIportal [25]) we used a list of homologous genes obtained via BLAST to select those candidates that were within genes with human homologues, as well as the human editing targets present in those homologues from the REDIportal database [25]. We then obtained the mRNA sequence of all the isoforms of genes that contained the candidate SNVs that had potential homology with humans and translated the sequences into the amino acid sequence. We also obtained all the amino acid sequences of the homologous human genes from Ensembl [26]. We then used MAFFT (v7.307) [19] to make the multiple alignments of all the homologous amino acid sequences (all amphioxus isoforms with each homologous human isoform). We then checked within the alignments the edited amino acids and selected those cases where the edited amino acids in human were within the accepted window size in the alignment of the candidate SNVs in the amphioxus and separated the cases in categories according to the conservation of the original and edited amino acids. We used accepted window sizes of 0, 1, 5, and infinite amino acids. Finally, we performed the aggregative analysis across window sizes, tissues and stages (Supplementary Table S1).
For the curation of the events in window size 0, class 1, we manually evaluated the quality of the alignment within the local region to the putative events. We also used the genomic sequence of the annotated genes to get the full pre-mRNA sequence of all the isoforms that include the putative events. We then got the predicted secondary structure of the pre-mRNA molecules using linearfold [27]. We considered a positive dsRNA result if the position of the putative editing was either in a dsRNA or in a nick (a ssRNA fragment of one nucleotide in length with dsRNA on both sides).
3. Results
3.1. The European Amphioxus has Two ADAR Genes
Using the available transcriptomic data from B. lanceolatum [18], we were able to retrieve two amphioxus genes belonging to the ADAR family. Analyses of these two genes by sequence alignment to homologous genes found in publicly available databases, allowed us to identify these sequences as Adar and Adarb (Figure 1A). As expected from its pre-duplicative condition, the amphioxus ADAR family was composed of only two members and lacks one of the vertebrate-specific paralogs.
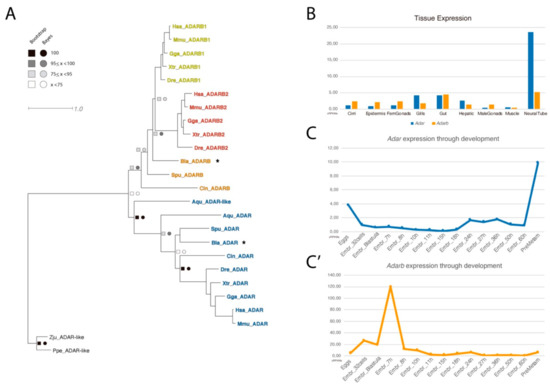
Figure 1.
Phylogeny and RNA-seq data of ADAR family. (A) Phylogenetic tree of the ADAR family. An asterisk marks the orthologous amphioxus ADAR genes identified, and the scale bar indicates the expected amino acid substitution rate per site. (B) Expression of Adar and Adarb1 in adult tissue and (C,C’) through development, measured in cRPKMs (corrected reads per kilo base per million mapped reads).
We further exploited available RNA-seq datasets to characterize the expression profile of Adar and Adarb in adult tissues (Figure 1B) and during embryonic development (Figure 1C,C’). For both paralogs, the highest expression was detected in the neural tube. Noticeably, Adar was expressed at a five times higher level than Adarb in the neural tube, while they were expressed at similar levels in the gut; moreover, Adar but not Adarb was also found in the gills. During development, Adar and Adarb were barely expressed in most of the stages analyzed, with the exception of an expression spike of Adarb at 7hpf (hours post fertilization) and the increase of Adar expression at pre-metamorphic stages.
3.2. ADAR and ADARB Expression through Development by HCR In Situ Hybridisation
We next took advantage of the high sensitivity of the hybridization chain reaction (HCR) method, to detect in situ the low levels of expression of the two Adar genes during the development of B. lanceolatum, on two developmental stages: mid-neurula and larva.
In the mid-neurula embryo, Adar and Adarb (Figure 2A–F) expression was restricted to specific cells in the developing neural tube (Figure 2B–E) and in the proximal half of all somites (Figure 2E,F), except in the first pair. Adarb was also expressed throughout the epidermis (best appreciated in Figure 2B). In the larva stage, both Adar and Adarb (Figure 2G–K) were expressed across the neural tube and in specific clusters of neurons (Figure 2H,I). Outside of the nervous system, Adar and Adarb transcripts were found in a few cells of the Hatchek’s pit (Figure 2J), in the distal half of the somites (Figure 2L) and in the nuclei of the developing muscle fibers (Figure 2M). In addition, we saw Adarb highly expressed in the endostyle (Figure 2J,K).
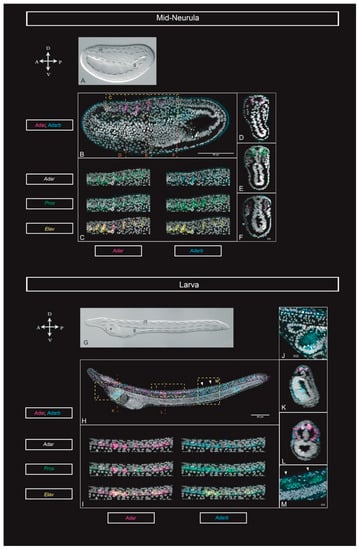
Figure 2.
Adar, Adarb, Elav, and Prox1 expression through development. (A) Bright field and (B) maximum projection for Adar and Adarb, from quadruple HCR (hybridization chain reaction) in situ hybridizations at the mid-neurula (18 hpf) stage. (C) Magnified view from D in B showing the co-expression of Adar, Adarb, Prox, and Elav in the neural tube of the mid-neurula. (D) Cross section at the level of C in B, showing expression of Adar and Adarb in the neural tube. (E) Cross section at the level of E in B, showing expression of co-expression of Adar and Prox in the neural tube and somites. (F) Cross section at the level of F in B, showing expression of Adar and Adarb in somites. (G) Bright field and (H) maximum projection for Adar and Adarb, from quadruple HCR in situ hybridizations at the early-larva (33 hpf) stage. (I) Magnified view of I in H showing the co-expression of Adar, Adarb, Prox, and Elav in the neural tube of the early larva. (J) Lateral view of the Hatscheck’s pit and the endostyle. (K) Cross-section at the level of K in H, showing co-expression of Adar and Adarb in the neural tube, and expression of Adarb in the endostyle. (L) Cross-section at the level of L in H showing expression in the neural tube and somites. (M) Detail at the level of M in H showing co-expression of Adar, Adarb, and Prox in the lateral muscle fibers. Arrowheads point at the center of the two somites shown. Scale bars: 80 µm and 10 µm for magnified views. Abbreviations: e: endostyle; g: gut; nt: neural tube.
To gain further insights into the cell types where ADAR-mediated RNA editing might be active, we co-registered both ADAR genes expression with that of Elav and Prox1, two genes with key roles in neurogenesis [28,29]
At mid-neurula stage, Adar and Adarb were co-expressed in the neural tube in Elav- and Prox1-positive cells (Figure 2C). Moreover, Adar and Prox1 colocalized in the proximal half of the somites (Figure 2E). In the larva Adar and Adarb were also co-expressed with Elav and Prox1 in the neural tube. However, while in the mid-neurula Adar genes were expressed exclusively in Elav- or Prox1-positive cells, at the larva stage several Elav- and Prox1-negative cells express one or both ADAR genes (Figure 2I). Additionally, in the mid-neurula and larva stage Adar and Prox1 were co-expressed in somites and muscles (Figure 2E,M).
3.3. A-to-I Editing: SNV Predictions
For each RNA-seq dataset, after looking for single nucleotide variants (SNVs) matching A-to-G (as inosine is detected as a guanosine by synthesis-based sequencing methods) or T-to-C (in the reverse strand) transitions, we filtered them using the genomic reads leaving only the SNVs that were not supported by the genomic reads available at the B. lanceolatum genome databases (see methods), hence not perceived as polymorphisms. This yielded a total of 184,163 unique putative RNA-editing events within 23,406 coding loci (from a total of 35,959 coding loci) with a mean 7, 87 SNVs per locus where more than half (109,237 ~59%) would result in a synonymous editing. A complete GO analysis of the genes harboring SNVs can be found in Figure S1. Focusing on tissue specificity, we selected the events appearing exclusively in one or two tissues and plotted them accumulatively relative to each tissue (e.g., tissue1, tissue1 + tissue2, tissue1 + tissue3, etc.). By this filtering, male gonads are the most enriched sample in putative RNA-editing events restricted to one tissue, while the gut is the most enriched overall. According to the expression data, neural tissue would be expected to be highly enriched in putative events, but surprisingly it ranked only in the 6th position. The tissue with the lowest occurrence of putative RNA-editing events was muscle. (Figure 3A). During embryonic development, the number of putative RNA-editing events did not change significantly, with a discrete increase at 10-hpf (Figure 3B).
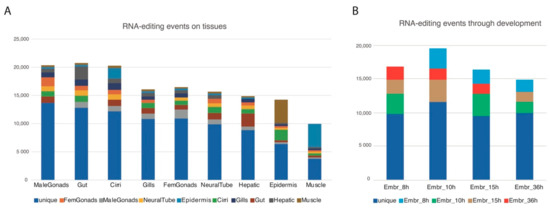
Figure 3.
Putative RNA-editing events by tissue and developmental stage. (A) Number of putative RNA-editing within coding regions by tissue, specific for one or two of the available tissues. (B) Number of putative RNA-editing events within coding loci by developmental stage, specific for one or two of the available stages.
3.4. Orthology-Based Approach to Infer RNA-Editing Conservation
The A-to-I SNV predictions, while being filtered by the genomic reads, could still harbor false positives that were not a result of RNA-editing events, but were due to genomic polymorphisms. We thus performed an orthology-based approach (see methods) to better infer the genuine RNA-editing events, identifying the changes that were conserved between two very distinct species (in this case human and amphioxus). As the homologous amino acid position can vary depending on the quality of the alignment, we implemented a window of homology and tested it at size 0, 1, 5, and infinite amino acids of aligned distance between the two edited amino acids. As the window size increases, so does the permissiveness of the approach, yielding more putative orthologous RNA-editing events. In addition, we separated the events into the following classes: class 0, when the editing resulted in a synonymous change in both interrogated species; class 1, where both the pre-edited and the edited amino acids were conserved between the interrogated species; class 2, where the pre-edited amino acids were not conserved between the interrogated species, but the edited ones were; class 3, where the pre-edited amino acids were conserved between the interrogated species, but the edited ones were not; class 4, where neither the pre-edited nor the edited amino acids were conserved between the interrogated species (Table 1). All the putative orthologous RNA-editing events obtained using the different window sizes except the infinite window size are available in the Supplementary Table S1. Interestingly, the tissue specificity for the different window size did not vary significantly except for the infinite window size, with gonads being the most enriched tissues in the other three window sizes (Figure 4).

Table 1.
Classification of putative orthologous RNA-editing events within coding loci for each range used. An event can belong to more than one class depending on the isoform analyzed.
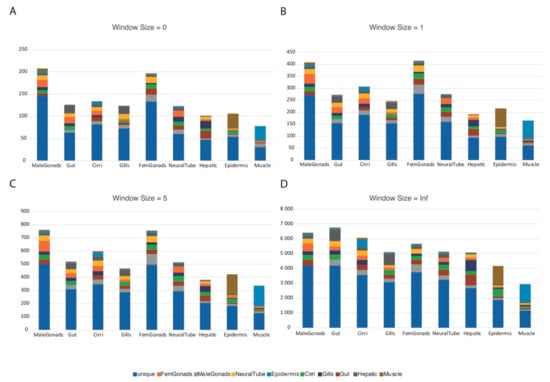
Figure 4.
Putative orthologous RNA-editing events by tissue using the tested window sizes. Number of putative orthologous RNA-editing events by tissue, specific for one or two of the available tissues; each graph corresponds to one of the four window sizes analyzed: (A) Window size 0; (B) Window size = 1; (C) Window size = 5; (D) Window size = Infinite.
A detailed list of selected class 1 events at window size 0 is also available (Table 2). We checked manually the quality of the alignments and whether the putative RNA-editing event was in a dsRNA for these events as ADAR proteins act on dsRNA. As a result, 30 events proved to have a good alignment quality with 9 candidates presenting both, a good alignment and the putatively edited nucleotide in a dsRNA region. This shows why a manual curation checking whether the alignments show conservation or have low quality and how is the secondary RNA structure of the gene at the SNV site was hereby needed as there can still be false positive events.

Table 2.
Selected windows size 0 class 1 RNA-editing events.
4. Discussion
4.1. Expression and Conservation of ADAR and ADARB in B. Lanceolatum
The most prominent cases of ADAR-dependent A-to-I editing have been reported in the nervous system and the brain of mammals and other vertebrates. Specifically, editing targets were found in key mediators of synaptic transmission [16]. More recently, a landmark study reported that RNA-editing in cephalopods was extremely abundant in the nervous system, and in particular in genes involved in neuronal excitability and morphology [3]. In mammals, Adar knockout mice were shown to be embryonically lethal and mice lacking Adarb1 died in a few days after birth due to the lack of editing in GluR-B receptor [11,12,30]. According to our expression analysis using a published B. lanceolatum RNA-seq dataset [18] (Figure 1B), Adar was highly expressed in the neural tube, whereas Adarb was found ubiquitously expressed at low levels in a variety of adult tissues. In vertebrates, however, Adarb1 was expressed more significantly in the neural and muscle tissues, Adarb2 was predominantly expressed in neural tissue, and Adar seemed to have a more pervasive expression [31]. Although a conserved microsyntenic cluster existed between amphioxus and human involving WDR37 and ADARB2, the tissue expression distribution seemed to not be conserved. It was remarkable how from the expected up to four vertebrate paralogs for each gene, Adar had only one ortholog without any clear synteny conserved with amphioxus, and Adarb had two, but one of them was enzymatically inactive. This could mean that an excess of A-to-I editing can be deleterious as very few organisms had more than two enzymatically active ADAR proteins [32].
The in situ data shown here was the first report of ADAR family expression in amphioxus during development, using the highly sensitive technique of in situ HCR [33], a recent technique still scarcely applied in invertebrates [21,34]. Our results proposed that the amphioxus ADAR family might have a role in gene regulation during the development of the nervous system, as this was where these genes are most prominently expressed. This is congruent with previously reported ADAR-mediated editing in the nervous system of other invertebrates such as Octopus bimaculoides, and in vertebrates such as mouse or human. Altogether, this suggests that RNA-editing regulation might have been instrumental for nervous system evolution.
Our results show that Adar and Adarb were both expressed in the developing neural tube and were initially co-expressed with Elav and Prox1. Both genes have established roles during neurogenesis: Elav is a well-known pan-neural marker [35] and was active in early stages of neural commitment [36,37], while Prox1 is a homeobox gene involved in many developmental processes, including neural progenitor differentiation and exit from the cell cycle [29,38,39]. The colocalization of ADAR paralogues with Elav and Prox1 at the mid-neurula stage suggests that RNA editing might play a role in neurogenesis in amphioxus. Interestingly, Takahashi and collaborators [40] demonstrated that the mRNA of Prox1, which can act as a tumour suppressor, experienced RNA-editing and loses tumour-suppressive functions in a subset of human cancers. This indicated that in the neural tube and muscles, where ADAR paralogues are co-expressed with Prox1, Adar may be editing Prox1, meaning that the edited isoform of Prox1 could have a role during the development of these particular tissues and cells.
4.2. An Orthology-Based Approach for Highly Polymorphic Species
The first report of an amphioxus genome, already indicated [41] that the amphioxus genome was highly polymorphic. This means that a simple SNV analysis of the RNA-seq reads against the genome, filtering the results for A-to-G and C-to-T (for the RNA-editing in the reverse strand) will not give direct data regarding editing events. A single RNA-seq analysis will result in a mixture of RNA-editing events and the DNA polymorphisms of the samples used for the analysis. One way of reducing the noise caused by the naturally occurring polymorphisms is to find the SNPs present in the genomic reads used to build the genome and filter them out [42], as we did to obtain the data shown in Figure 3. However, not all the polymorphisms actually occurring in an amphioxus population will be present in the available genomic reads, and the RNA-seq and genomic data usually come from different samples, hence some of the putative RNA-editing events could be false positives and be just polymorphisms. To increment the amount of bona-fide editing events, we chose to follow an orthology-based approach, taking advantage of the RNA-editing data of a distant but related species, the human [43], and the already reported gene orthology analysis between human and amphioxus [18,34]. Our approach of finding orthologous amino acids (Figure 4, Table 1, see methods) resulted in a strict filter, yielding 1588 putative conserved events. After we implemented a window of homology approach, however, the permissiveness of the pipeline increased and this resulted in 6291 putative events at window size 5, with 405 of them belonging to editing class 1 events (Table 1). An infinite window size was also used to compare edited orthologous pairs without considering whether the edited amino acid was conserved or not. The tissue distribution of the infinite window size (Figure 4D), however, was really similar to the tissue distribution of SNVs (Figure 1A), whereas the three other window sizes had a more similar tissue distribution. As the process is done automatically based on orthologies, a manual curation is needed as shown in Table 2 with the selection of window size 0, class 1 events. Although only nine of these events seem to have had both a good alignment and dsRNA in the edited region, the secondary structure predictions were still quite variable depending on the software used and the length of the sequence interrogated.
Although more analysis must be done to ensure that each of these putative RNA-editing events were not false positives, and problems, like the loss of non-conserved genes in the analysis need to be solved for a proper evolutionary analysis of the RNA-editing, we developed a tool that can be used between any two species to infer putative RNA-editing sites by homology. This method only required that data of RNA-editing exists for the reference species, and genomic and RNA-seq data is needed for the species interrogated.
5. Conclusions
We characterized experimentally for the first time the expression of the core RNA editing genes, Adar and Adarb in B. lanceolatum embryos, showing that both were expressed in the neural tube and muscles. We used a genomic filter for countering the polymorphic amphioxus genome and detected 184,163 unique cases of A-to-G and T-to-C variants that are putative RNA-editing events in several adult tissues and during embryonic development. Finally, we developed a pipeline for the assessment of RNA-editing conservation in which the astringency can be adjusted, and widely used with any species having RNA-seq and genomic datasets available.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/12/1440/s1, Supplementary Table S1: All putative orthologous RNA-editing events at range 0, 1 and 5; Figure S1: GO analysis of amphioxus SNVs; Pipeline available at https://github.com/mihizawi/Ortho-RNAediting.
Author Contributions
Conceptualization, J.G.-F., C.H.-Ú., and E.B.-G., methodology, M.Z.-Á., C.P.-C., G.G., and È.B.-G.; software, C.H.-Ú. and M.Z.-Á.; validation, C.H.-Ú. and M.Z.-Á.; formal analysis, C.H.-Ú., È.B.-G., and J.G.-F.; investigation, M.Z.-Á., C.P.-C., and G.G.; resources, J.G.-F. and È.B.-G.; data curation, C.H.-Ú. and M.Z.-Á.; writing—original draft preparation, C.H.-Ú., M.Z.-Á., C.P.-C., and J.G-F.; writing—review and editing, C.H.-Ú., M.Z.-Á., C.P.-C., G.G., È.B.-G., and J.G.-F. supervision, C.H.-Ú., J.G.-F., and È.B.-G.; project administration, J.G.-F.; funding acquisition, J.G.-F. and È.B.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grant BFU2017-86152-P (Ministerio de Ciencia, Innovación y Universidades, Spanish Government) to J.G.-F., C.H.-Ú.; by CRUK (C9545/A29580) to Elia Benito-Gutiérrez; by the Whitten Studentships to G.G.; M.Z.-Á. holds an UB-Santander predoctoral fellowship (University of Barcelona).
Acknowledgments
We wish to thank Stephanie Reichelt and her team at the Light Microscopy Facility at the Cancer Research UK Cambridge Institute for giving us access to the confocal during the difficult period of lockdown due to COVID-19.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Irimia, M.; Blencowe, B.J. Alternative splicing: Decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 2012, 24, 323–332. [Google Scholar] [CrossRef]
- Irimia, M.; Denuc, A.; Burguer, D.; Somorjai, I.; Martín-Durán, J.M.; Genikhovich, G.; Jimenez-Delgado, S.; Technau, U.; Roy, S.W.; Marfany, G.; et al. Stepwise assembly of the Nova-regulated alternative splicing network in the vertebrate brain. Proc. Natl. Acad. Sci. USA 2011, 108, 5319–5324. [Google Scholar] [CrossRef]
- Liscovitch-Brauer, N.; Alon, S.; Porath, H.T.; Elstein, B.; Unger, R.; Ziv, T.; Admon, A.; Levanon, E.Y.; Rosenthal, J.J.C.; Eisenberg, E. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 2017, 169, 191–202.e11. [Google Scholar] [CrossRef]
- Schmitz, J.F.; Zimmer, F.; Bornberg-Bauer, E. Mechanisms of transcription factor evolution in Metazoa. Nucleic Acids Res. 2016, 44, 6287–6297. [Google Scholar] [CrossRef]
- Savva, Y.A.; Rieder, L.E.; Reenan, R.A. The ADAR protein family. Genome Biol. 2012, 13, 252. [Google Scholar] [CrossRef] [PubMed]
- Porath, H.T.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 2017, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Härtel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Q.; He, Y.; Chen, L.; Hao, C.; Jiang, C.; Li, Y.; Dai, Y.; Kang, Z.; Xu, J.R. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016, 26, 499–509. [Google Scholar] [CrossRef]
- Thomas, J.M.; Beal, P.A. How do ADARs bind RNA? New protein-RNA structures illuminate substrate recognition by the RNA editing ADARs. BioEssays 2017, 39, 1600187. [Google Scholar] [CrossRef]
- Deffit, S.N.; Hundley, H.A. To edit or not to edit: Regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 2016, 7, 113–127. [Google Scholar] [CrossRef]
- Wang, Q.; Miyakoda, M.; Yang, W.; Khillan, J.; Stachura, D.L.; Weiss, M.J.; Nishikura, K. Stress-induced Apoptosis Associated with Null Mutation of ADAR1 RNA Editing Deaminase Gene. J. Biol. Chem. 2004, 279, 4952–4961. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; van den Hoogen, B.G.; Haagmans, B.L. ADAR1: “Editor-in-Chief” of Cytoplasmic Innate Immunity. Front. Immunol. 2019, 10, 1763. [Google Scholar] [CrossRef] [PubMed]
- Irimia, M.; Denuc, A.; Ferran, J.L.; Pernaute, B.; Puelles, L.; Roy, S.W.; Garcia-Fernàndez, J.; Marfany, G. Evolutionarily conserved A-to-I editing increases protein stability of the alternative splicing factor Nova1. RNA Biol. 2012, 9, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Pinto, Y.; Cohen, H.Y.; Levanon, E.Y. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 2014, 15, 31–33. [Google Scholar] [CrossRef]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 1–17. [Google Scholar] [CrossRef]
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Andrews, T.G.R.; Gattoni, G.; Busby, L.; Schwimmer, M.A.; Benito-Gutiérrez, È. Hybridization Chain Reaction for Quantitative and Multiplex Imaging of Gene Expression in Amphioxus Embryos and Adult Tissues. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2020; Volume 2148, pp. 179–194. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; D’Erchia, A.M.; Lo Giudice, C.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017, 45, D750–D757. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, H.; Deng, D.; Zhao, K.; Liu, K.; Hendrix, D.A.; Mathews, D.H. LinearFold: Linear-time approximate RNA folding by 5′-to-3′ dynamic programming and beam search. Bioinformatics 2019, 35, i295–i304. [Google Scholar] [CrossRef]
- Benito-Gutiérrez, È.; Illas, M.; Comella, J.X.; Garcia-Fernàndez, J. Outlining the nascent nervous system of Branchiostoma floridae (amphioxus) by the pan-neural marker AmphiElav. Brain Res. Bull. 2005, 66, 518–521. [Google Scholar] [CrossRef]
- Misra, K.; Mishra, K.; Gui, H.; Matise, M.P. Prox1 regulates a transitory state for interneuron neurogenesis in the spinal cord. Dev. Dyn. 2008, 237, 393–402. [Google Scholar] [CrossRef]
- Riedmann, E.M.; Schopoff, S.; Hartner, J.C.; Jantsch, M.F. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA 2008, 14, 1110–1118. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347. [Google Scholar] [CrossRef]
- Grice, L.F.; Degnan, B.M. The origin of the ADAR gene family and animal RNA editing. BMC Evol. Biol. 2015, 15, 4. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Schwarzkopf, M.; Fornace, M.E.; Acharya, A.; Artavanis, G.; Stegmaier, J.; Cunha, A.; Pierce, N.A. Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 2018, 145, dev165753. [Google Scholar] [CrossRef]
- Herrera-Úbeda, C.; Marín-Barba, M.; Navas-Pérez, E.; Gravemeyer, J.; Albuixech-Crespo, B.; Wheeler, G.N.; Garcia-Fernàndez, J. Microsyntenic Clusters Reveal Conservation of lncRNAs in Chordates Despite Absence of Sequence Conservation. Biology 2019, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Benito-Gutiérrez, È. A gene catalogue of the amphioxus nervous system. Int. J. Biol. Sci. 2006, 2, 149–160. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ratti, A.; Fallini, C.; Cova, L.; Fantozzi, R.; Calzarossa, C.; Zennaro, E.; Pascale, A.; Quattrone, A.; Silani, V. A role for the ELAV RNA-binding proteins in neural stem cells: Stabilization of Msi1 mRNA. J. Cell Sci. 2006, 119, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Perrone-Bizzozero, N.; Bolognani, F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 2002, 68, 121–126. [Google Scholar] [CrossRef]
- Kaltezioti, V.; Kouroupi, G.; Oikonomaki, M.; Mantouvalou, E.; Stergiopoulos, A.; Charonis, A.; Rohrer, H.; Matsas, R.; Politis, P.K. Prox1 regulates the notch1-mediated inhibition of neurogenesis. PLoS Biol. 2010, 8, e1000565. [Google Scholar] [CrossRef]
- Choksi, S.P.; Southall, T.D.; Bossing, T.; Edoff, K.; de Wit, E.; Fischer, B.E.; van Steensel, B.; Micklem, G.; Brand, A.H. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 2006, 11, 775–789. [Google Scholar] [CrossRef]
- Takahashi, M.; Yoshimoto, T.; Shimoda, M.; Kono, T.; Koizumi, M.; Yazumi, S.; Shimada, Y.; Doi, R.; Chiba, T.; Kubo, H. Loss of function of the candidate tumor suppressor prox1 by RNA mutation in human cancer cells. Neoplasia 2006, 8, 1003–1010. [Google Scholar] [CrossRef]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.K.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.-K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef]
- Pinto, Y.; Levanon, E.Y. Computational approaches for detection and quantification of A-to-I RNA-editing. Methods 2019, 156, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Kiran, A.; Baranov, P.V. DARNED: A DAtabase of RNA editing in humans. Bioinformatics 2010, 26, 1772–1776. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).