DNA Authentication of St John’s Wort (Hypericum perforatum L.) Commercial Products Targeting the ITS Region
Abstract
:1. Introduction
- Time, expense and expertise required for DNA sequencing and analysis.
- Inability to detect adulterant/contaminant material.
- Limited use with degraded DNA.
- Application to degraded samples due to the dramatically reduced amplicon size.
- No requirement to sequence the amplicons.
- No requirement to analyse sequencing data.
- All results from one machine, and one operator.
2. Materials and Methods
2.1. Sample Collection and Preparation
2.1.1. Commercial Products
2.1.2. Sample Preparation
2.2. DNA Extraction
2.3. DNA Quantitation
2.4. Conventional PCR and Gel Electrophoresis
2.5. qPCR Assays with Specific & Generic Primer Pairs
2.5.1. Protocol 1
2.5.2. Protocol 2
2.5.3. Analysis of qPCR Results
2.6. NGS Methods
2.7. Bioinformatics
2.7.1. Curated Database of the ITS Region of Significant Hypericum Species
- Close relatives of H. perforatum.
- Common in commercial trade as ornamental or medicinal plants.
- Reported to be found as adulterants of H. perforatum.
2.7.2. Next-Generation Sequencing Amplicon Processing
3. Results
3.1. Isolation and Characterisation of DNA Extracted from Commercial Products
3.2. New Primer Design for Specific PCR Testing
- Is the detection of fragmented DNA supported by other assays?
- Is it possible to identify the species of origin of these DNA fragments?
- Could this information be used to authenticate processed herbal products?
- The efficiency of these primers for qPCR was found to be poor, typically around 0.80.
- The specificity of the primers was broader than could be known when they were first designed. There are many more Hypericum ITS sequences now in GenBank than were present before 2009, and a BLAST search of the database with the FO2 and HRI-S primers gives 100% matches with accessions from several Hypericum species. In addition, the primers were found not to be a perfect match to the H. perforatum ITS haplotype 1, so could give both false positive and false negative signals.
3.2.1. Design of H. perforatum-Specific Primers
- They are close relatives of H. perforatum.
- They are common in commercial trade as ornamental or medicinal plants.
3.2.2. Development of a Quantitative Assay for H. perforatum DNA Fragments
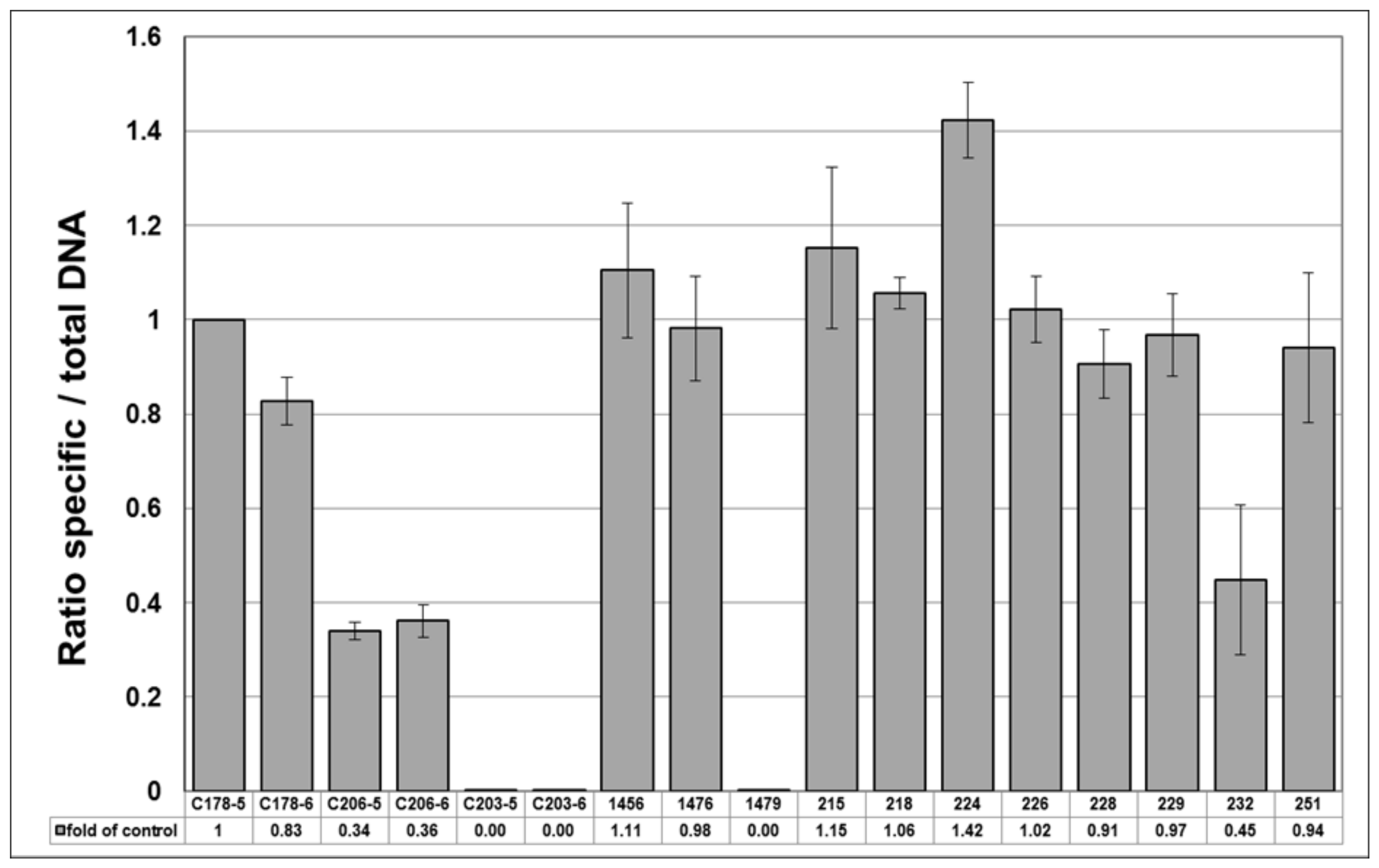
- An absolute method of ITS sequence quantitation with the HypG primers using a standard curve of a synthetic template of known copy number. The synthetic H. perforatum standard C22 was used to create a standard curve by serial 10-fold dilutions over the range S−3–S−8 (10–0.0001 pg µL−1). Direct conversion of mean Cq value for each sample from the standard curve allowed the number of ITS copies to be calculated (Table 3). The three reference standards C178, C203 and C206 each showed very good correspondence between the copy numbers calculated for the 0.01 and 0.001 pg µL−1 dilutions.
- The relative method of Pfaffl [38] was used to calculate the ratio of specific H. perforatum ITS sequences to total ITS sequences, with the generic HypG primers acting as the “reference gene” and the C178 standard used to calibrate the ratio to 1.0. As expected, the values for the two H. perforatum genomic DNA samples 1456 and 1476 were close to 1.0 (Table 3, Figure 3). The H. patulum standard C203 and genomic DNA 1479 gave ratios of 0.00, indicating that none of the ITS sequences present were from H. perforatum. However, the H. maculatum standard C206 showed a ratio of 0.37–0.39. This could be indicative of a significant cross-reaction with the 460–650 primers, suggesting that the reaction requires further optimisation to ensure that mismatch priming does not occur with these primers.
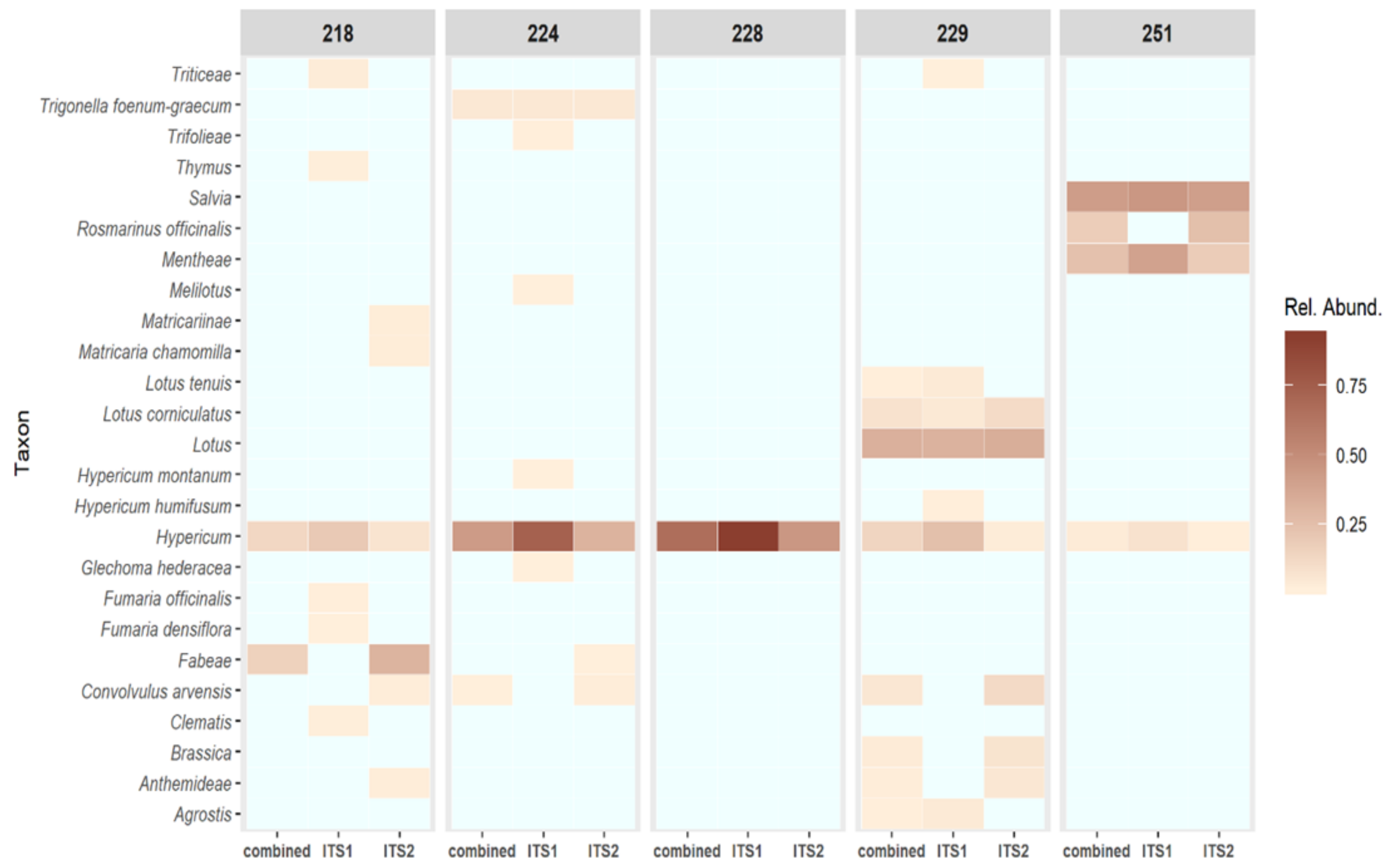
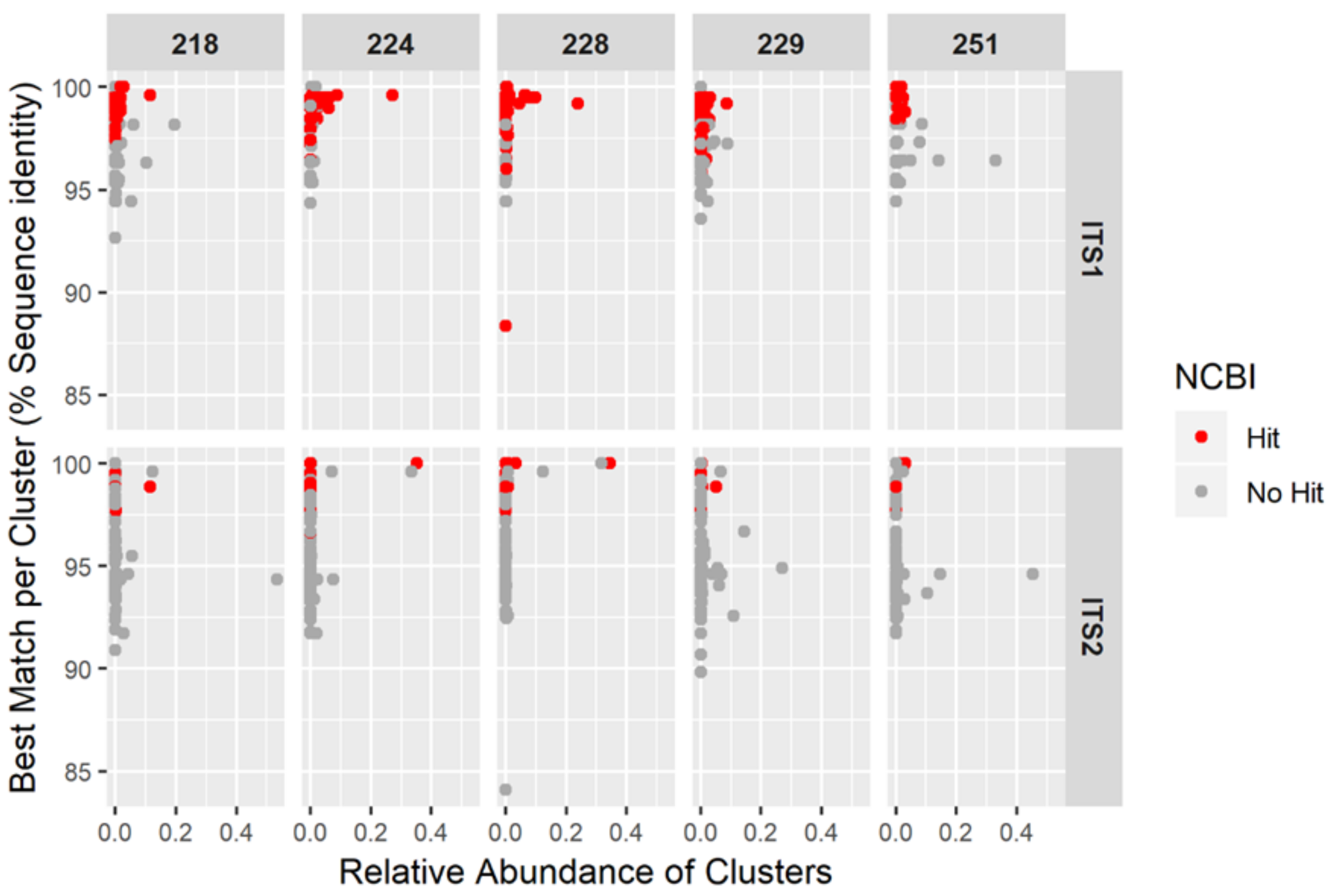
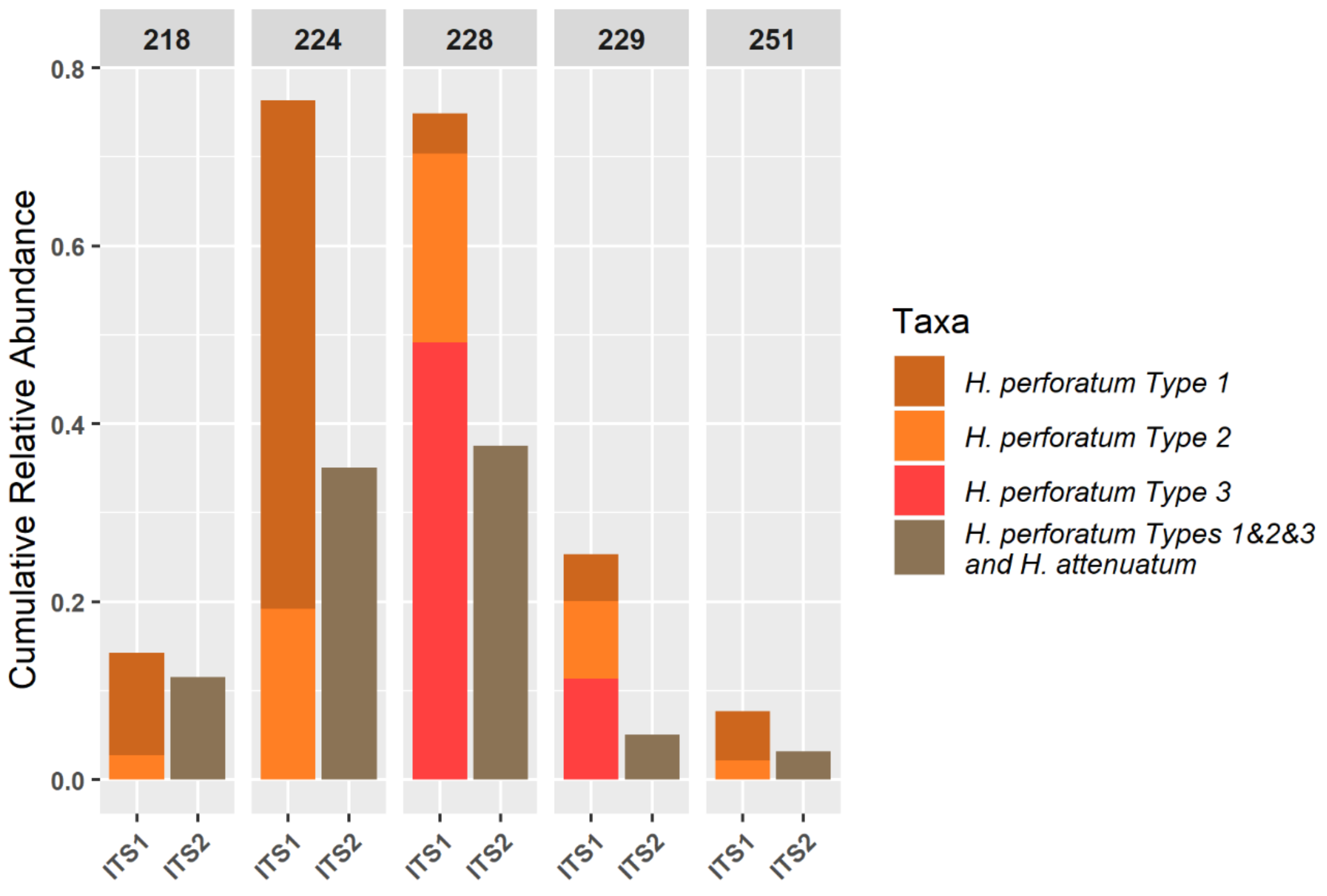
3.3. NGS Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, T.; Kawa, K.; Eckl, V.; Morton, C.; Stredney, R. Herbal Supplement Sales in US Increased 8.5% in 2017, Topping $8 Billion. HerbalGram 2018, 119, 62–71. [Google Scholar]
- Bilia, A.R. Science meets regulation. J. Ethnopharmacol. 2014, 158, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Coghlan, M.L.; Haile, J.; Houston, J.; Murray, D.C.; White, N.E.; Moolhuijzen, P.; Bellgard, M.I.; Bunce, M. Deep sequencing of plant and animal DNA contained within traditional Chinese medicines reveals legality issues and health safety concerns. PLoS Genet. 2012, 8, e1002657. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Leon, C.; Yu-Lin, L. Chinese Medicinal Plants, Herbal Drugs and Substitutes an Identification Guide; Royal Botanic Gardens: Richmond, UK, 2017. [Google Scholar]
- Heinrich, M. Quality and safety of herbal medical products: Regulation and the need for quality assurance along the value chains. Br. J. Clin. Pharmacol. 2015, 80, 62–66. [Google Scholar] [CrossRef]
- Agapouda, A.; Booker, A.; Kiss, T.; Hohmann, J.; Heinrich, M.; Csupor, D. Quality control of Hypericum perforatum L. analytical challenges and recent progress. J. Pharm. Pharmacol. 2017, 71, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.; Anderson, L.A.; Phillipson, J.D. St John’s wort (Hypericum perforatum L.): A review of its chemistry, pharmacology and clinical properties. J. Pharm. Pharmacol. 2001, 53, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Berghöfer, R.; Hölzl, J. Johanniskraut (Hypericum perforatum L.)–Prüfung auf Verfälschung. Dtsch. Apoth. Ztg. 1986, 47, 2569–2573. [Google Scholar]
- Costa, J.; Campos, B.; Amaral, J.S.; Nunes, M.E.; Oliveira, M.B.; Mafra, I. HRM analysis targeting ITS1 and matK loci as potential DNA mini-barcodes for the authentication of Hypericum perforatum and Hypericum androsaemum in herbal infusions. Food Control 2016, 61, 105–114. [Google Scholar]
- Crockett, S.L.; Douglas, A.W.; Scheffler, B.E.; Khan, I.A. Genetic profiling of Hypericum (St. John’s Wort) species by nuclear ribosomal ITS sequence analysis. Planta Med. 2004, 70, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Crockett, S.L.; Robson, N.K.B. Taxonomy and Chemotaxonomy of the Genus Hypericum. Med. Aromat. Plant Sci. Biotechnol. 2011, 5, 1–13. [Google Scholar]
- Howard, C.; Bremner, P.D.; Fowler, M.R.; Isodo, B.; Scott, N.W.; Slater, A. Molecular identification of Hypericum perforatum by PCR amplification of the ITS and 5.8 S rDNA region. Planta Med. 2009, 75, 864–869. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Kuzmina, M.L.; Braukmann, T.W.; Borisenko, A.V.; Zakharov, E.V. Authentication of Herbal Supplements Using Next-Generation Sequencing. PLoS ONE 2016, 11, e0156426. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, M. Hypericum Perforatum Botanical Adulterants Bulletin January 2017. Available online: www.botanicaladulterants.org (accessed on 6 April 2019).
- Raclariu, A.C.; Paltinean, R.; Vlase, L.; Labarre, A.; Manzanilla, V.; Ichim, M.C.; Crisan, G.; Brysting, A.K.; de Boer, H. Comparative authentication of Hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci. Rep. 2017, 7, 1291. [Google Scholar] [CrossRef] [PubMed]
- Commission, B.P. Deoxyribonucleic acid (DNA) based identification techniques for herbal drugs. British Pharmacopoeia Appendix XI V. In British Pharmacopoeia; TSO: London, UK, 2016. [Google Scholar]
- Kazi, T.; Hussain, N.; Bremner, P.; Slater, A.; Howard, C. The application of a DNA-based identification technique to over-the-counter herbal medicines. Fitoterapia 2013, 87, 27–30. [Google Scholar] [CrossRef]
- Sgamma, T.; Lockie-Williams, C.; Kreuzer, M.; Williams, S.; Scheyhing, U.; Koch, E.; Slater, A.; Howard, C. DNA barcoding for industrial quality assurance. Planta Med. 2017, 14, 8–21. [Google Scholar] [CrossRef]
- Masiero, E.; Banik, D.; Abson, J.; Greene, P.; Slater, A.; Sgamma, T. Genus-Specific Real-Time PCR and HRM Assays to Distinguish Liriope from Ophiopogon Samples. Plants 2017, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- De Boer, H.J.; Ichim, M.C.; Newmaster, S.G. DNA Barcoding and Pharmacovigilance of Herbal Medicines. Drug Saf. 2015, 38, 611–620. [Google Scholar] [CrossRef]
- Raclariu, A.C.; Heinrich, M.; Ichim, M.C.; de Boer, H. Benefits and Limitations of DNA Barcoding and Metabarcoding in Herbal Product Authentication. Phytochem. Anal. 2018, 29, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Xu, Z.; Jia, J.; Leon, C.; Hu, S.; Lin, Y.; Ragupathy, S.; Song, J.; Newmaster, S.G. Biomonitoring for traditional herbal medicinal products using DNA metabarcoding and single molecule, real-time sequencing. Acta Pharm. Sin. B 2018, 8, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore! Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 6 April 2019).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 April 2019).
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R. UCHIME2: Improved chimera prediction for amplicon sequencing. bioRxiv 2016, 2016, 074252. [Google Scholar]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wickham, H. Tidyverse: Easily Install and Load “Tidyverse” Packages; R Package Version 1; RStudio: Boston, MA, USA, 2017. [Google Scholar]
- Mitra, S.; Kannan, R. A note on unintentional adulterations in Ayurvedic herbs. Ethnobot. Leaflets 2007, 2007, 3. [Google Scholar]
- Nürk, N.M.; Madriñán, S.; Carine, M.A.; Chase, M.W.; Blattner, F.R. Molecular phylogenetics and morphological evolution of St. John’s wort (Hypericum; Hypericaceae). Mol. Phylogenet. Evol. 2013, 66, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Sgamma, T.; Masiero, E.; Mali, P.; Mahat, M.; Slater, A. Sequence-Specific Detection of Aristolochia DNA—A Simple Test for Contamination of Herbal Products. Front. Plant Sci. 2018, 9, 1828. [Google Scholar] [CrossRef]
- Sgamma, T.; Masiero, E.; Mali, P.; Mahat, M.; Slater, A. Forensic botany and forensic chemistry working together: Application of plant DNA barcoding as a complement to forensic chemistry—A case study in Brazil, 2019. Genome 2019, 62, 11–18. [Google Scholar]
- Mohammed Abubakar, B.; Mohd Salleh, F.; Shamsir Omar, M.S.; Wagiran, A. Review: DNA barcoding and chromatography fingerprints for the authentication of botanicals in herbal medicinal products, 2017. Evid.-Based Complement. Altern. Med. 2017, 2017, 1352948. [Google Scholar] [CrossRef]
- Frigerio, J.; Gorini, T.; Galimberti, A.; Bruni, I.; Tommasi, N.; Mezzasalma, V.; Labra, M. DNA barcoding to trace Medicinal and Aromatic Plants from the field to the food supplement. J. Appl. Bot. Food Qual. 2019, 92, 33–38. [Google Scholar]
- Newmaster, S.G.; Shanmughanandhan, D.; Kesanakurti, P.; Shehata, H.; Faller, A.; Noce, I.D.; Lee, J.Y.; Rudzinski, P.; Lu, Z.; Zhang, Y.; et al. Recommendations for Validation of Real-Time PCR Methods for Molecular Diagnostic Identification of Botanicals. J. AOAC Int. 2019, 102. [Google Scholar] [CrossRef] [PubMed]
- Seethapathy, G.S.; Raclariu-Manolica, A.C.; Anmarkrud, J.A.; Wangensteen, H.; De Boer, H.J. DNA Metabarcoding Authentication of Ayurvedic Herbal Products on the European Market Raises Concerns of Quality and Fidelity. Front. Plant Sci. 2019, 10, 68. [Google Scholar] [CrossRef] [PubMed]
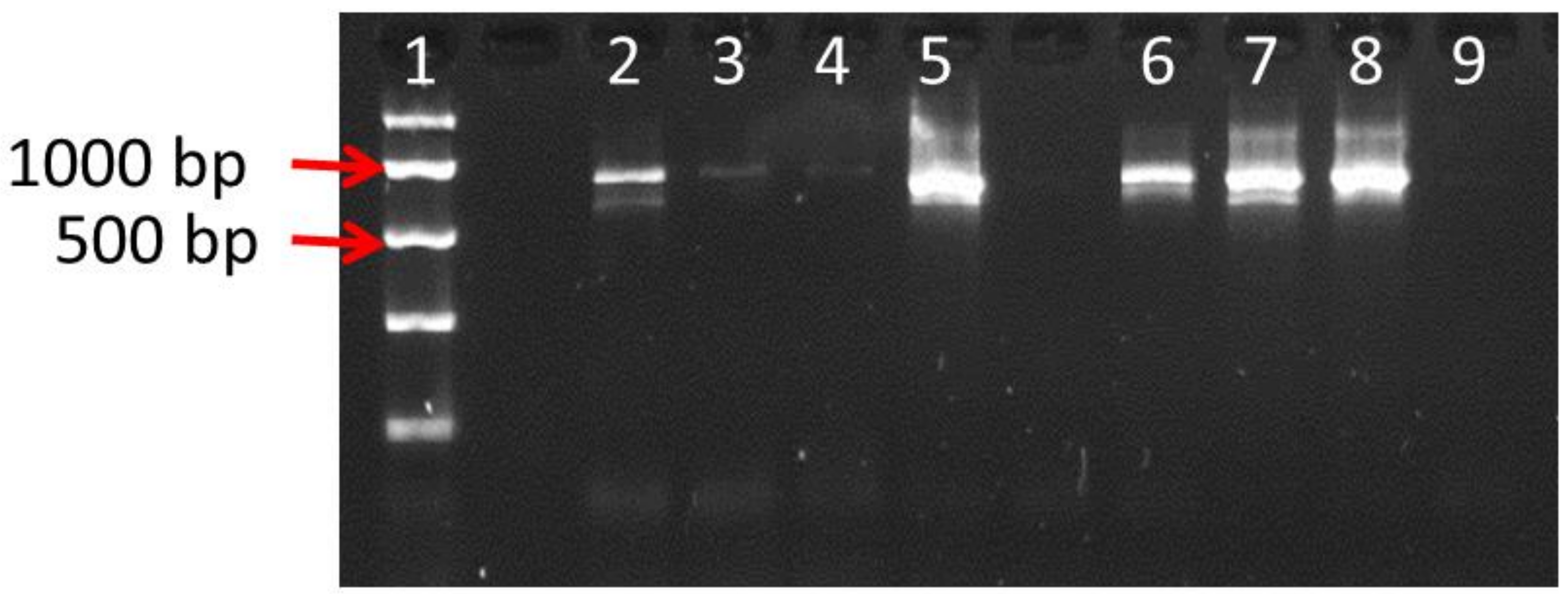

| Sample Number | Type | Amount/Unit | Drug Extract Ratio and Information | THR |
|---|---|---|---|---|
| 211 | Tab | 425 mg extract | 5–7:1 (equivalent to 2135–2975 mg SJW). 60% Ethanol extraction. | Y |
| 213 | Tab | 334 mg extract | 5–7:1 (equivalent to 1670–2338 mg SJW). 60% Ethanol extraction. | Y |
| 215 | Cap | 142 mg extract | Equivalent to 711–995 mg SJW. 60% Ethanol extraction. | Y |
| 218 | Tea bag | 1.5 g SJW | ||
| 221 | Cap | 450 mg | Standardised to 0.3% hypericin (1.35 mg) | |
| 223 | Tab | 500 mg SJW powder | Standardised to 0.3% total hypericins | |
| 224 | Cap | 200 mg SJW powder | (Brown rice flour filler stated) | |
| 226 | Cap | 300 mg | Standardised to 0.3% hypericin (0.9 mg) | |
| 228 | Cap | 500 mg SJW | 0.02% hypericin | |
| 229 | Tea bag | 2 g SJW | ||
| 230 | Tab | 425 mg extract | 3.5–6:1 (equivalent to 1490–2550 mg SJW). 60% ethanol extraction. | Y |
| 232 | Tab | 40–73 mg extract | 3.1–4.0:1. 60% ethanol extraction. | Y |
| 233 | Tab | 333 mg | Standardised to 0.3% hypericin (1000 µg) | |
| 234 | Tab | 1000 mg | 5:1 = 5000 mg dried leaf | |
| 3000 µg hypericin | ||||
| 235 | Tab | 300 mg SJW | ||
| 244 | Cap | 350 mg SJW | ||
| 245 | Cap | 300 mg | Standardised to 0.3% hypericin (0.9 mg) | |
| 250 | Cap | 300 mg | Standardised to 0.3% hypericin (0.9 mg) | |
| 251 | Tab | 315 mg powder | Standardised to 0.3% hypericin | |
| 222 | Tab | 34 mg motherwort extract | Leonorus cardiac, Humulus lupulus, Passifloroa incarnata, Lactuca virosa, Valeriana officinalis. 60% ethanol extraction. |
| Sample Number | ng DNA per mg Sample | Traditional Barcoding | qPCR Cq Values | ||
|---|---|---|---|---|---|
| ITS Band | ITS BLAST ID | HypGF/HypGR | FO2/HRI-S | ||
| 211 * | 21.0 | - | 35 | 35 | |
| 213 * | 14.5 | - | 37 | 35 | |
| 215 | 37.5 | + | 35 | 34 | |
| 218 | 36.5 | ++ | H. perf (99%) | 18 | 17 |
| 221 * | 18.5 | - | 34 | 30 | |
| 223 * | 23.0 | - | 37 | 37 | |
| 224 | 29.6 | + | 19 | 17 | |
| 226 | 13.5 | - | 27 | 25 | |
| 228 | 158.5 | ++ | H. perf (99%) | 16 | 17 |
| 229 | 100.5 | ++ | H. perf (99%) | 16 | 16 |
| 230 * | 44.5 | - | 35 | 34 | |
| 232 | 0.0 | - | 32 | 31 | |
| 233 * | 108.0 | - | 38 | N/A | |
| 234 * | 160.5 | - | N/A | N/A | |
| 235 * | 46.5 | - | 37 | N/A | |
| 244 * | 150.0 | - | N/A | N/A | |
| 245 | 3.0 | - | 37 | 33 | |
| 250 | 3.5 | - | 28 | N/A | |
| 251 | 1.5 | - | 28 | 25 | |
| 222 | 120.5 | + | Mixed sample | 28 | 36 |
| Sample Number | Sequence Origin | Type | 460–650 | SD | HypG | SD | ITS Copies/µL × 103 | Ratio Spec/Tot |
|---|---|---|---|---|---|---|---|---|
| C178-5 | H. perforatum (type 1) | Std | 19.96 | 0.017 | 17.43 | 0.022 | 31.3 | 1.00 |
| C178-6 | Std | 23.84 | 0.068 | 20.87 | 0.084 | 3.2 | 0.83 | |
| C206-5 | H. maculatum | Std | 20.27 | 0.069 | 16.22 | 0.047 | 69.6 | 0.34 |
| C206-6 | Std | 23.95 | 0.176 | 19.82 | 0.078 | 6.4 | 0.36 | |
| C203-5 | H. patulum | Std | 35.8 | 1.748 | 16.77 | 0.503 | 50.4 | 0.00 |
| C203-6 | Std | >40 | ND | 20.18 | 0.082 | 5.1 | 0.00 | |
| 1456 | H. perforatum | Plant | 16.27 | 0.108 | 14.03 | 0.234 | 296.8 | 1.11 |
| 1476 | H. perforatum | Plant | 13.68 | 0.025 | 11.40 | 0.152 | 1692.1 | 0.98 |
| 1479 | H. patulum | Plant | 29.65 | 0.329 | 12.60 | 0.053 | 764.1 | 0.00 |
| Sample Number | 460–650 | SD | HypG | SD | ITS Copies/µL × 103 | Ratio Spec/Tot |
|---|---|---|---|---|---|---|
| 215 | 29.70 | 0.248 | 26.92 | 0.094 | 0.059 | 1.15 |
| 218 | 19.86 | 0.075 | 17.41 | 0.031 | 31.7 | 1.06 |
| 224 | 19.30 | 0.080 | 17.29 | 0.055 | 34.2 | 1.42 |
| 226 | 25.85 | 0.147 | 23.08 | 0.175 | 0.747 | 1.02 |
| 228 | 15.42 | 0.110 | 12.95 | 0.009 | 603.0 | 0.91 |
| 229 | 20.79 | 0.130 | 18.17 | 0.061 | 19.2 | 0.97 |
| 232 | 35.14 | 0.416 | 30.75 | 0.425 | 0.005 | 0.45 |
| 245 | 34.59 * | 0.011 | 32.60 | 1.679 | 0.002 | ND |
| 250 | 36.19 * | ND | 34.68 * | 3.452 | 0.001 | ND |
| 251 | 23.92 | 0.172 | 21.11 | 0.103 | 2.74 | 0.94 |
| Sample | Region | Total Reads in Clusters | Number of Clusters | Reads per DNA Barcode Not Assigned (%) | Reads per Sample Not Assigned (%) | Hypericum Reads per Sample (%) |
|---|---|---|---|---|---|---|
| 218 | ITS1 | 147,659 | 243 | 54.80 | 0.49 | 14.22 |
| ITS2 | 162,586 | 134 | 42.87 | |||
| 224 | ITS1 | 63,271 | 147 | 2.54 | 0.34 | 44.27 |
| ITS2 | 112,617 | 257 | 45.73 | |||
| 228 | ITS1 | 133,583 | 333 | 1.12 | 0.26 | 67.93 |
| ITS2 | 158,767 | 469 | 46.34 | |||
| 229 | ITS1 | 154,781 | 260 | 8.28 | 0.10 | 16.42 |
| ITS2 | 150,085 | 217 | 12.63 | |||
| 251 | ITS1 | 75,880 | 121 | 2.24 | 0.03 | 4.34 |
| ITS2 | 58,531 | 61 | 3.71 |
| Sample Number | qPCR Ratio Specific: Total | NGS Results | ITS Copies/µL × 103 |
|---|---|---|---|
| 215 | 1.15 | n/a | 0.059 |
| 226 | 1.02 | n/a | 0.747 |
| 232 | 0.45 | n/a | 0.005 |
| 245 | n/a | n/a | 0.002 |
| 250 | n/a | n/a | 0.001 |
| 218 | 1.06 | Good quality, some background | 31.7 |
| 224 | 1.42 | Good quality, some background | 34.2 |
| 228 | 0.91 | Pure | 603.0 |
| 229 | 0.97 | Some Hypericum—strong Lotus signal | 19.2 |
| 251 | 0.94 | Some Hypericum—strong Salvia, Menthae and Rosmarinus signals | 2.74 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howard, C.; Hill, E.; Kreuzer, M.; Mali, P.; Masiero, E.; Slater, A.; Sgamma, T. DNA Authentication of St John’s Wort (Hypericum perforatum L.) Commercial Products Targeting the ITS Region. Genes 2019, 10, 286. https://doi.org/10.3390/genes10040286
Howard C, Hill E, Kreuzer M, Mali P, Masiero E, Slater A, Sgamma T. DNA Authentication of St John’s Wort (Hypericum perforatum L.) Commercial Products Targeting the ITS Region. Genes. 2019; 10(4):286. https://doi.org/10.3390/genes10040286
Chicago/Turabian StyleHoward, Caroline, Eleanor Hill, Marco Kreuzer, Purvi Mali, Eva Masiero, Adrian Slater, and Tiziana Sgamma. 2019. "DNA Authentication of St John’s Wort (Hypericum perforatum L.) Commercial Products Targeting the ITS Region" Genes 10, no. 4: 286. https://doi.org/10.3390/genes10040286
APA StyleHoward, C., Hill, E., Kreuzer, M., Mali, P., Masiero, E., Slater, A., & Sgamma, T. (2019). DNA Authentication of St John’s Wort (Hypericum perforatum L.) Commercial Products Targeting the ITS Region. Genes, 10(4), 286. https://doi.org/10.3390/genes10040286





