Rad21 Haploinsufficiency Prevents ALT-Associated Phenotypes in Zebrafish Brain Tumors
Abstract
1. Introduction
2. Materials and Methods
2.1. Maintenance of Zebrafish
2.2. Genotyping
2.3. Cell Culture and Cell Lines
2.4. Rad21 Immunoblotting
2.5. C-Circle Assay and Telomeric qPCR
2.6. Q-TRAP Assay
2.7. TERRA Dot Blot
2.8. Analysis of Gene Expression by qPCR
2.9. Immunofluorescence Combined with Quantitative Fluorescence In-Situ Hybridization (Q-FISH) on Interphase Nuclei
2.10. Statistics
3. Results
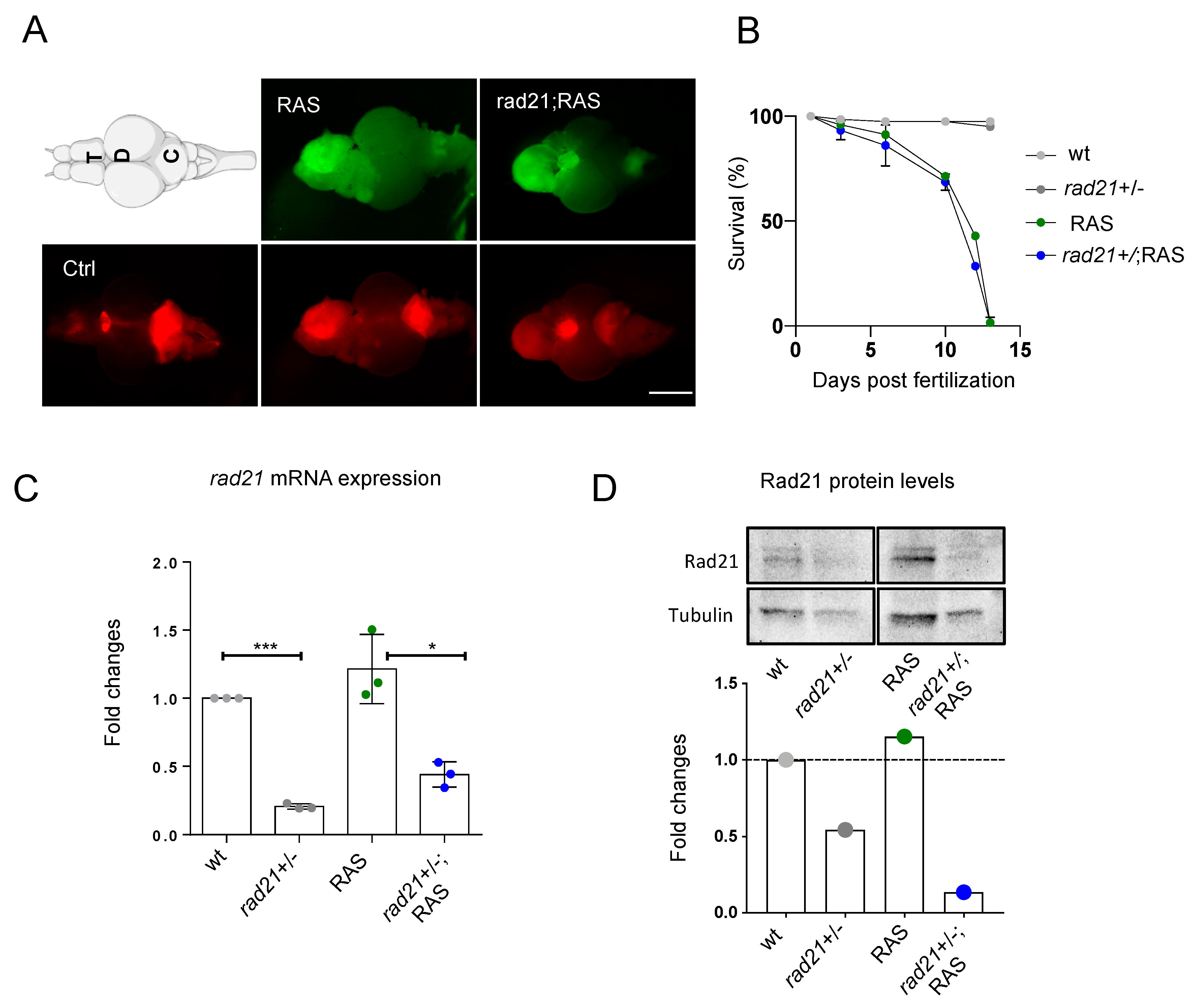
3.1. Rad21a Haploinsufficiency Does Not Worsen Brain Tumor Development
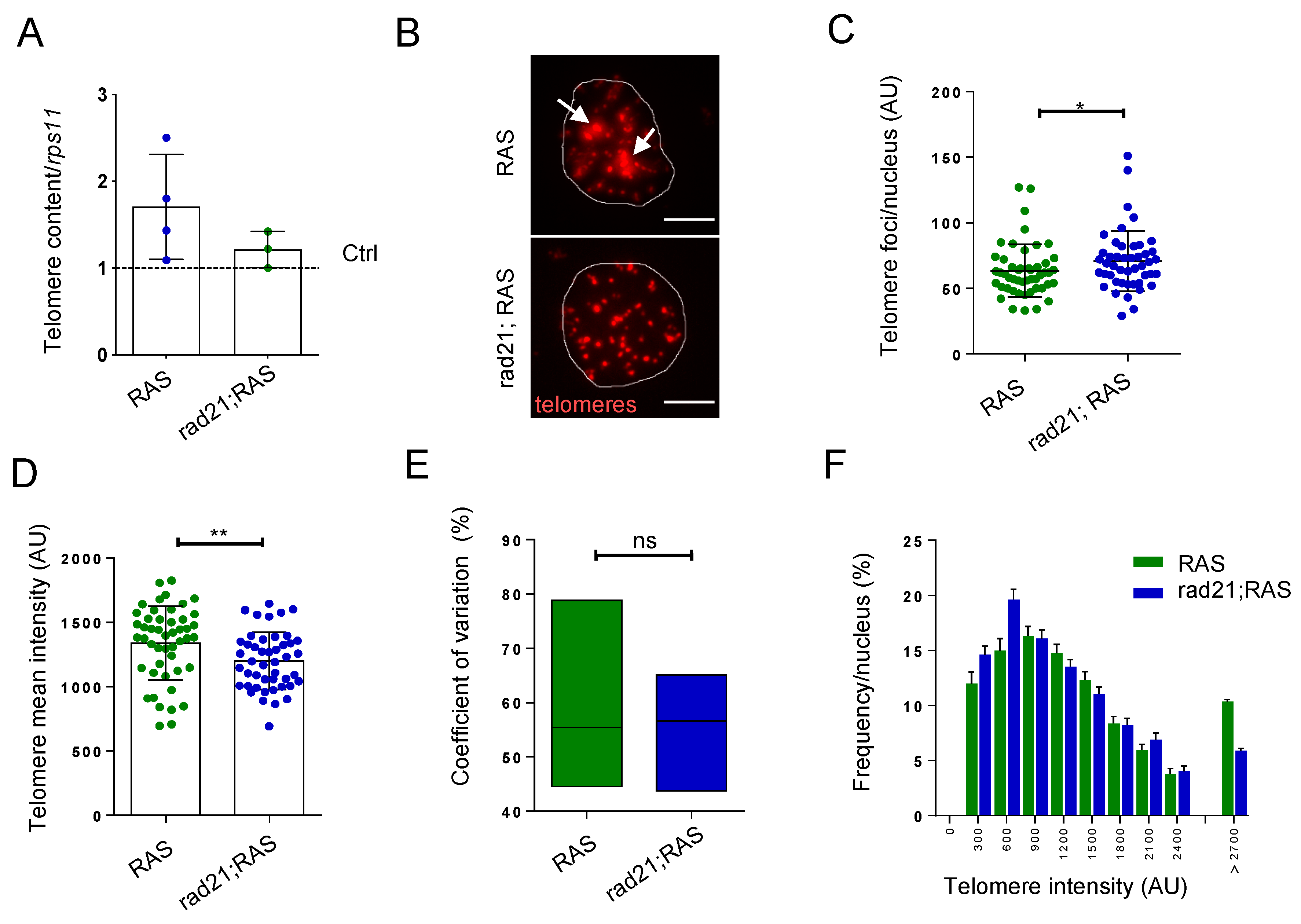
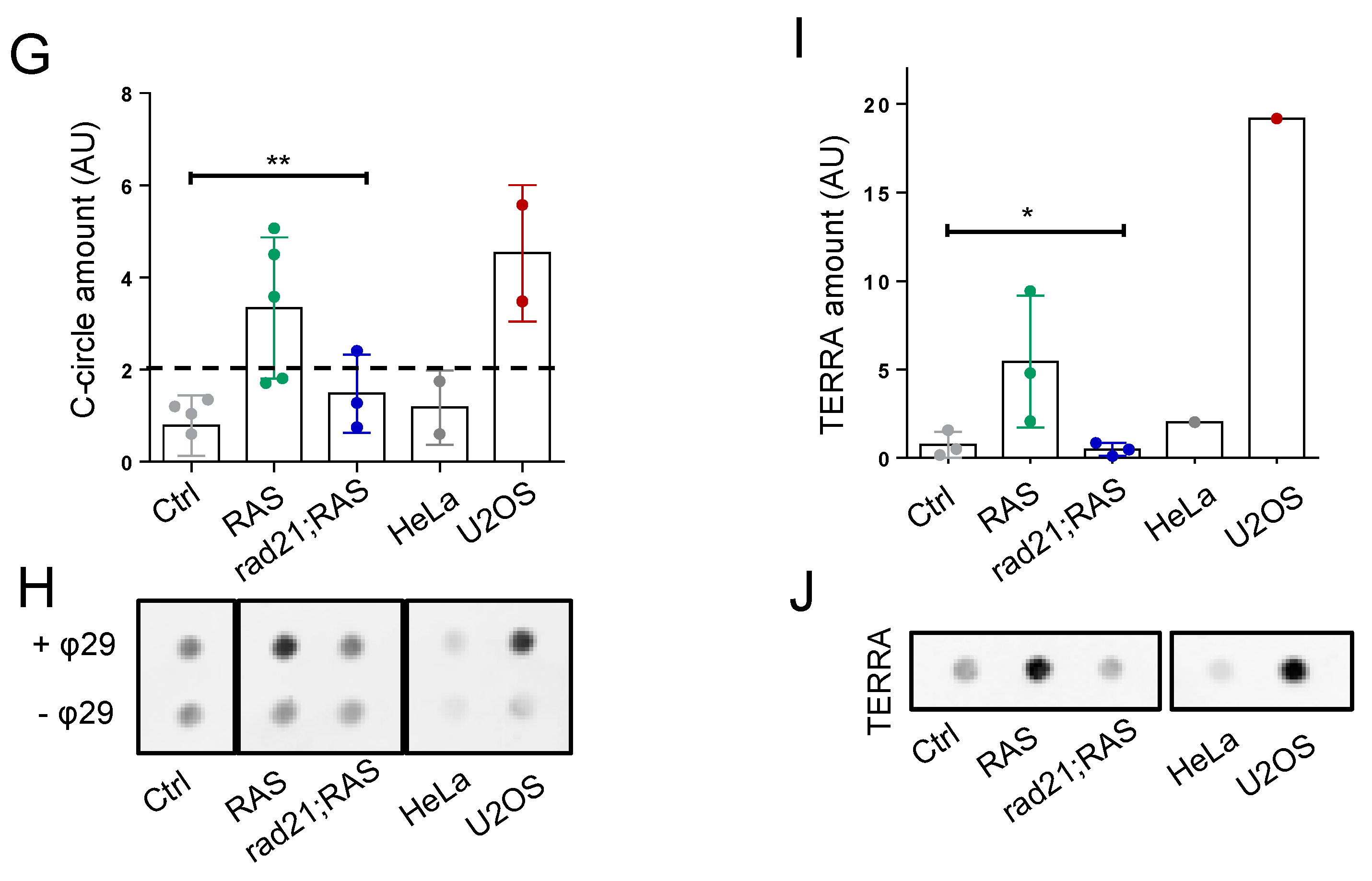
3.2. Rad21a Haploinsufficiency Prevents ALT
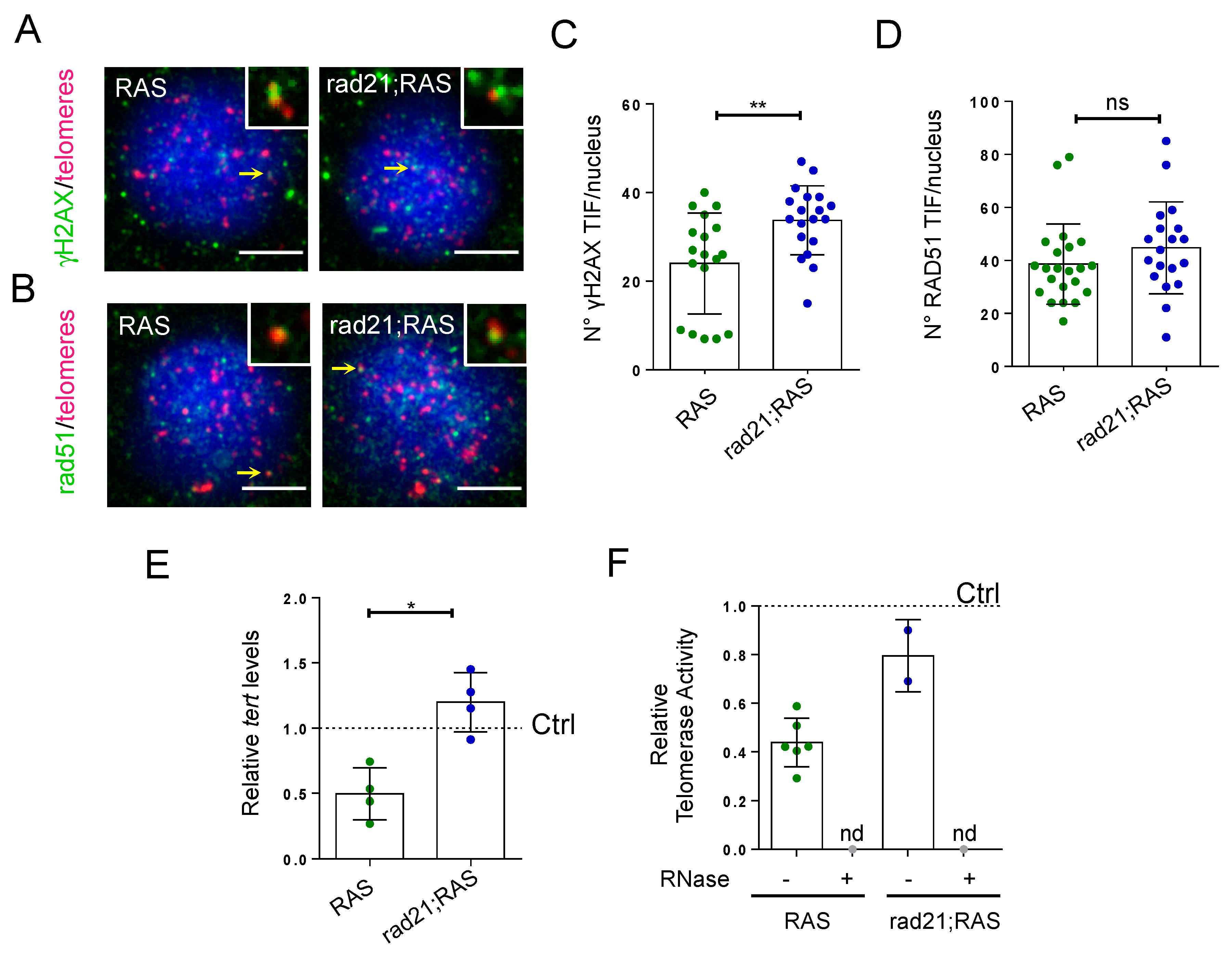
3.3. Rad21;RAS Brain Tumors Show an Increase in γH2AX TIFs, but Have Normal Tert Levels and Rad51 Foci
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mehta, G.D.; Kumar, R.; Srivastava, S.; Ghosh, S.K. Cohesin: Functions beyond sister chromatid cohesion. FEBS Lett. 2013, 587, 2299–2312. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Yu, Z.L.X.L.H. The Smc complexes in DNA damage response. Cell Biosci. 2012, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ju, X. New roles of hRAD21 in alternative lengthening of telomeres in cancer genesis. Cancer Biol. Ther. 2010, 9, 984–985. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, D. Cohesin: Genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 2011, 21, 199–206. [Google Scholar] [CrossRef]
- Wendt, K.S.; Yoshida, K.; Itoh, T.; Bando, M.; Koch, B.; Schirghuber, E.; Tsutsumi, S.; Nagae, G.; Ishihara, K.; Mishiro, T.; et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nat. Cell Biol. 2008, 451, 796–801. [Google Scholar] [CrossRef]
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef]
- Busslinger, G.A.; Stocsits, R.R.; Van Der Lelij, P.; Axelsson, E.; Tedeschi, A.; Galjart, N.; Peters, J.-M. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nat. Cell Biol. 2017, 544, 503–507. [Google Scholar] [CrossRef]
- Hsieh, T.-H.S.; Cattoglio, C.; Slobodyanyuk, E.; Hansen, A.S.; Rando, O.J.; Tjian, R.; Darzacq, X. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 2020, 78, 539–553.e8. [Google Scholar] [CrossRef]
- Litwin, I.; Pilarczyk, E.; Wysocki, R. The Emerging Role of Cohesin in the DNA Damage Response. Genes 2018, 9, 581. [Google Scholar] [CrossRef]
- Li, X.; Heyer, W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- Bauerschmidt, C.; Arrichiello, C.; Burdak-Rothkamm, S.; Woodcock, M.; Hill, M.A.; Stevens, D.L.; Rothkamm, K. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 2009, 38, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Balakrishnan, K.; Malaterre, J.; Beasley, M.; Yan, Y.; Essers, J.; Appeldoorn, E.; Tomaszewski, J.M.; Vazquez, M.; Verschoor, S.; et al. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- De Vitis, M.; Berardinelli, F.; Sgura, A. Telomere Length Maintenance in Cancer: At the Crossroad between Telomerase and Alternative Lengthening of Telomeres (ALT). Int. J. Mol. Sci. 2018, 19, 606. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.-M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.L.; Westra, W.H.; et al. Prevalence of the Alternative Lengthening of Telomeres Telomere Maintenance Mechanism in Human Cancer Subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Sobinoff, A.P.; Pickett, H.A. Alternative Lengthening of Telomeres: DNA Repair Pathways Converge. Trends Genet. 2017, 33, 921–932. [Google Scholar] [CrossRef]
- Mayrhofer, M.; Gourain, V.; Reischl, M.; Affaticati, P.; Jenett, A.; Joly, J.-S.; Benelli, M.; Demichelis, F.; Poliani, P.L.; Sieger, D.; et al. A novel brain tumour model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. Dis. Model. Mech. 2017, 10, 15–28. [Google Scholar] [CrossRef]
- Idilli, A.I.; Cusanelli, E.; Pagani, F.; Berardinelli, F.; Bernabé, M.; Cayuela, M.L.; Poliani, P.L.; Mione, M.C. Expression of tert Prevents ALT in Zebrafish Brain Tumors. Front. Cell Dev. Biol. 2020, 8, 65. [Google Scholar] [CrossRef]
- Westerfield, M.; Zon, L.I.; Detrich, W.H., III. Essential Zebrafish Methods: Cell and Developmental Biology, 1st ed.; Academic Press: Oxford, UK, 2009. [Google Scholar]
- Santoriello, C.; Gennaro, E.; Anelli, V.; Distel, M.; Kelly, A.; Köster, R.W.; Hurlstone, A.; Mione, M.C. Kita Driven Expression of Oncogenic HRAS Leads to Early Onset and Highly Penetrant Melanoma in Zebrafish. PLoS ONE 2010, 5, 15170. [Google Scholar] [CrossRef]
- Amsterdam, A.; Nissen, R.M.; Sun, Z.; Swindell, E.C.; Farrington, S.; Hopkins, N. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 2004, 101, 12792–12797. [Google Scholar] [CrossRef]
- Rhodes, J.M.; Bentley, F.K.; Print, C.G.; Dorsett, D.; Misulovin, Z.; Dickinson, E.J.; Crosier, K.E.; Crosier, P.S.; Horsfield, J.A. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev. Biol. 2010, 344, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.D.; Lau, L.M.; Koch, S.; La Rotta, N.M.; Dagg, R.A.; Reddel, R.R. The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 2017, 114, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Stone, R.C.; Hunt, S.C.; Skurnick, J.H.; Lu, X.; Cao, X.; Harley, C.B.; Aviv, A. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 2010, 5, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Hanish, J.P.; Yanowitz, J.L.; De Lange, T. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 1994, 91, 8861–8865. [Google Scholar] [CrossRef]
- Lau, L.M.S.; Dagg, R.A.; Henson, J.D.; Au, A.Y.M.; Royds, J.A.; Reddel, R.R. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 2013, 41, 34. [Google Scholar] [CrossRef]
- Gilles, J.-F.; Dos Santos, M.; Boudier, T.; Bolte, S.; Heck, N. DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 2017, 115, 55–64. [Google Scholar] [CrossRef]
- Deardorff, M.A.; Wilde, J.J.; Albrecht, M.; Dickinson, E.; Tennstedt, S.; Braunholz, D.; Mönnich, M.; Yan, Y.; Xu, W.; Gil-Rodríguez, M.C.; et al. RAD21 Mutations Cause a Human Cohesinopathy. Am. J. Hum. Genet. 2012, 90, 1014–1027. [Google Scholar] [CrossRef]
- Dunham, M.A.; Neumann, A.A.; Fasching, C.L.; Reddel, R.R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000, 26, 447–450. [Google Scholar] [CrossRef]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef]
- Henson, J.D.; Cao, Y.; Huschtscha, L.I.; Chang, A.C.; Au, A.Y.M.; Pickett, H.A.; Reddel, R.R. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 2009, 27, 1181–1185. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Lingner, J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015, 25, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Episkopou, H.; Draskovic, I.; Van Beneden, A.; Tilman, G.; Mattiussi, M.; Gobin, M.; Arnoult, N.; Londoño-Vallejo, A.; Decottignies, A. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014, 42, 4391–4405. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; De Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. Loss of ATRX, Genome Instability, and an Altered DNA Damage Response Are Hallmarks of the Alternative Lengthening of Telomeres Pathway. PLoS Genet. 2012, 8, 1002772. [Google Scholar] [CrossRef] [PubMed]
- Doksani, Y. The Response to DNA Damage at Telomeric Repeats and Its Consequences for Telomere Function. Genes 2019, 10, 318. [Google Scholar] [CrossRef]
- Dagg, R.A.; Pickett, H.A.; Neumann, A.A.; Napier, C.E.; Henson, J.D.; Teber, E.T.; Arthur, J.W.; Reynolds, C.P.; Murray, J.; Haber, M.; et al. Extensive Proliferation of Human Cancer Cells with Ever-Shorter Telomeres. Cell Rep. 2017, 19, 2544–2556. [Google Scholar] [CrossRef]
- Potts, P.R.; Porteus, M.H.; Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006, 25, 3377–3388. [Google Scholar] [CrossRef]
- Atienza, J.M.; Roth, R.B.; Rosette, C.; Smylie, K.J.; Kammerer, S.; Rehbock, J.; Ekblom, J.; Denissenko, M.F. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 2005, 4, 361–368. [Google Scholar]
- Birkenbihl, R.P.; Subramani, S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992, 20, 6605–6611. [Google Scholar] [CrossRef]
- Koshland, D.E.; Guacci, V. Sister chromatid cohesion: The beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 2000, 12, 297–301. [Google Scholar] [CrossRef]
- Hellmuth, S.; Gutiérrez-Caballero, C.; Llano, E.; Pendas, A.M.; Stemmann, O. Local activation of mammalian separase in interphase promotes double-strand break repair and prevents oncogenic transformation. EMBO J. 2018, 37, 99184. [Google Scholar] [CrossRef]
- Benedict, B.; Van Schie, J.J.; Oostra, A.B.; Balk, J.A.; Wolthuis, R.M.F.; Riele, H.T.; De Lange, J. WAPL-Dependent Repair of Damaged DNA Replication Forks Underlies Oncogene-Induced Loss of Sister Chromatid Cohesion. Dev. Cell 2020, 52, 683–698.e7. [Google Scholar] [CrossRef] [PubMed]
- Muntoni, A.; Neumann, A.A.; Hills, M.; Reddel, R.R. Telomere elongation involves intra-molecular DNA replication in cells utilizing alternative lengthening of telomeres. Hum. Mol. Genet. 2008, 18, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; Yadav, T.; Ouyang, J.; Lan, L.; Zou, L. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 2019, 26, 955–968.e3. [Google Scholar] [CrossRef] [PubMed]
- Wijchers, P.J.; De Laat, W. Genome organization influences partner selection for chromosomal rearrangements. Trends Genet. 2011, 27, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Zuin, J.; Dixon, J.; Van Der Reijden, M.I.J.A.; Ye, Z.; Kolovos, P.; Brouwer, R.; Van De Corput, M.; Van De Werken, H.; Knoch, T.; Van Ijcken, W.; et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Ing-Simmons, E.; Seitan, V.C.; Faure, A.J.; Flicek, P.; Carroll, T.; Dekker, J.; Fisher, A.G.; Lenhard, B.; Merkenschlager, M. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015, 25, 504–513. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Z.; Stong, N.; Plasschaert, R.; Moczan, A.; Chen, H.-S.; Hu, S.; Wikramasinghe, P.; Davuluri, R.V.; Bartolomei, M.S.; et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012, 31, 4165–4178. [Google Scholar] [CrossRef]
- Nanavaty, V.; Abrash, E.W.; Hong, C.; Park, S.; Fink, E.E.; Li, Z.; Sweet, T.J.; Bhasin, J.M.; Singuri, S.; Lee, B.H.; et al. DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex. Mol. Cell 2020, 78, 752–764.e6. [Google Scholar] [CrossRef]
- Lee, D.D.; Leão, R.R.; Komosa, M.; Gallo, M.; Zhang, C.H.; Lipman, T.; Remke, M.; Heidari, A.; Nunes, N.M.; Apolónio, J.D.; et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J. Clin. Investig. 2018, 129, 223–229. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Idilli, A.I.; Pazzi, C.; dal Pozzolo, F.; Roccuzzo, M.; Mione, M.C. Rad21 Haploinsufficiency Prevents ALT-Associated Phenotypes in Zebrafish Brain Tumors. Genes 2020, 11, 1442. https://doi.org/10.3390/genes11121442
Idilli AI, Pazzi C, dal Pozzolo F, Roccuzzo M, Mione MC. Rad21 Haploinsufficiency Prevents ALT-Associated Phenotypes in Zebrafish Brain Tumors. Genes. 2020; 11(12):1442. https://doi.org/10.3390/genes11121442
Chicago/Turabian StyleIdilli, Aurora Irene, Cecilia Pazzi, Francesca dal Pozzolo, Michela Roccuzzo, and Maria Caterina Mione. 2020. "Rad21 Haploinsufficiency Prevents ALT-Associated Phenotypes in Zebrafish Brain Tumors" Genes 11, no. 12: 1442. https://doi.org/10.3390/genes11121442
APA StyleIdilli, A. I., Pazzi, C., dal Pozzolo, F., Roccuzzo, M., & Mione, M. C. (2020). Rad21 Haploinsufficiency Prevents ALT-Associated Phenotypes in Zebrafish Brain Tumors. Genes, 11(12), 1442. https://doi.org/10.3390/genes11121442





