Characterization of Solanum melongena Thioesterases Related to Tomato Methylketone Synthase 2
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and Treatment Conditions
2.2. Identification of SmMKS2 Genes in the Eggplant Genome
2.3. cDNA Isolation and Phylogenetic Analysis of SmMKS2 Sequences
2.4. Gene Expression Analysis by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.5. Expression of SmMKS2-1, SmMKS2-2 and SmMKS2-3 in E. coli
2.6. Accession Numbers
2.7. Data Analysis
3. Results
3.1. Identification of MKS2 Genes in S. melongena
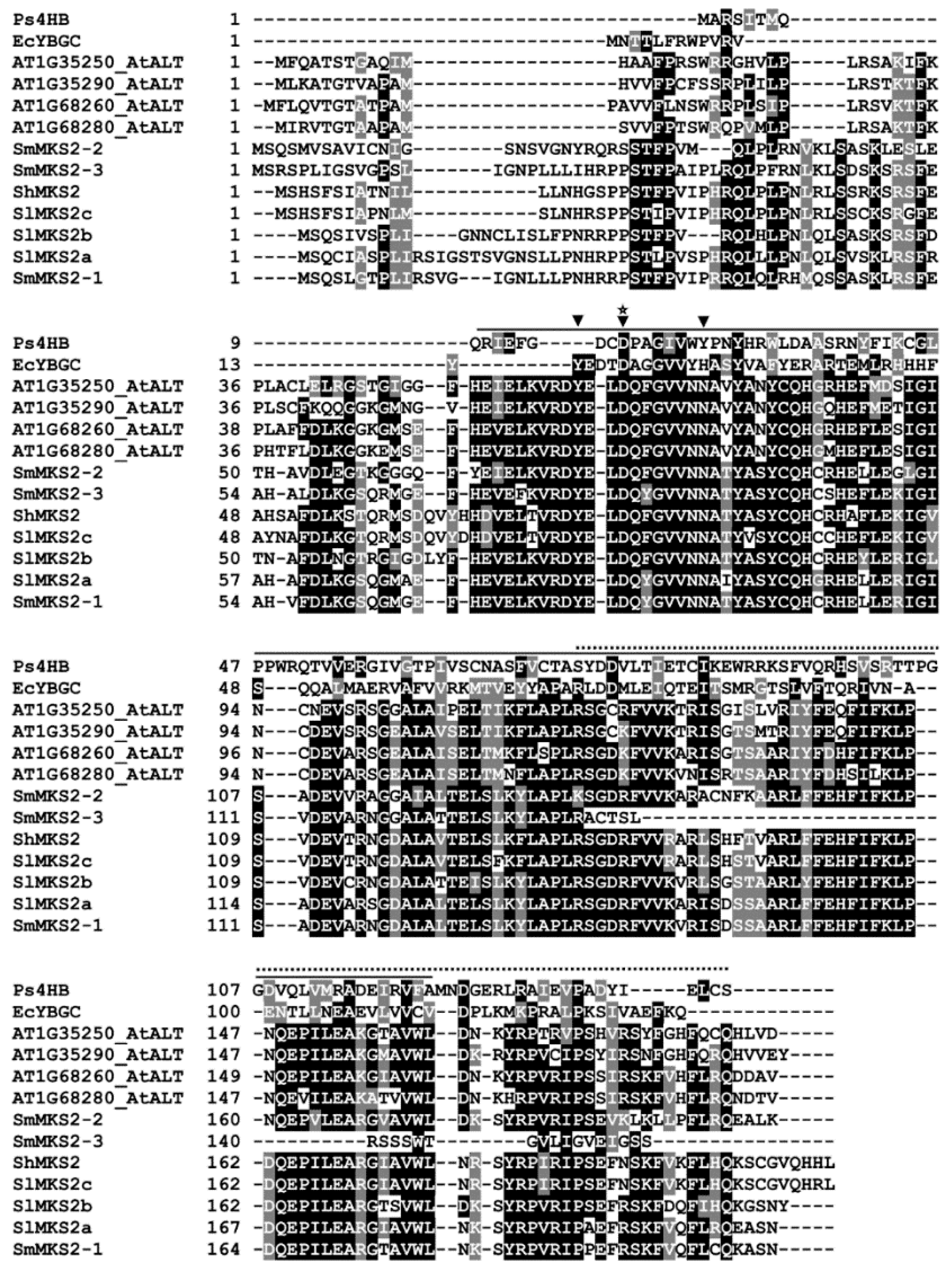
3.2. Phylogenetic Analysis of the Eggplant SmMKS2s
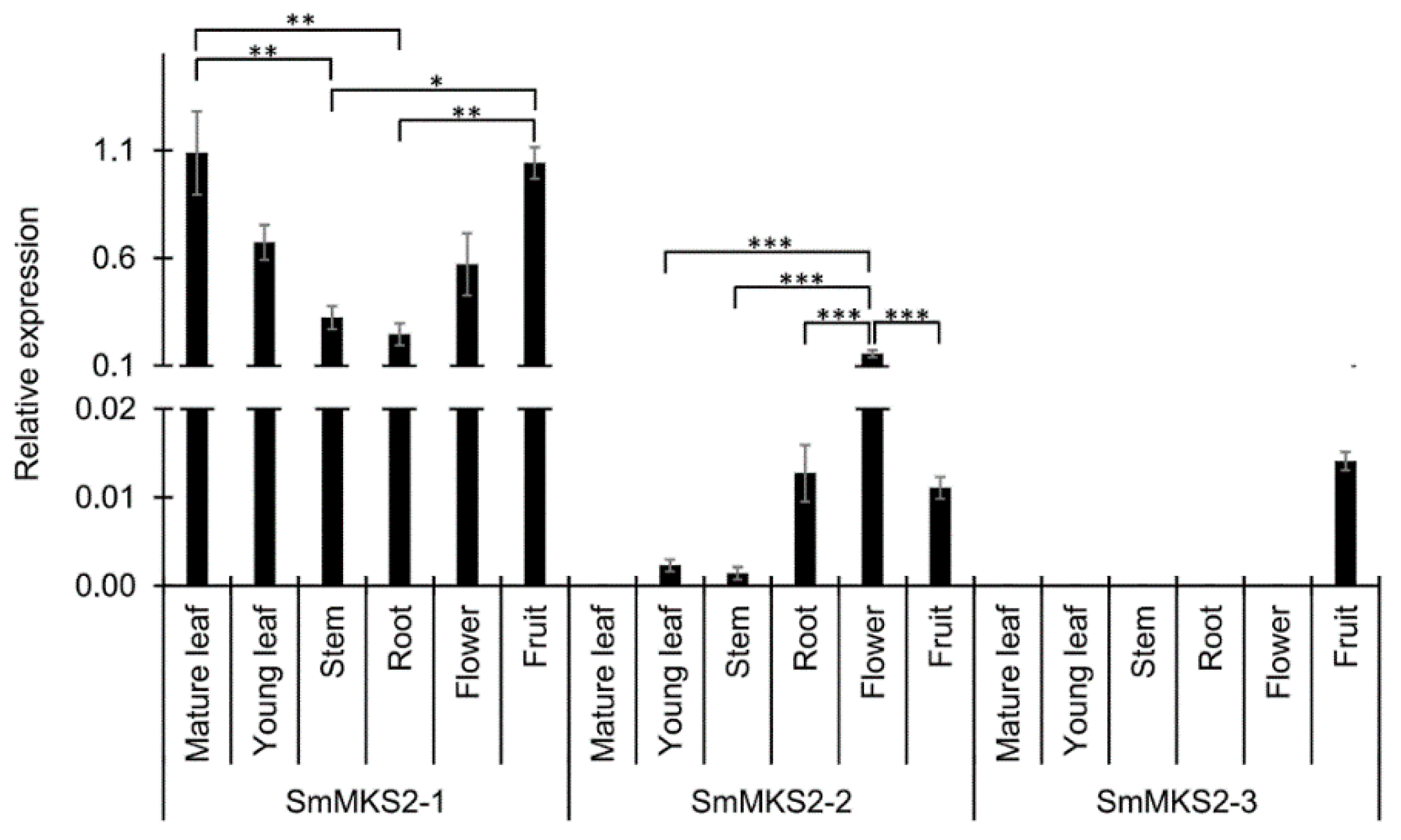
3.3. SmMKS2 Genes are Differently Expressed in Various Eggplant Organs
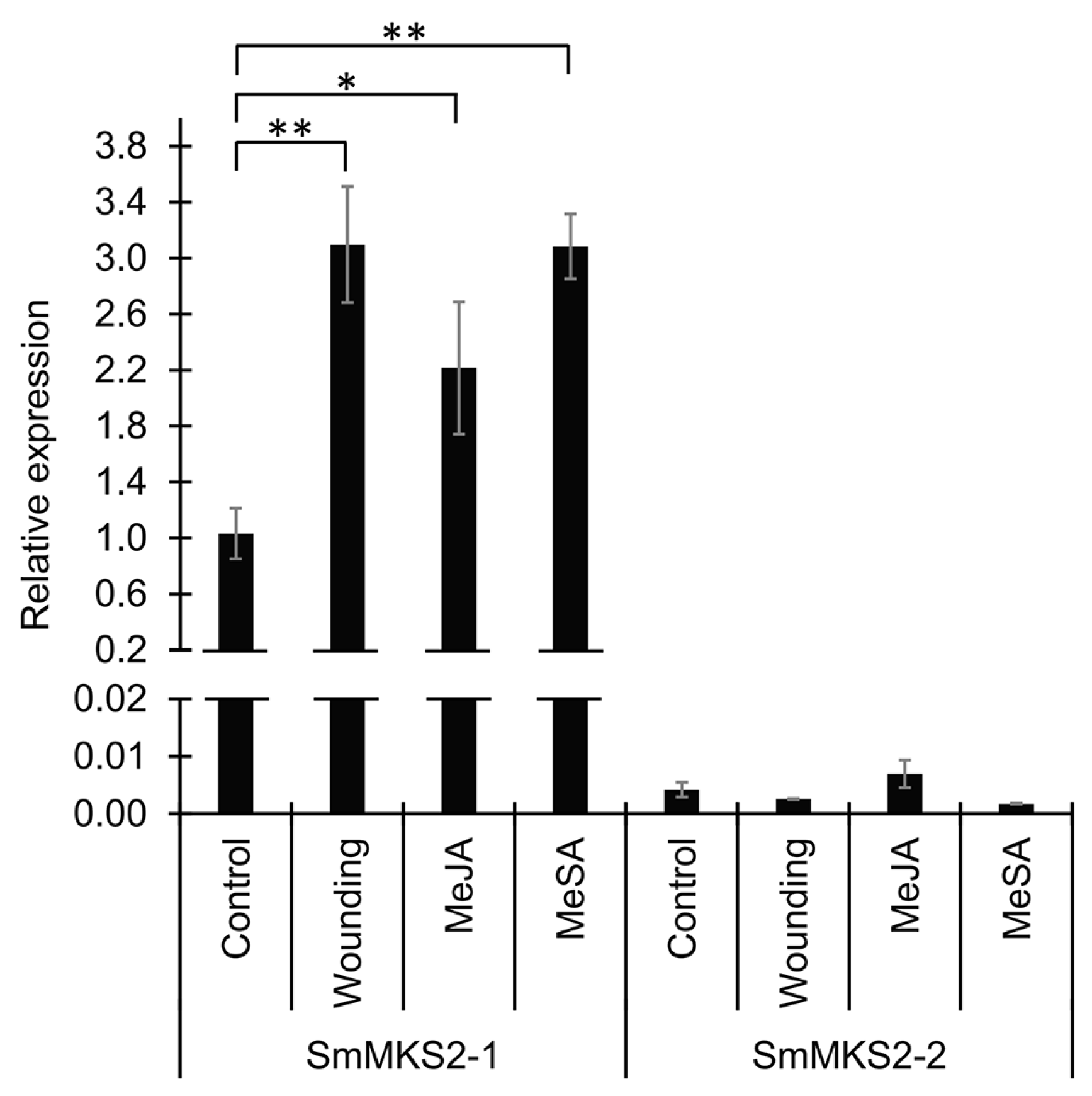
3.4. SmMKS2-1 but Neither SmMKS2-2 Nor SmMKS2-3 is Induced by Artificial Wounding, Methyl Jasmonate, and Methyl Salicylate
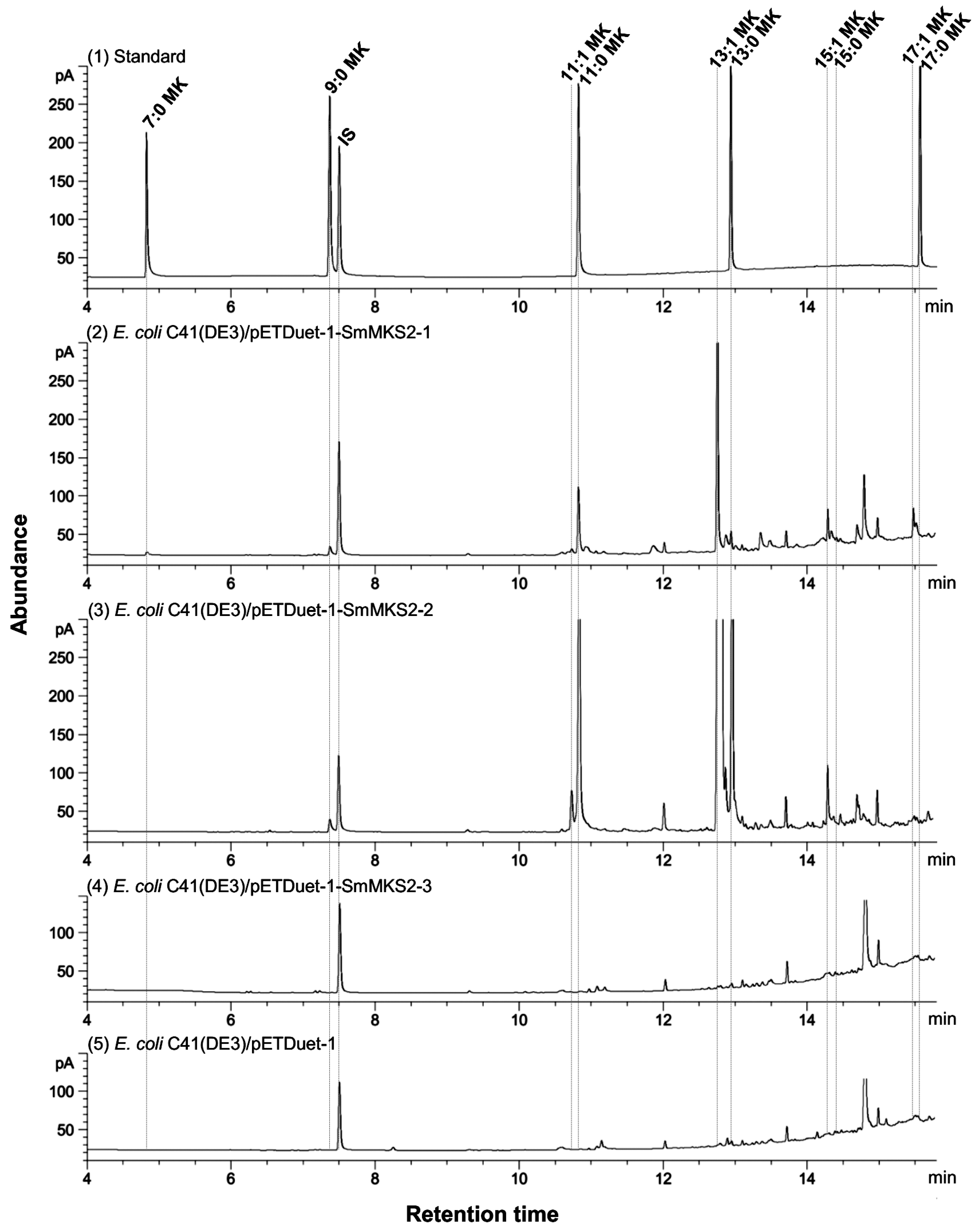
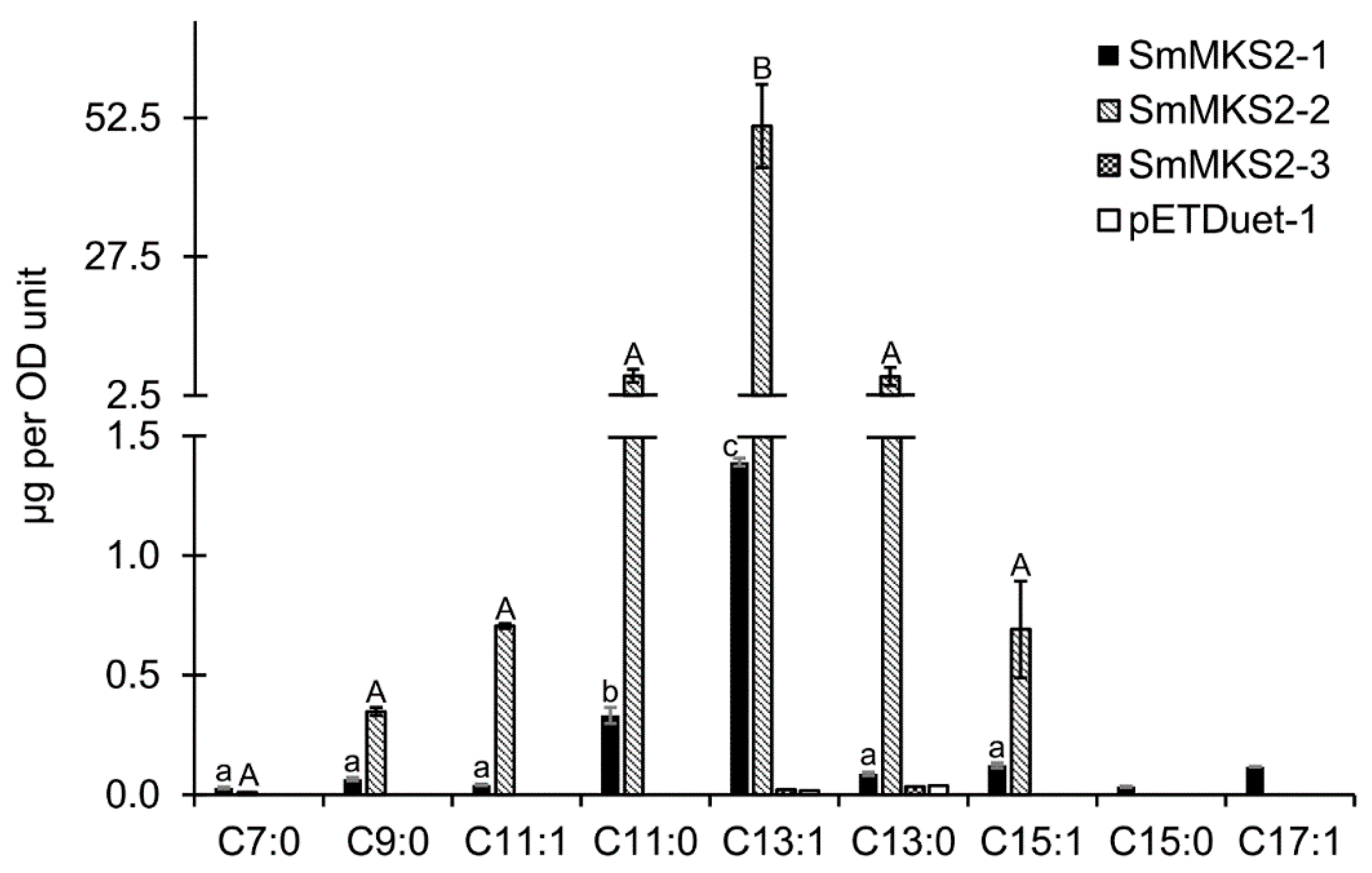
3.5. Expression of SmMKS2 Genes in E. coli Led to the Production of β-Ketoacids
4. Discussion
4.1. Biochemical Activities of SmMKS2s
4.2. Induction of SmMKS2-1 by Wounding, MeJA and MeSA Implies a Possible Biological Role for This Gene in Plant Defense
4.3. Evolution of SmMKS2 Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Antonious, G.F. Persistence of 2-tridecanone on the leaves of seven vegetables. Bull. Environ. Contam. Toxicol. 2004, 73, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Strohalm, H.; Dregus, M.; Wahl, A.; Engel, K.-H. Enantioselective Analysis of Secondary Alcohols and Their Esters in Purple and Yellow Passion Fruits. J. Agric. Food Chem. 2007, 55, 10339–10344. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.L.; Monte, F.J.Q.; Maria da Conceição, F.; de Mattos, M.C.; Lemos, T.L.; Gotor-Fernández, V.; de Gonzalo, G.; Gotor, V. Bioreduction of aromatic aldehydes and ketones by fruits’ barks of Passiflora edulis. J. Mol. Catal. B Enzym. 2008, 54, 130–133. [Google Scholar] [CrossRef]
- Goh, E.B.; Baidoo, E.E.K.; Keasling, J.D.; Beller, H.R. Engineering of bacterial methyl ketone synthesis for biofuels. Appl. Environ. Microbiol. 2012, 78, 70–80. [Google Scholar] [CrossRef]
- Hanko, E.K.R.; Denby, C.M.; Sànchez i Nogué, V.; Lin, W.; Ramirez, K.J.; Singer, C.A.; Beckham, G.T.; Keasling, J.D. Engineering β-oxidation in Yarrowia lipolytica for methyl ketone production. Metab. Eng. 2018, 48, 52–62. [Google Scholar] [CrossRef]
- Yu, G.; Nguyen, T.T.H.; Guo, Y.; Schauvinhold, I.; Auldridge, M.E.; Bhuiyan, N.; Ben-Israel, I.; Iijima, Y.; Fridman, E.; Noel, J.P.; et al. Enzymatic Functions of Wild Tomato Methylketone Synthases 1 and 2. Plant Physiol. 2010, 154, 67–77. [Google Scholar] [CrossRef]
- Pulsifer, I.P.; Lowe, C.; Narayaran, S.A.; Busuttil, A.S.; Vishwanath, S.J.; Domergue, F.; Rowland, O. Acyl-lipid thioesterase1-4 from Arabidopsis thaliana form a novel family of fatty acyl-acyl carrier protein thioesterases with divergent expression patterns and substrate specificities. Plant Mol. Biol. 2014, 84, 549–563. [Google Scholar] [CrossRef]
- Kornberg, A.; Ochoa, S.; Mehler, A.H. Spectrophotometric studies on the decarboxylation of β-keto acids. J. Biol. Chem. 1948, 174, 159–172. [Google Scholar]
- Kalinger, R.S.; Pulsifer, I.P.; Rowland, O. Elucidating the substrate specificities of acyl-lipid thioesterases from diverse plant taxa. Plant Physiol. Biochem. 2018, 127, 104–118. [Google Scholar] [CrossRef]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [PubMed]
- Hirakawa, H.; Shirasawa, K.; Miyatake, K.; Nunome, T.; Negoro, S.; Ohyama, A.; Yamaguchi, H.; Sato, S.; Isobe, S.; Tabata, S.; et al. Draft genome sequence of eggplant (Solanum melongena L.): The representative Solanum species indigenous to the old world. DNA Res. 2014, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- Aubriot, X.; Knapp, S.; Syfert, M.M.; Poczai, P.; Buerki, S. Shedding new light on the origin and spread of the brinjal eggplant (Solanum melongena L.) and its wild relatives. Am. J. Bot. 2018, 105, 1175–1187. [Google Scholar] [CrossRef]
- Weese, T.L.; Bohs, L. Eggplant origins: Out of Africa, into the Orient. Taxon 2010, 59, 49–56. [Google Scholar] [CrossRef]
- Fowler, J.H.; Narvaez-Vasquez, J.; Aromdee, D.N.; Pautot, V.; Holzer, F.M.; Walling, L.L. Leucine Aminopeptidase Regulates Defense and Wound Signaling in Tomato Downstream of Jasmonic Acid. Plant Cell Online 2009, 21, 1239–1251. [Google Scholar] [CrossRef]
- Falara, V.; Alba, J.M.; Kant, M.R.; Schuurink, R.C.; Pichersky, E. Geranyllinalool Synthases in Solanaceae and Other Angiosperms Constitute an Ancient Branch of Diterpene Synthases Involved in the Synthesis of Defensive Compounds. Plant Physiol. 2014, 166, 428–441. [Google Scholar] [CrossRef]
- Ament, K.; Van Schie, C.C.; Bouwmeester, H.J.; Haring, M.A.; Schuurink, R.C. Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 2006, 224, 1197–1208. [Google Scholar] [CrossRef]
- Yao, H.; Guo, L.; Fu, Y.; Borsuk, L.A.; Wen, T.-J.; Skibbe, D.S.; Cui, X.; Scheffler, B.E.; Cao, J.; Emrich, S.J.; et al. Evaluation of five ab initio gene prediction programs for the discovery of maize genes. Plant Molecular Biology 2005, 57, 445–460. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Benning, M.M.; Wesenberg, G.; Liu, R.; Taylor, K.L.; Dunaway-Mariano, D.; Holden, H.M. The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3. J. Biol. Chem. 1998, 273, 33572–33579. [Google Scholar] [CrossRef]
- Cantu, D.C.; Chen, Y.; Reilly, P.J. Thioesterases: A new perspective based on their primary and tertiary structures. Protein Sci. 2010, 19, 1281–1295. [Google Scholar] [CrossRef]
- Angelini, A.; Cendron, L.; Goncalves, S.; Zanotti, G.; Terradot, L. Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori. Proteins Struct. Funct. Genet. 2008, 72, 1212–1221. [Google Scholar] [CrossRef]
- Antonious, G.F.; Dahlman, D.L.; Hawkins, L.M. Insecticidal and Acaricidal Performance of Methyl Ketones in Wild Tomato Leaves. Bull. Environ. Contam. Toxicol. 2003, 71, 400–407. [Google Scholar] [CrossRef]
- Gonçalves, M.I.F.; Maluf, W.R.; Gomes, L.A.A.; Barbosa, L.V. Variation of 2-tridecanone level in tomato plant leaflets and resistance to two mite species (Tetranychus sp.). Euphytica 1998, 104, 33–38. [Google Scholar]
- Williams, W.G.; Kennedy, G.G.; Yamamoto, R.T.; Thacker, J.D.; Bordner, J. 2-Tridecanone: A naturally occurring insecticide from the wild tomato Lycopersicon hirsutum f. glabratum. Science 1980, 207, 888–889. [Google Scholar] [CrossRef]
- Ben-Israel, I.; Yu, G.; Austin, M.B.; Bhuiyan, N.; Auldridge, M.; Nguyen, T.; Schauvinhold, I.; Noel, J.P.; Pichersky, E.; Fridman, E. Multiple Biochemical and Morphological Factors Underlie the Production of Methylketones in Tomato Trichomes. Plant Physiol. 2009, 151, 1952–1964. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Guo, Y.; Austin, M.B.; Ramsey, J.; Fridman, E.; Pichersky, E.; Noel, J.P. Emergent Decarboxylase Activity and Attenuation of α/β-Hydrolase Activity during the Evolution of Methylketone Biosynthesis in Tomato. Plant Cell 2012, 24, 1596–1607. [Google Scholar] [CrossRef]
- Yu, G.; Pichersky, E. Heterologous Expression of Methylketone Synthase1 and Methylketone Synthase2 Leads to Production of Methylketones and Myristic Acid in Transgenic Plants. Plant Physiol. 2014, 164, 612–622. [Google Scholar] [CrossRef]
- Van Poecke, R.M.P.; Posthumus, M.A.; Dicke, M. Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: Chemical, behavioral, and gene-expression analysis. J. Chem. Ecol. 2001, 27, 1911–1928. [Google Scholar] [CrossRef]
- Mayer, K.M.; Shanklin, J. Identification of amino acid residues involved in substrate specificity of plant acyl-ACP thioesterases using a bioinformatics-guided approach. BMC Plant Biol. 2007, 7, 1. [Google Scholar] [CrossRef]
- Jones, A.; Davies, H.M.; Voelker, T.A. Palmitoyl-Acyl Carrier Protein (ACP) Thioesterase and the Evolutionary Origin of Plant Acyl-ACP Thioesterases. Plant Cell 2007, 7, 359. [Google Scholar]
- Choudhary, B.; Gaur, K. The development and regulation of Bt Brinjal in India (Eggplant/Aubergine). ISAAA Brief 38 2009. [Google Scholar]
- Knudsen, J.T.; Tollsten, L.; Bergström, L.G. Floral scents-a checklist of volatile compounds isolated by head-space techniques. Phytochemistry 1993, 33, 253–280. [Google Scholar] [CrossRef]
- Flath, R.A.; Ohinata, K. Volatile components of the orchid Dendrobium superbum Rchb. f. J. Agric. Food Chem. 1982, 30, 841–842. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khuat, V.L.U.; Bui, V.T.T.; Tran, H.T.D.; Truong, N.X.; Nguyen, T.C.; Mai, P.H.H.; Dang, T.L.A.; Dinh, H.M.; Pham, H.T.A.; Nguyen, T.T.H. Characterization of Solanum melongena Thioesterases Related to Tomato Methylketone Synthase 2. Genes 2019, 10, 549. https://doi.org/10.3390/genes10070549
Khuat VLU, Bui VTT, Tran HTD, Truong NX, Nguyen TC, Mai PHH, Dang TLA, Dinh HM, Pham HTA, Nguyen TTH. Characterization of Solanum melongena Thioesterases Related to Tomato Methylketone Synthase 2. Genes. 2019; 10(7):549. https://doi.org/10.3390/genes10070549
Chicago/Turabian StyleKhuat, Vy Le Uyen, Vi Thi Tuong Bui, Huong Thi Diem Tran, Nuong Xuan Truong, Thien Chi Nguyen, Phuc Huynh Hanh Mai, Tuan Le Anh Dang, Hiep Minh Dinh, Hong Thi Anh Pham, and Thuong Thi Hong Nguyen. 2019. "Characterization of Solanum melongena Thioesterases Related to Tomato Methylketone Synthase 2" Genes 10, no. 7: 549. https://doi.org/10.3390/genes10070549
APA StyleKhuat, V. L. U., Bui, V. T. T., Tran, H. T. D., Truong, N. X., Nguyen, T. C., Mai, P. H. H., Dang, T. L. A., Dinh, H. M., Pham, H. T. A., & Nguyen, T. T. H. (2019). Characterization of Solanum melongena Thioesterases Related to Tomato Methylketone Synthase 2. Genes, 10(7), 549. https://doi.org/10.3390/genes10070549



