Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. cDNA Cloning and Sequence Analysis
2.3. Tissue Expression Analysis of Sheep SPP1, LAP3, and LCORL
2.4. SNP Identification
2.5. Association Analysis
3. Results
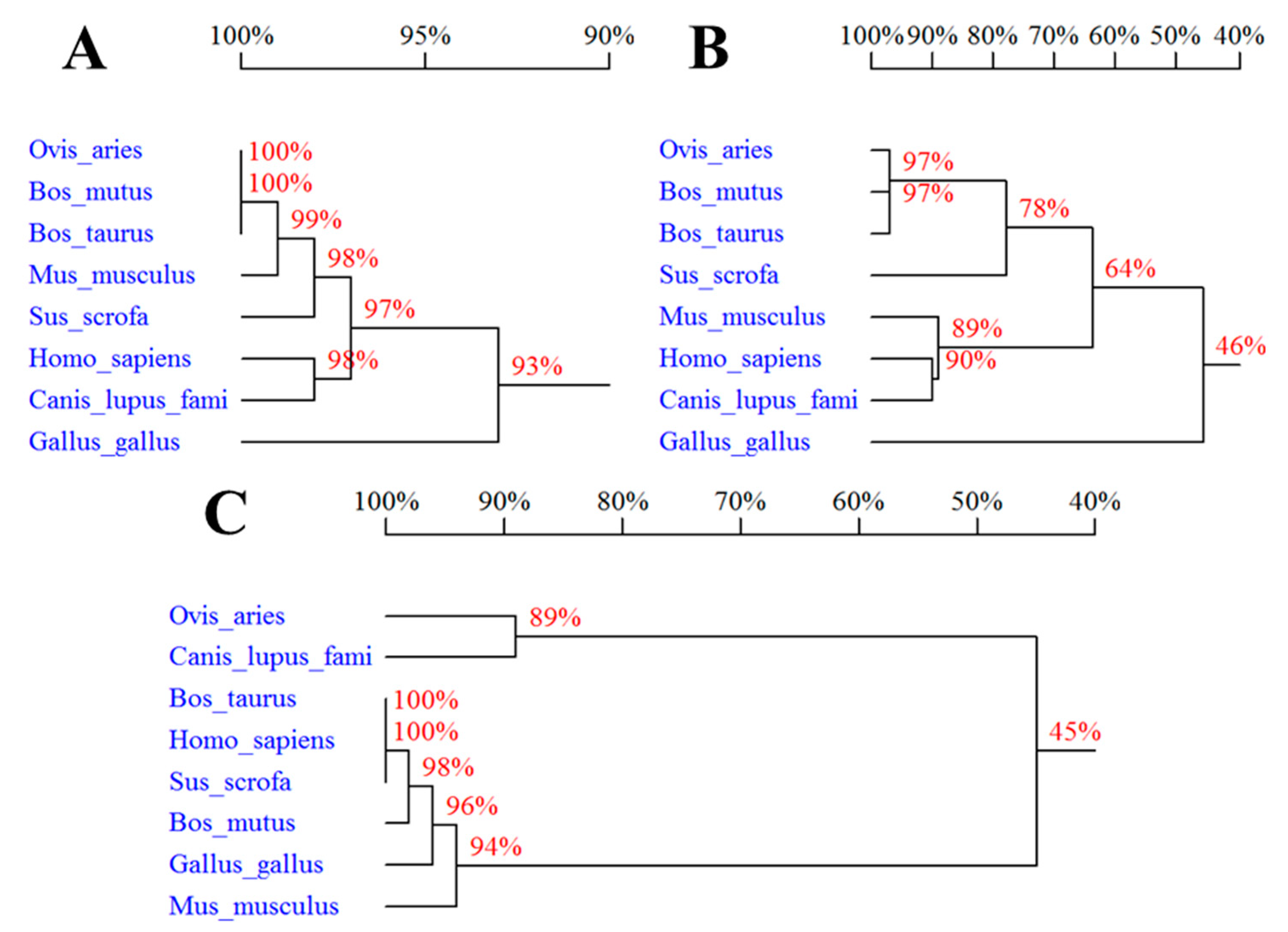
3.1. Molecular Cloning and Sequence Analysis of Sheep SPP1, LAP3, and LCORL
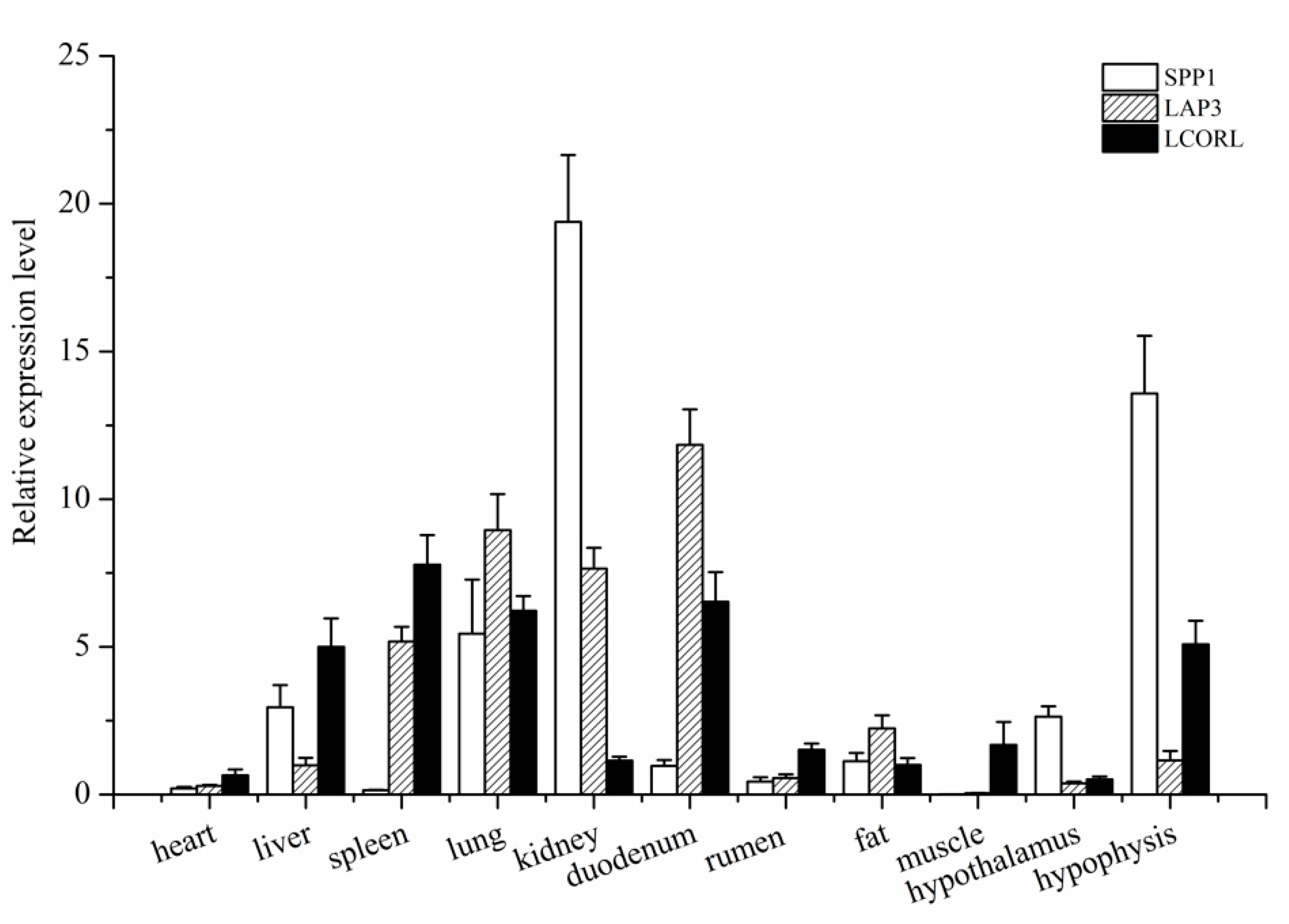
3.2. Expression Profile Analysis
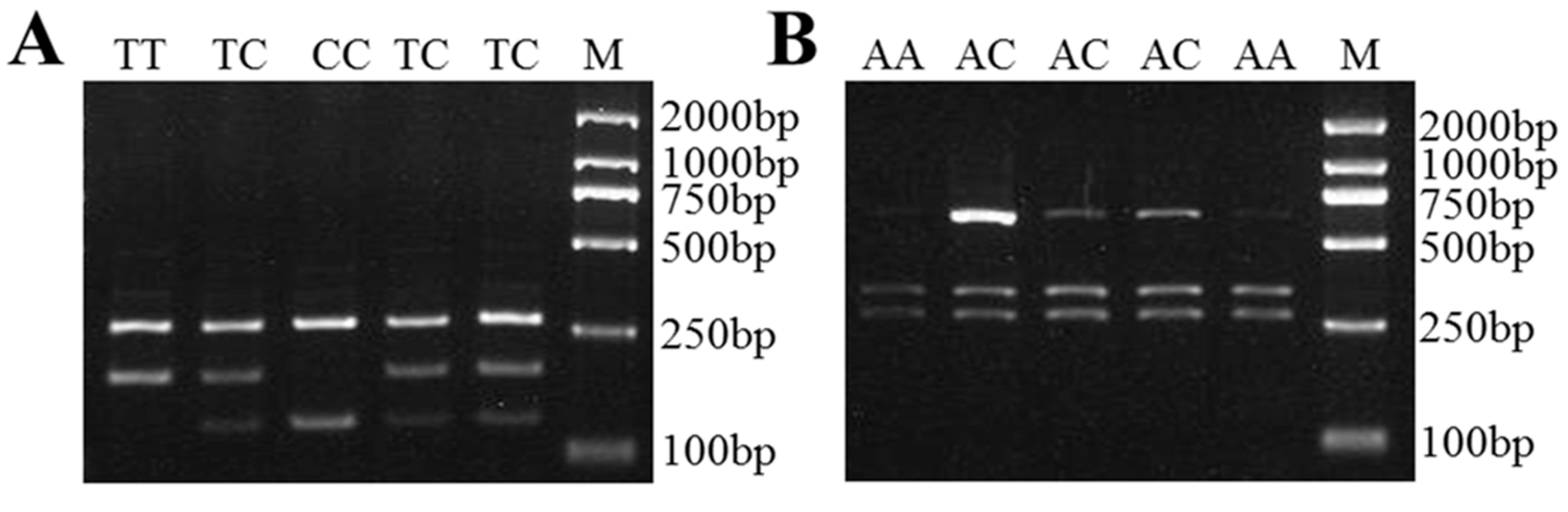
3.3. SNPs of Sheep SPP1, LAP3, and LCORL
3.4. Association of Sheep SPP1, LAP3, and LCORL with BW
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Almamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef] [PubMed]
- Prince, C.W.; Oosawa, T.; Butler, W.T.; Tomana, M.; Bhown, A.S.; Bhown, M.; Schrohenloher, R. Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone. J. Biol. Chem. 1987, 262, 2900–2907. [Google Scholar] [PubMed]
- Sodek, J.; Ganss, B.; Mckee, M.D. Osteopontin. Crit. Rev. Oral Biol. Med. 2000, 11, 279–303. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, R.D.; Kim, J.J.; Ashwell, M.S.; Sonstegard, T.S.; Van Tassell, C.P.; Connor, E.E.; Taylor, J.F. Fine-mapping milk production quantitative trait loci on BTA6: Analysis of the bovine osteopontin gene. Proc. Natl. Acad. Sci. USA 2005, 102, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- White, S.N.; Asas, E.; Allan, M.F.; Keele, J.W.; Snelling, W.N.; Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M.; Smith, T.P. Evaluation in beef cattle of six deoxyribonucleic acid markers developed for dairy traits reveals an osteopontin polymorphism associated with postweaning growth. Anim. Sci. 2007, 85, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ranqaswami, H.; Bulbule, A.; Kundu, G.C. Osteopontin: Role in cell signaling and cancer progression. Trends Cell Biol. 2006, 16, 79. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Lin, X.; Itskovich, W.; Aquinaldi, J.G.; Chaplin, W.F.; Denhardt, D.T.; Fayad, Z.A. Prenatal detection of embryo resorption in osteopontin-deficient mice using serial noninvasive magnetic resonance microscopy. Pediatr. Res. 2004, 55, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, H.T.; van Loon-Klaassen, L.A.; Eqberts, W.T.; de Jonq, W.W.; Bloemendal, H. The primary structure of leucine aminopeptidase from bovine eye lens. J. Biol. Chem. 1982, 257, 7077–7085. [Google Scholar]
- Matsui, M.; Fowler, J.H.; Wallinq, L.L. Leucine aminopeptidases: Diversity in structure and function. Biol. Chem. 2006, 55, 1535–1544. [Google Scholar] [CrossRef]
- Mural, T. Leucine aminopeptidase (LAP). Rinsho Byori 2001, 11, 303–306. [Google Scholar]
- Olsen, H.G.; Lien, S.; Gautier, M.; Nilsen, H.; Roseth, A.; Berq, P.R.; Sundsaasen, K.K.; Svendsen, M.; Meuwissen, T.H. Mapping of a milk production quantitative trait locus to a 420-kb region on bovine chromosome 6. Genetics 2005, 169, 275–383. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Zinder, M.; Seroussi, E.; Larkin, D.M.; Loor, J.J.; Everts-van der Wind, A.; Lee, J.H.; Drackley, J.K.; Band, M.R.; Hernandez, A.G.; Shani, M.; et al. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 2005, 15, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Lindholm-Perry, A.K.; Kuehn, L.A.; Oliver, W.T.; Sexten, A.K.; Miles, J.R.; Rempel, L.A.; Cushman, R.A.; Freetly, H.C. Adipose and muscle tissue gene expression of two genes (NCAPG and LCORL) located in a chromosomal region associated with cattle feed intake and gain. PLoS ONE 2013, 8, e80882. [Google Scholar] [CrossRef] [PubMed]
- Lango, A.H.; Estrada, K.; Lettre, G.; Berndt, S.L.; Weedon, M.N.; Rivadeneira, F.; Willer, C.J.; Jackson, A.U.; Vedantam, S.; Raychaudhuri, S.; et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010, 467, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Vaysse, A.; Ratnakumar, A.; Derrien, T.; Axelsson, E.; Pielberq, G.R.; Siqurdsson, S.; Fall, T.; Seppala, E.H.; Hansen, M.S.; Lawley, C.T. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011, 7, 1002316. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.E.; Hayes, B.J.; Bolormaa, S.; Goddard, M.E. Polymorphic regions affecting human height also control stature in cattle. Genetics 2011, 187, 981–984. [Google Scholar] [CrossRef]
- Siqner-Hasler, H.; Flury, C.; Haase, B.; Burqer, D.; Simianer, H.; Leeb, T.; Rieder, S. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS ONE 2012, 7, e37282. [Google Scholar] [CrossRef] [PubMed]
- Rubin, C.J.; Megens, H.J.; Barrio, A.M.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, Ö.; Jern, P.; Jørgensen, C.B.; et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lim, W.; Jeong, W.; Kim, J.; Ka, H.; Bazer, F.W.; Han, J.Y.; Song, G. Differential expression of secreted phosphoprotein 1 in response to estradiol-17β and in ovarian tumors in chickens. Biochem. Biophys. Res. Commun. 2012, 422, 494–500. [Google Scholar] [CrossRef]
- Broad, L.M.; Sanger, H.E.; Mogg, A.J.; Colvin, E.M.; Zwart, R.; Evans, D.A.; Pasqui, F.; Sher, E.; Wishart, G.N.; Barth, V.N. Identification and pharmacological profile of SPP1, a potent, functionally selective and brain penetrant agonist at muscarinic M1 receptors. Br. J. Pharmacol. 2019, 176, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Asaumi, S.; Takemoto, M.; Yokote, K.; Ridall, A.L.; Butler, W.T.; Fujimoto, M.; Kobayashi, K.; Kawamura, H.; Take, A.; Saito, Y. Identification and characterization of high glucose and glucosamine responsive element in the rat osteopontin promoter. J. Diabetes Complicat. 2003, 17, 34–38. [Google Scholar] [CrossRef]
- Hijiya, N.; Setoguchi, M.; Matsuura, K.; Higuchi, Y.; Akizuki, S.; Yamamoto, S. Cloning and characterization of the human osteopontin gene and its promoter. Biochem. J. 1994, 303, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Yokosuka, O.; Kanda, T.; Fukai, K.; Imazeki, F.; Muramatsu, M.; Seki, N.; Miyazaki, M.; Ochiai, T.; Hirasawa, H. Serum osteopontin levels in patients with acute liver dysfunction. Scand. J. Gastroenterol. 2006, 41, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, P.A.; Riley, L.G.; Raadsma, H.W.; Williamson, P.; Wynn, P.C. A functional genomics approach to evaluate candidate genes located in a QTL interval for milk production traits on BTA6. Anim. Genet. 2010, 40, 492–498. [Google Scholar] [CrossRef]
- Allan, M.F.; Thallman, R.M.; Cushman, R.A.; Echternkamp, S.E.; White, S.N.; Kuehn, L.A.; Casas, E.; Smith, T.P. Association of a single nucleotide polymorphism in SPP1 with growth traits and twinning in a cattle population selected for twinning rate. Anim. Sci. 2007, 85, 341–347. [Google Scholar] [CrossRef]

| Primer Name | Primer Sequence (5′-3′) | Annealing Temperature (°C) | Size (bp) |
|---|---|---|---|
| SPP1-CDS-S | CATCAGCATCACAGGGGACT | 57 | 1122 |
| SPP1-CDS-A | GGAAAGAACATAGACTAAACCCT | ||
| SPP1-expression-S | ATGACTCCGACGATGCTGAAC | 57 | 135 |
| SPP1-expression-A | CGTAGGGAAAGGTGGAGTG | ||
| SPP1-SNP-S | GGACAGAGGCTGAAGGAATAC | 58 | 885 |
| SPP1-SNP-A | CATCCAAAGCAGGTCTTAT | ||
| LAP3-CDS-S | TCGGTGGAGGGCGGTACG | 55 | 567 |
| LAP3-CDS-A | GAAGATAAGGAACCTCAT | ||
| LAP3-expression-S | TGCCCATCAACATTGTAGGT | 60 | 170 |
| LAP3-expression-A | AGTGTGAGCGTAGCAGAGCG | ||
| LAP3-SNP1-S | GGCACTGCTTTCTATCATTG | 55 | 351 |
| LAP3- SNP1-A | ATAGGTGTTCACTGAGGGTT | ||
| LAP3-SNP2-S | CTTTTAGTCTTTTGACCTTC | 55 | 407 |
| LAP3-SNP2–A | GCTTTGTATCATTTTTAGCT | ||
| LCORL-CDS-S | AACTGACCAAACCGACAT | 54 | 1543 |
| LCORL-CDS-A | TATCCAAGCACCTGTCCC | ||
| LCORL-expression-S | CTGCTTACCTCCTTTAGA | 52 | 280 |
| LCORL-expression-A | GTCCTCCTGACTTTTACC | ||
| LCORL–SNP1-S | AGAGTCTCAGAATCCCCTAA | 52 | 495 |
| LCORL–SNP1-A | TTGCTTATTTCTGCTGGTGT | ||
| LCORL-SNP2-S | GAACCCATTGAAAACGATAA | 55 | 594 |
| LCORL-SNP2-A | AGGTGGGAAAATAAACTGAT | ||
| GAPDH-expression-S | GGGGTCTACACTCCCAACTGC | 58 | 379 |
| GAPDH-expression-A | CAGAAGGCGGCGATGGAA |
| Gene Name | Locus | Genotype | n | Live Weight | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Birth | 1-month | 2-month | 3-month | 4-month | 6-month | 8-month | 10-month | 12-month | ||||
| SPP1 | NM_001009224.1:c.132A>C | AA | 143 | 3.499 ± 0.061a | 11.227 ± 0.275 | 18.041 ± 0.351 | 23.231 ± 0.387 | 28.135 ± 0.419 | 33.656 ± 0.472 | 35.036 ± 0.494 | 41.245 ± 0.769 | 46.384 ± 0.829a |
| AC | 102 | 3.354 ± 0.074b | 11.082 ± 0.331 | 17.825 ± 0.423 | 22.999 ± 0.466 | 28.223 ± 0.506 | 34.610 ± 0.569 | 35.408 ± 0.595 | 40.345 ± 0.927 | 44.818 ± 1.000b | ||
| CC | 44 | 3.569 ± 0.103a | 11.507 ± 0.462 | 17.791 ± 0.591 | 23.300 ± 0.651 | 27.800 ± 0.706 | 33.422 ± 0.794 | 34.817 ± 0.831 | 41.667 ± 1.294 | 47.383 ± 1.396a | ||
| LAP3 | XM_012179698.3: c.232C>G | CC | 104 | 3.449 ± 0.067a | 11.257 ± 0.295a | 18.029 ± 0.372A | 23.275 ± 0.415a | 28.075 ± 0.451 | 33.792 ± 0.511 | 35.144 ± 0.533 | 41.252 ± 0.830 | 46.067 ± 0.901 |
| CG | 139 | 3.500 ± 0.074a | 11.251 ± 0.327a | 18.034 ± 0.412A | 23.393 ± 0.459a | 28.429 ± 0.499 | 34.163 ± 0.566 | 35.019 ± 0.590 | 40.791 ± 0.919 | 46.124 ± 0.998 | ||
| GG | 46 | 3.285 ± 0.099b | 10.865 ± 0.441b | 17.535 ± 0.556B | 22.799 ± 0.619b | 28.466 ± 0.673 | 34.778 ± 0.763 | 36.051 ± 0.795 | 41.114 ± 1.240 | 45.764 ± 1.345 | ||
| XM_012179698.3: c.1154C>T | CC | 86 | 3.334 ± 0.072Aa | 10.195 ± 0.338a | 16.457 ± 0.434a | 21.671 ± 0.460 | 26.294 ± 0.526 | 31.857 ± 0.601 | 33.359 ± 0.609 | 38.990 ± 1.001 | 43.528 ± 1.082 | |
| TC | 129 | 3.268 ± 0.054ABa | 9.652 ± 0.252ab | 15.862 ± 0.323ab | 21.510 ± 0.343 | 25.795 ± 0.392 | 31.432 ± 0.448 | 33.007 ± 0.454 | 38.021 ± 0.747 | 42.794 ± 0.807 | ||
| TT | 74 | 3.080 ± 0.071Bb | 9.135 ± 0.332b | 15.088 ± 0.426b | 21.097 ± 0.452 | 25.850 ± 0.516 | 31.819 ± 0.590 | 33.042 ± 0.598 | 38.489 ± 0.983 | 43.516 ± 1.062 | ||
| LCORL | XM_027970888.1: c.-1096T>C | CC | 132 | 3.241 ± 0.056 | 9.414 ± 0.261 | 15.496 ± 0.338 | 21.131 ± 0.357 | 25.478 ± 0.408 | 31.569 ± 0.458 | 33.278 ± 0.460 | 38.874 ± 0.765 | 43.804 ± 0.841 |
| TC | 118 | 3.214 ± 0.054 | 9.740 ± 0.253 | 15.896 ± 0.328 | 21.495 ± 0.346 | 26.090 ± 0.396 | 31.507 ± 0.444 | 32.873 ± 0.446 | 37.835 ± 0.741 | 42.529 ± 0.815 | ||
| TT | 39 | 3.269 ± 0.117 | 10.145 ± 0.546 | 16.798 ± 0.708 | 22.138 ± 0.748 | 26.663 ± 0.885 | 31.811 ± 0.959 | 32.512 ± 0.963 | 37.827 ± 1.602 | 43.112 ± 1.760 | ||
| XM_027970888.1: c.2162A>C | AA | 225 | 3.219 ± 0.040 | 9.529 ± 0.187 | 15.930 ± 0.244 | 21.424 ± 0.255 | 26.010 ± 0.291 | 31.872 ± 0.327 | 33.322 ± 0.372 | 38.401 ± 0.556 | 43.261 ± 0.608 | |
| AC | 64 | 3.241 ± 0.072 | 9.732 ± 0.337 | 15.561 ± 0.441 | 21.198 ± 0.462 | 25.247 ± 0.526 | 30.770 ± 0.592 | 32.228 ± 0.592 | 37.121 ± 1.005 | 41.911 ± 1.098 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La, Y.; Zhang, X.; Li, F.; Zhang, D.; Li, C.; Mo, F.; Wang, W. Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep. Genes 2019, 10, 616. https://doi.org/10.3390/genes10080616
La Y, Zhang X, Li F, Zhang D, Li C, Mo F, Wang W. Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep. Genes. 2019; 10(8):616. https://doi.org/10.3390/genes10080616
Chicago/Turabian StyleLa, Yongfu, Xiaoxue Zhang, Fadi Li, Deyin Zhang, Chong Li, Futao Mo, and Weimin Wang. 2019. "Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep" Genes 10, no. 8: 616. https://doi.org/10.3390/genes10080616
APA StyleLa, Y., Zhang, X., Li, F., Zhang, D., Li, C., Mo, F., & Wang, W. (2019). Molecular Characterization and Expression of SPP1, LAP3 and LCORL and Their Association with Growth Traits in Sheep. Genes, 10(8), 616. https://doi.org/10.3390/genes10080616





