Abstract
Plant genomes are punctuated by repeated bouts of proliferation of transposable elements (TEs), and these mobile bursts are followed by silencing and decay of most of the newly inserted elements. As such, plant genomes reflect TE-related genome expansion and shrinkage. In general, these genome activities involve two mechanisms: small RNA-mediated epigenetic repression and long-term mutational decay and deletion, that is, genome-purging. Furthermore, the spatial relationships between TE insertions and genes are an important force in shaping gene regulatory networks, their downstream metabolic and physiological outputs, and thus their phenotypes. Such cascading regulations finally set up a fitness differential among individuals. This brief review demonstrates factual evidence that unifies most updated conceptual frameworks covering genome size, architecture, epigenetic reprogramming, and gene expression. It aims to give an overview of the impact that TEs may have on genome and adaptive evolution and to provide novel insights into addressing possible causes and consequences of intimidating genome sizes (20–30 Gb) in a taxonomic group, conifers.
1. Introduction
Gymnosperms today comprise a little more than 1000 species that are two to three orders of magnitude lower than c. 352,000 species of extant angiosperms [1]. Yet, gymnosperms have a long and extensive fossil record that dates back to the Carboniferous (c. 290 million years ago (mya)) [2,3]. By contrast, morphologically recognizable fossil angiosperms first appeared more recently (in the lower Cretaceous; c. 130 mya), and fossil evidence of their rise to ecological dominance emerged in the mid-Cretaceous (c. 100 mya) [4,5]. The sudden appearance and rapid diversification of angiosperms in the fossil records were, to C. Darwin, a “perplexing phenomenon” and an “abominable mystery” (letter from Darwin to Hooker 1887; [6]). In the ancient and widespread plant lineages of gymnosperms, two-thirds are conifers (Coniferales or Pinophyta), mainly including Pinaceae and Cupressophytes of 546–615 species [7,8]. This lineage plays an important role in global carbon, nutrient, and atmospheric cycles and is of great ecological and economic importance worldwide. The intimidating genome sizes of conifers have been a major bottleneck, constraining the exploration of the astonishing rise of angiosperms from gymnosperms at the molecular level. However, recent years have witnessed fascinating strides in our understanding of the genome evolution and adaptation in conifer species, owing to next-generation sequencing technologies and interdisciplinary developments, such as bioinformatics, quantitative and population genetics, and evolutionary biology. In the past decade, complete draft assemblies have been obtained for four conifer genomes: Norway spruce (Picea abies; [9]), white spruce (Picea glauca; [10,11]), loblolly pine (Pinus taeda; [12,13,14]), and sugar pine (Pinus lambertiana; [15]). More conifer genome sequencing is underway (e.g., Pseudotsuga menziesii and Larix sibirica; http://pinegenome.org/). From a genomic evolutionary perspective, the different propensities for genome expansion through polyploidy and/or repeat amplification vs. genome contraction through epigenetics suppression and various recombination-based mechanisms have led to contrasting genome sizes of organisms. For the genome of Picea abies and Pinus taeda, high proportion (c. 60%) of long-terminal repeat retrotransposons (LTR-RTs) [9,12,13] and unique non-coding RNA (ncRNA) generation features [16,17] have shed novel insights into genome evolution and adaptation in conifers. Here, we review some of the most updated examples on the roles of transposable elements (TEs) in plant genome evolution and adaptation through epigenetics mechanisms, whereby we discuss how TEs and ncRNAs (and DNA methylation) dynamics contribute to conifer genomic and adaptive evolution.
2. Transposable Elements: A Source of Genetic Innovation
TEs are stretches of DNA sequences that can move and amplify their copy number within a host genome [18,19]. TEs are a major source of genomic mutations [20], and TE insertions provide a potent mutagenic mechanism for the evolution of new genes and their functionalities [21,22,23,24]. For instance, the Ac/Ds family insertion into a starch synthase gene created Mendel’s wrinkled peas [25], a disruptive insertion of a phytochrome A paralog promoted adaptation to high latitudes [26,27], and the hopscotch insertion helped shape the genomic architecture of the modern maize plant [28]. TEs also provide the raw material from which novel regulatory sequences are derived [22,29], such as promoters and enhancers [30,31]. Such changes in cis regulatory element change the structure and the regulatory and/or epigenetic environment, leading to modified gene expression (see review [32]). In addition, TEs affect the genome architecture by facilitating chromosomal sequence rearrangements due to their potential to arouse chromosomal mutations, for example, in maize [33]. TE-mediated genome rearrangement has been generalized throughout all organisms, including plant species [34,35].
Studies using Drosophila melanogaster lines have demonstrated that fitness is negatively correlated with TE numbers [36,37]. In plants, a positive correlation between genome size and the extinction probability has been documented [38,39,40]. TEs often proliferate faster than they can be removed, thus contributing to genome growth or even genome obesity [41,42]. Generally, plant genome size variation is mainly due to polyploidization and TE proliferation and/or deletion; thus, variable TEs insertion and deletion rates constitute an essential driving force in genome size evolution [34]. Recent advances in our understanding of the centrality of TEs to genome size and genic evolution provide insights into the mechanistic underpinnings of biased fractionation and polyploidy events [21,22,42,43], depicted in Figure 1. Biased fractionation may result from changes in epigenetic landscape near genes mediated by TEs. These epigenetic changes result in unequal gene expression between duplicates, establishing different fitness, which leads to biased gene loss with respect to ancestral genomes, typically termed “biased fractionation” [44]. It has been proposed that the differential loss of ancestral genome is explained by two observations [45]. First, homologs of retained duplicated genes in the most highly fractionated genome are often expressed at lower levels than their counterparts in the more intact genome [44,46]. Second, epigenetically silenced TEs are often more physically adjacent to the homolog with lower expression, indicative of a positive-effect repression of gene transcription [47,48,49].
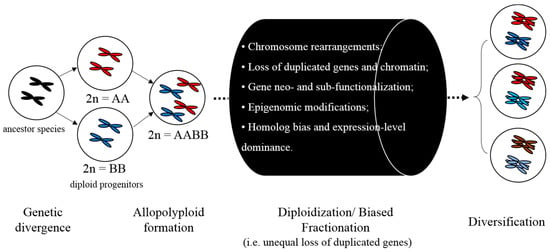
Figure 1.
Conceptual illustration of key realizations in plant genome architecture. The merger of two diploids via hybridization and/or allopolyploidy has novel evolutionary consequences [21], enumerated in the black cylinder.
Moreover, some TEs fold into stem-loop secondary structures and thus potentially contribute to the formation of ncRNAs [50,51]. Because TEs serve as an important resource for ncRNA generation, biased fractionation can lead to quantitative and qualitative mismatches between silencing machineries, as described in [30,52]. These regulatory mismatches likely give rise to various perturbations in the ncRNA populations (e.g., production of novel ncRNAs), with corresponding effects on gene expression, which has cascading influences on individual fitness.
3. Transposable Elements–Epigenetics Components Interplay: An Evolutionary Force in Adaptation
In the past decade, major advances in understanding the molecular mechanisms underpinning adaptation (e.g., responses to stress and developmental process) have been made [53], notably by highlighting the relevance of two environment-sensitive molecular elements: TEs and epigenetic components (e.g., [54,55]). While TEs provide agents of adaptive evolution and adaptation by creating genetic variants and diversity [56,57] (or see review [58]), epigenetic mechanisms play regulatory roles by the alteration of epigenetic landscapes (see review [59,60]). Epigenetic components comprise a molecular network that can affect complex traits—sometimes transmitted across generations—in the absence of genetic variation [61,62,63]. For instance, patterns of DNA methylation can persist across generations and produce heritable phenotypic changes [64,65]. Cortijo et al. [65] demonstrated that specific heritable methylation patterns in Arabidopsis thaliana experimental strains accounted for 60%–90% of the heritability for flowering time and primary root length. Interestingly, TEs and epigenetic components are intimately connected through the environment, potentially amplifying their actions on phenotypes and genotypes [54,66,67,68], thus playing a central role in an organism’s adaptation [69,70].
TE activities can be triggered by environmental cues, accelerate mutation rates, and rewire regulatory networks [71,72]. As first put forth by McClintock [73], TEs comprise a significant adaptive response of the genome to environmental challenges. In particular, stress may stimulate bursts of TE activity [74,75], as may hybridization and polyploidy [35,76,77]. Stressors triggering TE activities have been instantiated in plants [42,78]. For instance, thermal stress induces genome-wide modifications of methylation patterns in the grass Leymus chinensis [79]; DNA methylation under drought stress in Populus trichocarpa occurs in TEs [80]. Field evidence also indicates that environmental conditions affect the epigenomes in wild populations [63]. For instance, invasive populations of Japanese knotweed (Fallopia japonica) established in various habitats in northeastern America exhibit massive epigenetic differentiation, largely exceeding the observed genetic differentiation, and some of the epigenetic patterns might respond to local habitat conditions [63]. Moreover, TEs can regulate gene expression by attracting repressive marks. In plants, TEs associated with long intergenic ncRNAs are tissue-specific and expressed under specific stressful conditions [81]. TEs can also modify gene expression in response to stress to which the elements are themselves responsive [82,83]. For instance, In some plants (e.g., wheat, tobacco, oat), specific DNA elements in the 5′ LTR-RTs were identified in relation to stress responses, such as phytohormones, heat, light, or salinity [84,85,86,87,88]. In maize and rice, stress-sensitive TEs were shown to be inserted in the flanking regions of some genes, inducing specific stress-responsive regulation of these genes [82,83]. These discoveries highlight that TEs and epigenetic components tightly interact through numerous pathways and suggest their joint implication in organisms’ responses to stress. Nonetheless, TEs do not always have a positive fitness effect under stress conditions (see review [89]), and a negative relationship between stress and TEs have been recently documented in plants [81,90]. As such, the TEs–stress relationship is complex, although evidence has shown that TEs are activated under stress in most studies performed so far, and TE insertions have been shown to have a beneficial effect in some cases (e.g., [91,92]).
A priori, epigenetic components play a major role in storing genetic information for particular phenotypes in a silent state as long as epigenetic marks are faithfully transmitted across generations, hence generating so-called hidden genetic variation [93]. Environmentally induced epigenetic changes can reveal hidden genetic variation, which provides a mechanism for rapid adaptation [94]. Moreover, under stress, TEs can be activated in somatic cells either directly or through the arousal of their epigenetic control, thus producing non-heritable phenotypic variation among somatic cells within an organism [95]. This mechanism, termed genetic mosaicism, can instantaneously generate adaptive phenotypic variation in response to stress, especially in long-lived organisms [95]. In addition, mobile ncRNAs may be involved in coordinated responses to stimuli perceived in different organs. For instance, small interfering RNAs (siRNAs) expressed in shoot cells (i.e., from photosynthetic organs) can move to root cells (i.e., water-providing organs) and modify DNA methylation profiles in the latter cells, hence providing a coordinating system between functional organs [96].
Epigenetics components are key in repressing TE activities, thus making TEs “well behaved” and protecting genome integrity against TEs’ disruptive mobility [54]. Many TEs are targeted by DNA methyltransferases, and the arousal of epigenetic silencing is often associated with the activation of TEs [97,98], thus partly accounting for the sensitivity of TEs to the environment [78]. Conversely, TEs contribute to the evolution of genetic and epigenetic regulatory networks [66,72]. ncRNAs are integral parts of the epigenetic regulatory machinery through interaction with enzymes involved in DNA methylation and histone modifications. Research has shown that several ncRNAs are encoded by TEs or by endogenous genes that are likely to be derived from TEs [99,100]. Thus, TEs are essential genomic components encoding elements involved in the epigenetic machinery [100]. Another aspect of ncRNAs encoded by TEs is that they can act to repress the proliferation of the TEs from which they originate through a sequence complementary match (i.e., TE silencing) [101]. Thus, epigenetically induced controls on the activities of ncRNAs constitute a pathway to modulate TE activities [100]. Collectively, ncRNAs are key elements in TEs–epigenetics components interactions.
In addition, recent research has also advanced our understanding of genome size evolution from an organismal adaptation perspective. Under stressful environments, TEs may be under purifying selection, resulting in a compact genome [102,103]. TE-mediated genome instability could be accelerated in the face of environmental insults, such as high salinity and strong UV exposure [104,105,106], leading to enhanced selection pressures on TEs. Even within the same species, different environmental exposures may lead to the variation of genome size and TE contents. For example, a study of independent adaptations to high altitude in Zea mays showed that genome size experiences parallel pressures from natural selection, causing a reduction in genome size with increasing altitude [107]. This study also highlighted that genome size evolution may be environmentally dependent and that correlated changes in genome size may be mediated through, for instance, flowering time [108,109,110].
4. Genome-Purging Mechanism: Another Evolutionary Force to Counterbalance Transposable Element Increase
The explosion of selfish genetic elements is typically thought to be controlled by unconditionally deleterious effects, such as the harmful effects of gene disruption, meiotic defects on pollen viability and seed set [111], and ectopic recombination events causing chromosomal rearrangements [112,113]. It was thought that plants were on a path to obesity through continual DNA bloating, but recent research supports that most plants actively purge DNAs [114]. Genome-purging mechanisms include illegitimate or unequal combination between LTRs and other types of deletions, which facilitates to counterbalance the ever increasing TEs [115]. Intra-strand homologous recombination between directly repeated LTRs deletes the sequences between LTRs, leaving solo-LTRs [116]. Analysis of solo-LTRs and comparison of internally deleted RTs has indicated that illegitimate intra-strand homologous recombination may be the driving force in maintaining slim genomes of Arabidopsis [117] and rice [118]. Recently inserted TEs are often removed from the genome (i.e., genome contraction), resulting in rapid genomic turnover. For example, the rice genome underwent several bursts of LTR-RTs during the last 5 million years but ultimately removed over half of the inserted LTR-RT DNA [119,120,121]. A three-fold increase in the genome size of diploid members of Gossypium is due to the accumulation of LTR-RTs over the past 5–10 mya [122].
Another mechanism that prevents genomes from uncontrolled expansion is through non-homologous end joining (NHEJ) after deletion-biased double-strand break (DSB), which leads to massive genomic restructuring by purging LTR-RTs in a small genome [123]. This pathway is frequently invoked as the possible cause of genome shrinkage (see review [124]). The genome of Oryza brachyantha is 60% smaller than its close relative cultivating species, Oryza sativa, and 50% of their size difference was found to be due to the amplification and deletion of recent LTR-RTs [123]. In c. 32,000 protein-coding genes of O. brachyantha, only 70% of them were in collinear positions as the rice genome [123]. It is therefore reasonable to argue that the low LTR-RT activity and massive internal deletions of the LTRs by NHEJ after DSB have shaped the current O. brachyantha genome. Likewise, NHEJ after DSB was found to be a common pathway in the genomic reduction of A. thaliana [125].
5. Transposable Elements in Conifer Genome Architecture
The conifer genomes are elusively large (20–30 Gb) [126] and abundant, and diverse RTs are the main component of non-genic portions [127,128]. It is estimated that 62% of Pinus taeda genome is composed of RTs, of which 70% are LTRs, mainly Pseudoviridae (also known as Ty1/Copia elements) and Metaviridae (Ty3/Gypsy) ([9,12,13]; also see summary of conifer TEs in a conifer TE database, ConTEdb [129] based on more than 0.4 million TEs of three sequenced conifer genomes). Similarly, the total representation of all TE classes was estimated as 69% in P. abies with similar types of LTRs [9]. The comparative analysis of LTR-RTs in conifers also indicated that the accumulation of retro-elements in conifer genomes is very ancient and has occurred over a very long timeframe spanning tens to hundreds of millions of years [9].
The large conifer genomes contain a huge wealth of divergent and ancient repeats, including RTs, which have most likely arisen through a combination of ongoing repeat amplification over long periods of evolutionary time [130,131], combined with a lack of efficient and/or slower rates of repeat elimination via recombination-based processes [9,132]. They have the lowest recombination rates relative to any eukaryotic lineages reported so far [132,133,134]. The possible reason for this is that, when genome size grows above a certain threshold, large tracts of heterochromatin are formed from stretches of repeats, which become “locked down” into highly condensed chromatin through epigenetic activities. This leads to the reduction of the potentially negative impact of TE activity and limits the accessibility of the recombination and hence the potential of recombination-based processes to eliminate DNA [66]. Surprisingly, LTR-RTs of the Norway spruce genome are mostly low-copy LTRs with 80% singletons [9]. The intimidating genome size seems to have resulted from a slow but steady accumulation of diverse LTR-RTs, which might be due to the lack of efficient elimination mechanisms, as evidenced by the fact that pine species also have low-copy LTRs [127].
In addition to TE characteristics, conifers share other common features, making them distinct from other spermatophytes. For instance, conifer genome sizes have a 5.5-fold difference (1C = 6.6 to 36 pg), and most conifers have a remarkable constancy in chromosome numbers of 2n = 24 [135]. Natural polyploids are exceedingly rare in conifers (and gymnosperms in general) [136,137], indicating that WGD is not an important evolutionary driving force in conifer genomes. Moreover, conifers have particularly low average linkage disequilibrium [138,139], which is often confined within genes [140]. These distinct conifer genomic features indicate largely independent evolution of individual genes [141], favoring efficient natural selection instead of genetic drift, leading to genetically based clines (see review [142]). Finally, conifer genes tend to accumulate long introns, with the largest introns spanning 60 kb in spruce [9] and 120 kb in pine [13]. One or a few long introns of several kb are found in conifer genes and conserved across species [143]. However, no evidence has shown that very long introns in conifers would reduce the level of gene expression [9,143]. This is in contrast to findings in mammalians, which report high expression in genes with shorter introns [144].
As outlined for the nine conifers most frequently distributed worldwide, the genome size variation is around two-fold (Figure 2A), comparable with the c. 5.5-fold variation within all conifers [135]. The genomes of the four sequenced conifers are significantly larger than those of non-conifer tree species (p = 0.0035; Figure 2B). However, the genome size variation seems to be independent of the number of protein-coding genes, and there is no significant difference in gene numbers between conifers and non-conifer trees (Figure 2C; also see [145]). Conifers have similar numbers of unigenes, less than 40,000 as in many non-conifer trees (p = 0.98; Figure 2C). Moreover, conifer genomes have more class I TEs compared with non-conifer trees (p = 0.005) but have very low content of class II TEs compared with non-conifer trees (p = 0.94; Figure 2D) and most angiosperms [9,143]. Nonetheless, a high ratio of class I TEs is not a distinct feature between conifers and angiosperms as class I TEs make up as high as 76% of maize genome, leaving another 10% for other types of TEs [146], and constitute over 65% of 17 Gb wheat genome [147]. In addition, variation in genome size and variation in chromosome number are not correlated in trees (Figure 2C and Table S1), which is in agreement with findings in flowering plants in general [148].
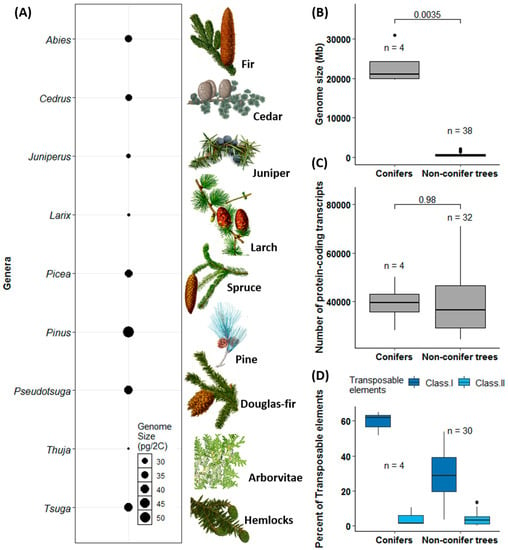
Figure 2.
Comparison of genome size, protein-coding transcripts, and transposable elements between conifers and other sequenced tree species. In (A), estimated mean genome sizes of major conifer species is based on the Plant DNA C-values Database (http://data.kew.org/cvalues/) as of February 2019. All botanical illustrations are from the public domain (Note S1). In (B–D), the number of species (n) used in each calculation is different due to incomplete information available in original publications. The p-values based on t-tests are given on the top of each panel. p-values for the comparisons of class I and II TEs between conifers and non-conifer trees were 0.005 and 0.94, respectively. The data used for these plots were extracted from the publications listed in Table S1.
6. Non-Coding RNA Features in Conifers
Excessive TEs can destroy the genome, and many organisms have developed diverse mechanisms to inhibit TE activities, including RNA-based silencing pathways [149,150]. Insights into the role of epigenetic pathways, including the siRNA-directed DNA methylation (RdDM) in plants, to suppress activity of TEs as well as other counterbalancing mechanisms is an active area of research [52]. Endogenous 24-nt-long ncRNAs are enriched in intergenic and repetitive genomic regions [151,152], and more generally, most plant ncRNA loci are mapped to intergenic segments [50]. The feature of exceptionally large amount of excessive genomic DNA in conifers implies a rich source of TE-derived ncRNAs, thus building a distinct landscape of ncRNA populations targeting TEs and genes involved in various biological processes. In particular, 24-nt-long ncRNAs act as “genome guardians”, providing multigenerational protections against invasive TEs. The lack of regulation of 24-nt-long ncRNAs, which occurs at late seed set in conifers, alters developmental programs and may lead to increased TE activities and exceptionally large genome size [16]. The trade-off between large amount of TEs and novel ncRNAs, which are originated from foldback TEs to restrain TE expansion, may steer the direction and speed of genome evolution.
In addition, genome-wide average DNA methylation levels are positively correlated with genome size and TE contents [153]. Conspicuous 24-nt-long ncRNAs [9,16] and a very high level of DNA methylation [153] uniquely yielded at the reproductive period in conifers. This pattern of ncRNA dynamics prompts interesting investigations in linking ncRNAs to methylation dynamics to unravel the roles of epigenetics in genomic and adaptive evolution.
7. Outstanding Questions
With updated knowledge of TEs and ncRNAs in mind and considering the genomic features of conifers and several incipient studies on conifer epigenetics, we put up the following four questions and hypotheses for further testing:
- To what extent do TEs contribute to conifer genetic variation and population divergence? Based on TE polymorphisms, what are TE dynamics in natural populations and how are these loci under selection?
- ⇨
- Given the availability of reference genome and TE consensus sequences, computational approaches (e.g., [154]) provide a possibility of addressing this question.
- How does the landscape of ncRNAs and DNA methylation alter over time (e.g., reproduction vs. vegetative growth) in the region of TEs (or different types of TEs), genes, and introns of focus conifers?
- ⇨
- We could additionally focus on polymorphic TE loci identified in the previous question and genes involved in known pathways to explore how epigenetic components work in synergy in those “hotspots”.
- What are the features of TEs–epigenetics components (ncRNA and DNA methylation) interactions when individuals are exposed to stressful vs. benign environmental conditions? How are these features associated with local adaptation?
- ⇨
- By identifying polymorphic TEs among populations (e.g., [155]), we are able to infer whether different types of TEs have undergone differential expansion or contraction, hence playing a role in adaptive evolution. Then, contrasting these results with their DNA methylation landscape permits testing whether and how the two mechanisms jointly contribute to adaptation.
- Is there evidence supporting genome-purging mechanisms in conifers?
- ⇨
- We could look for answer by, for example, estimating death rates of LTR-RTs (e.g., prediction by [156,157]) by counting solo-LTRs and truncated elements in sequenced conifers and their close relatives.
8. Closing Remarks
Upon the discovery of transposable elements in the 1950s, Barbara McClintock envisioned that these sequences might operate to control gene expression and play a major role in evolution. Her prophetic remarks are finally receiving growing empirical support after several paradigm shifts in the past decades. We now know that a large amount of so-call ”junk” or ”parasite” or ”selfish” DNAs in plant taxa, especially in conifers, are not true junks but ubiquitous and influential genetic elements, and this has posited a meaningful conundrum in evolutionary genetics and genomics. They are now considered as drivers in evolution and are the focus of numerous genomic studies. The activity of LTR-RTs is under the control of epigenetic suppressing mechanisms. Epigenetic regulations of the transposable elements are the first line of defense against uncontrolled transposable element proliferation. Also, genome-purging mechanisms have been adopted to counterbalance the genome size amplification. Thus, the current genome evolution of organisms may be driven by a long battle of repeat sequence amplification and genome-purging systems in which conifers should represent an outstanding node for comparative studies. In addition, TEs can generate a broad range of genetic variation in natural populations by interplaying with epigenetic components, highlighting the interactive roles of TEs, epigenetic components, and the environment in adaptive evolution and adaptation. Thus, TEs in conjunction with epigenetics landscapes (ncRNAs and DNA methylation) provide a novel avenue to unravel the molecular underpinnings of local adaptation in long-lived perennials, such as conifers.
9. Terminology
- Biased fractionation: the unequal loss of genes from ancestral progenitor genomes, which is a frequent event after polyploidy in many lineages.
- Class I elements of TEs: retrotransposons, which use reverse transcriptase to copy an RNA genome into the host DNA (i.e., “copy and paste”); see a unified TE classification system [158,159] and illustration by Chénais et al. [24].
- Class II elements of TEs: DNA transposons; the DNA genome of the element itself serves as the template for transposition either by a “cut and paste” mechanism or using a rolling circle process.
- Long terminal repeats (LTRs): identical DNA sequence that can be repeated at the ends of retrotransposons.
- Non-coding RNAs (ncRNAs): randomly grouped into short (<200 nt) and long (>200 nt) types [160]. Members of short ncRNAs involved in plant transcriptional (indirect and low) and post-transcriptional (major) regulations have been well documented (e.g., review in [52]). These short ncRNAs chiefly consist of microRNAs (prevalence of 21 or 22 nt long in suppressing target mRNAs), heterochromatic small interfering RNAs (hc-siRNAs; 24 nt mediators in silencing DNA methylation and histone modifications), and trans-acting siRNAs (tasiRNAs or phasiRNAs; 22 (or 21) nt with a phased configuration, playing similar roles as microRNAs or other uncharacterized functions).
- Whole-genome duplication (WGD) or polyploidization: an event in which the entire genome of an organism is copied once or multiple times. A widely accepted hypothesis for WGD events is based on a hexaploidization of all eudicots (ancestral γ events), first put forth by Jaillon et al. [161].
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/3/228/s1, Table S1: Details of sequenced tree genomes (as of February 2019).
Author Contributions
Y.L. conceived this study, compiled data, performed analyses, and wrote the manuscript; Y.A.E.-K. coordinated the project.
Funding
This project was funded by the Johnson’s Family Forest Biotechnology Endowment and the National Science and Engineering Research Council of Canada Discovery and Industrial Research Chair to Y.A.E.
Acknowledgments
We thank two anonymous referees for providing concrete, helpful comments.
Conflicts of Interest
We have no competing interests.
References
- Paton, A.J.; Brummitt, N.; Govaerts, R.; Harman, K.; Hinchcliffe, S.; Allkin, B.; Lughadha, E.N. Towards target 1 of the global strategy for plant conservation: A working list of all known plant species — progress and prospects. Taxon 2008, 57, 602–611. [Google Scholar] [CrossRef]
- Willis, K.J.; McElwain, J.C. The Evolution of Plants; Oxford University Press: New York, NY, USA, 2002. [Google Scholar]
- Hilton, J.; Bateman, R.M. Pteridosperms are the backbone of seed-plant phylogeny. J. Torrey Bot. Soc. 2006, 133, 119–168. [Google Scholar] [CrossRef]
- Bateman, R.M.; Hilton, J.; Rudall, P.J. Morphological and molecular phylogenetic context of the angiosperms: Contrasting the ’top-down’ and ’bottom-up’ approaches used to infer the likely characteristics of the first flowers. J. Exp. Bot. 2006, 57, 3471–3503. [Google Scholar] [CrossRef]
- Friis, E.M.; Pedersen, K.R.; Crane, P.R. Diversity in obscurity: Fossil flowers and the early history of angiosperms. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 369–382. [Google Scholar] [CrossRef]
- Friedman, W.E. The meaning of Darwin’s “abominable mystery”. Am. J. Bot. 2009, 96, 5–21. [Google Scholar] [CrossRef]
- Farjón, A. A Handbook of the World Conifers; Brill Academic Publishers: Leiden, The Netherlands, 2010. [Google Scholar]
- Eckenwalder, J.E. Conifers of the World: The Complete Reference; Timber Press: London, UK, 2009. [Google Scholar]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Birol, I.; Raymond, A.; Jackman, S.D.; Pleasance, S.; Coope, R.; Taylor, G.A.; Yuen, M.M.; Keeling, C.I.; Brand, D.; Vandervalk, B.P.; et al. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 2013, 29, 1492–1497. [Google Scholar] [CrossRef]
- Warren, R.L.; Keeling, C.I.; Yuen, M.M.; Raymond, A.; Taylor, G.A.; Vandervalk, B.P.; Mohamadi, H.; Paulino, D.; Chiu, R.; Jackman, S.D.; et al. Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J. 2015, 83, 189–212. [Google Scholar] [CrossRef]
- Neale, D.B.; Wegrzyn, J.L.; Stevens, K.A.; Zimin, A.V.; Puiu, D.; Crepeau, M.W.; Cardeno, C.; Koriabine, M.; Holtz-Morris, A.E.; Liechty, J.D.; et al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 2014, 15, R59. [Google Scholar] [CrossRef]
- Wegrzyn, J.L.; Liechty, J.D.; Stevens, K.A.; Wu, L.S.; Loopstra, C.A.; Vasquez-Gross, H.A.; Dougherty, W.M.; Lin, B.Y.; Zieve, J.J.; Martinez-Garcia, P.J.; et al. Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 2014, 196, 891–909. [Google Scholar] [CrossRef]
- Zimin, A.; Stevens, K.A.; Crepeau, M.W.; Holtz-Morris, A.; Koriabine, M.; Marcais, G.; Puiu, D.; Roberts, M.; Wegrzyn, J.L.; de Jong, P.J.; et al. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics 2014, 196, 875–890. [Google Scholar] [CrossRef]
- Stevens, K.A.; Wegrzyn, J.L.; Zimin, A.; Puiu, D.; Crepeau, M.; Cardeno, C.; Paul, R.; Gonzalez-Ibeas, D.; Koriabine, M.; Holtz-Morris, A.E.; et al. Sequence of the sugar pine megagenome. Genetics 2016, 204, 1613–1626. [Google Scholar] [CrossRef]
- Liu, Y.; El-Kassaby, Y.A. Landscape of fluid sets of hairpin-derived 21-/24-nt-long small RNAs at seed set uncovers special epigenetic features in Picea glauca. Genome Biol. Evol. 2017, 9, 82–92. [Google Scholar] [CrossRef]
- Xia, R.; Xu, J.; Arikit, S.; Meyers, B.C. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 2015, 32, 2905–2918. [Google Scholar] [CrossRef]
- Casacuberta, E.; González, J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013, 22, 1503–1517. [Google Scholar] [CrossRef]
- Bukhari, A.I.; Shapiro, J.A.; Adhya, S.L.; Cold Spring Harbor Laboratory. DNA Insertion Elements, Plasmids, and Episomes; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1977; p. 782. [Google Scholar]
- Lynch, M. The Origins of Genome Architecture; Sinauer Associates: Sunderland, MA, USA, 2007. [Google Scholar]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploid plant genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Ferguson, A.A.; Jiang, N. What makes up plant genomes: The vanishing line between transposable elements and genes. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2016, 1859, 366–380. [Google Scholar] [CrossRef]
- Chénais, B.; Caruso, A.; Hiard, S.; Casse, N. The impact of transposable elements on eukaryotic genomes: From genome size increase to genetic adaptation to stressful environments. Gene 2012, 509, 7–15. [Google Scholar] [CrossRef]
- Bhattacharyya, M.K.; Smith, A.M.; Ellis, T.H.N.; Hedley, C.; Martin, C. The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 1990, 60, 115–122. [Google Scholar] [CrossRef]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef]
- Kanazawa, A.; Liu, B.H.; Kong, F.J.; Arase, S.; Abe, J. Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia-like retrotransposon in soybean. J. Mol. Evol. 2009, 69, 164–175. [Google Scholar] [CrossRef]
- Studer, A.; Zhao, Q.; Ross-Ibarra, J.; Doebley, J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 2011, 43, 1160–1163. [Google Scholar] [CrossRef]
- Lisch, D.R. Transposons in plant gene regulation. In Plant Transposons and Genome Dynamics in Evolution; Fedoroff, N.V., Ed.; Willey-Blackwell: Ames, IA, USA, 2013; pp. 93–116. [Google Scholar]
- Springer, N.M.; Lisch, D.; Li, Q. Creating order from chaos: Epigenome dynamics in plants with complex genomes. Plant Cell 2016, 28, 314–325. [Google Scholar] [CrossRef]
- Diez, C.M.; Roessler, K.; Gaut, B.S. Epigenetics and plant genome evolution. Curr. Opin. Plant Biol. 2014, 18, 1–8. [Google Scholar] [CrossRef]
- Mita, P.; Boeke, J.D. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 2016, 37, 90–100. [Google Scholar] [CrossRef]
- McClintock, B. The Discovery and Characterization of Transposable Elements: The Collected Papers of Barbara Mcclintock; Garland: New York, NY, USA, 1987. [Google Scholar]
- Bennetzen, J.L. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005, 15, 621–627. [Google Scholar] [CrossRef]
- Parisod, C.; Alix, K.; Just, J.; Petit, M.; Sarilar, V.; Mhiri, C.; Ainouche, M.; Chalhoub, B.; Grandbastien, M.A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010, 186, 37–45. [Google Scholar] [CrossRef]
- Houle, D.; Nuzhdin, S.V. Mutation accumulation and the effect of copia insertions in Drosophila melanogaster. Genet. Res. 2004, 83, 7–18. [Google Scholar] [CrossRef]
- Papaceit, M.; Avila, V.; Aguadé, M.; García-Dorado, A. The dynamics of the roo transposable element in mutation-accumulation lines and segregating populations of Drosophila melanogaster. Genetics 2007, 177, 511–522. [Google Scholar] [CrossRef]
- Vinogradov, A.E. Genome size and extinction risk in vertebrates. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004, 271, 1701–1705. [Google Scholar] [CrossRef]
- Vinogradov, A.E. Evolution of genome size: Multilevel selection, mutation bias or dynamical chaos? Curr. Opin. Genet. Dev. 2004, 14, 620–626. [Google Scholar] [CrossRef]
- Vinogradov, A.E. Selfish DNA is maladaptive: Evidence from the plant Red List. Trends Genet. 2003, 19, 609–614. [Google Scholar] [CrossRef]
- Brookfield, J.F. The ecology of the genome - Mobile DNA elements and their hosts. Nat. Rev. Genet. 2005, 6, 128–136. [Google Scholar] [CrossRef]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.S. The genomes and transposable elements in plants: Are they friends or foes? Genes Genom. 2017, 39, 359–370. [Google Scholar] [CrossRef]
- Schnable, J.C.; Springer, N.M.; Freeling, M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 2011, 108, 4069–4074. [Google Scholar] [CrossRef]
- Steige, K.A.; Slotte, T. Genomic legacies of the progenitors and the evolutionary consequences of allopolyploidy. Curr. Opin. Plant Biol. 2016, 30, 88–93. [Google Scholar] [CrossRef]
- Woodhouse, M.R.; Cheng, F.; Pires, J.C.; Lisch, D.; Freeling, M.; Wang, X.W. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc. Natl. Acad. Sci. USA 2014, 111, 5283–5288. [Google Scholar] [CrossRef]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef]
- Hollister, J.D.; Smith, L.M.; Guo, Y.L.; Ott, F.; Weigel, D.; Gaut, B.S. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 2011, 108, 2322–2327. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Meyers, B.C.; Cai, C.; Xu, W.; Ma, J. Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell 2015, 27, 546–562. [Google Scholar] [CrossRef] [PubMed]
- Piriyapongsa, J.; Jordan, I.K. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 2008, 14, 814–821. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, M.; Mao, Z.T.; Li, C.X. Characterization and evolution of microRNA genes derived from repetitive elements and duplication events in plants. PLoS ONE 2012, 7, e34092. [Google Scholar] [CrossRef]
- Wendel, J.F.; Jackson, S.A.; Meyers, B.C.; Wing, R.A. Evolution of plant genome architecture. Genome Biol. 2016, 17, 37. [Google Scholar] [CrossRef]
- Galhardo, R.S.; Hastings, P.J.; Rosenberg, S.M. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 399–435. [Google Scholar] [CrossRef]
- Zeh, D.W.; Zeh, J.A.; Ishida, Y. Transposable elements and an epigenetic basis for punctuated equilibria. Bioessays 2009, 31, 715–726. [Google Scholar] [CrossRef]
- Rey, O.; Danchin, E.; Mirouze, M.; Loot, C.; Blanchet, S. Adaptation to global change: A transposable element–epigenetics perspective. Trends Ecol. Evol. 2016, 31, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Schrader, L.; Kim, J.W.; Ence, D.; Zimin, A.; Klein, A.; Wyschetzki, K.; Weichselgartner, T.; Kemena, C.; Stokl, J.; Schultner, E.; et al. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nature Commun. 2014, 5, 5495. [Google Scholar] [CrossRef]
- Stapley, J.; Santure, A.W.; Dennis, S.R. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol. Ecol. 2015, 24, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Schrader, L.; Schmitz, J. The impact of transposable elements in adaptive evolution. Mol. Ecol. 2018, 0, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 2009, 60. [Google Scholar] [CrossRef] [PubMed]
- Cubas, P.; Vincent, C.; Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999, 401, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Schrey, A.W.; Pigliucci, M. Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol. Lett. 2012, 15, 1016–1025. [Google Scholar] [CrossRef]
- Reinders, J.; Wulff, B.B.H.; Mirouze, M.; Marí-Ordóñez, A.; Dapp, M.; Rozhon, W.; Bucher, E.; Theiler, G.; Paszkowski, J. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009, 23, 939–950. [Google Scholar] [CrossRef]
- Cortijo, S.; Wardenaar, R.; Colome-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.M.; Wincker, P.; et al. Mapping the epigenetic basis of complex traits. Science 2014, 343, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Fablet, M.; Vieira, C. Evolvability, epigenetics and transposable elements. Biomol. Concepts 2011, 2, 333–341. [Google Scholar] [CrossRef]
- Oliver, K.R.; Greene, W.K. Mobile DNA and the TE-Thrust hypothesis: Supporting evidence from the primates. Mobil. DNA 2011, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.R.; Greene, W.K. Transposable elements: Powerful facilitators of evolution. Bioessays 2009, 31, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Ito, H.; Kim, J.M.; Matsunaga, W.; Saze, H.; Matsui, A.; Endo, T.A.; Harukawa, Y.; Takagi, H.; Yaegashi, H.; Masuta, Y.; et al. A stress-activated transposon in Arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 2016, 6, srep23181. [Google Scholar] [CrossRef]
- Grandbastien, M.A. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2015, 1849, 403–416. [Google Scholar] [CrossRef]
- Kawakami, T.; Strakosh, S.C.; Zhen, Y.; Ungerer, M.C. Different scales of Ty1/copia-like retrotransposon proliferation in the genomes of three diploid hybrid sunflower species. Heredity 2010, 104, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Senerchia, N.; Felber, F.; Parisod, C. Genome reorganization in F1 hybrids uncovers the role of retrotransposons in reproductive isolation. P. Roy. Soc. B Biol. Sci. 2015, 282, 20142874. [Google Scholar] [CrossRef]
- Miousse, I.R.; Chalbot, M.C.G.; Lumen, A.; Ferguson, A.; Kavouras, I.G.; Koturbash, I. Response of transposable elements to environmental stressors. Mutat. Res. Rev. Mut. Res. 2015, 765, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.J.; Yang, X.J.; Wang, H.Y.; Shi, F.X.; Liu, Y.; Liu, J.S.; Li, L.F.; Wang, D.L.; Liu, B. Cytosine methylation alteration in natural populations of Leymus chinensis induced by multiple abiotic stresses. PLoS ONE 2013, 8, e55772. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Zhang, Z.; Wu, H.; Huang, C.; Shuai, P.; Ye, C.Y.; Tang, S.; Wang, Y.; Yang, L.; Wang, J.; et al. Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 2014, 15 Suppl 1, S9. [Google Scholar] [CrossRef]
- Wang, D.; Qu, Z.; Yang, L.; Zhang, Q.; Liu, Z.H.; Do, T.; Adelson, D.L.; Wang, Z.Y.; Searle, I.; Zhu, J.K. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 2017, 90, 133–146. [Google Scholar] [CrossRef]
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 2009, 461, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Woodrow, P.; Pontecorvo, G.; Ciarmiello, L.F.; Fuggi, A.; Carillo, P. Ttd1a promoter is involved in DNA-protein binding by salt and light stresses. Mol. Biol. Rep. 2011, 38, 3787–3794. [Google Scholar] [CrossRef] [PubMed]
- Suoniemi, A.; Anamthawat-Jónsson, K.; Arna, T.; Schulman, A.H. Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgare L) genome. Plant Mol. Biol. 1996, 30, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Beguiristain, T.; Grandbastien, M.A.; Puigdomènech, P.; Casacuberta, J.M. Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol. 2001, 127, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999, 18, 383–393. [Google Scholar] [CrossRef]
- Egue, F.; Chénais, B.; Tastard, E.; Marchand, J.; Hiard, S.; Gateau, H.; Hermann, D.; Morant-Manceau, A.; Casse, N.; Caruso, A. Expression of the retrotransposons Surcouf and Blackbeard in the marine diatom Phaeodactylum tricornutum under thermal stress. Phycologia 2015, 54, 617–627. [Google Scholar] [CrossRef]
- Horváth, V.; Merenciano, M.; González, J. Revisiting the relationship between transposable elements and the eukaryotic stress response. Trends Genet. 2017, 33, 832–841. [Google Scholar] [CrossRef]
- Mao, H.D.; Wang, H.W.; Liu, S.X.; Li, Z.; Yang, X.H.; Yan, J.B.; Li, J.S.; Tran, L.S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nature Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
- Hunter, R.G.; Murakami, G.; Dewell, S.; Seligsohn, M.a.; Baker, M.E.R.; Datson, N.A.; McEwen, B.S.; Pfaff, D.W. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc. Natl. Acad. Sci. USA 2012, 109, 17657–17662. [Google Scholar] [CrossRef]
- Secco, D.; Wang, C.; Shou, H.X.; Schultz, M.D.; Chiarenza, S.; Nussaume, L.; Ecker, J.R.; Whelan, J.; Lister, R. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 2015, 4, e09343. [Google Scholar] [CrossRef]
- Rutherford, S.L.; Henikoff, S. Quantitative epigenetics. Nat. Genet. 2003, 33, 6–8. [Google Scholar] [CrossRef]
- True, H.L.; Berlin, I.; Lindquist, S.L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature 2004, 431, 184–187. [Google Scholar] [CrossRef]
- Folse, H.J.; Roughgarden, J. Direct benefits of genetic mosaicism and intraorganismal selection: Modeling coevolution between a long-lived tree and a short-lived herbivore. Evolution 2012, 66, 1091–1113. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef]
- Fultz, D.; Choudury, S.G.; Slotkin, R.K. Silencing of active transposable elements in plants. Curr. Opin. Plant Biol. 2015, 27, 67–76. [Google Scholar] [CrossRef]
- Friedli, M.; Trono, D. The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell. Dev. Biol. 2015, 31, 429–451. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- McCue, A.D.; Nuthikattu, S.; Reeder, S.H.; Slotkin, R.K. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 2012, 8, e1002474. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Vaury, C. RNAi: A defensive RNA-silencing against viruses and transposable elements. Heredity 2006, 96, 195–202. [Google Scholar] [CrossRef]
- Hu, T.T.; Pattyn, P.; Bakker, E.G.; Cao, J.; Cheng, J.F.; Clark, R.M.; Fahlgren, N.; Fawcett, J.A.; Grimwood, J.; Gundlach, H.; et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011, 43, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Laclette, E.; Lyons, E.; Hernández-Guzmán, G.; Pérez-Torres, C.A.; Carretero-Paulet, L.; Chang, T.-H.; Lan, T.; Welch, A.J.; Juárez, M.J.A.; Simpson, J.; et al. Architecture and evolution of a minute plant genome. Nature 2013, 498, 94. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, P.; Goedecke, W.; Obe, G. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 2000, 15, 289–302. [Google Scholar] [CrossRef]
- Schuermann, D.; Molinier, J.; Fritsch, O.; Hohn, B. The dual nature of homologous recombination in plants. Trends Genet. 2005, 21, 172–181. [Google Scholar] [CrossRef]
- Argueso, J.L.; Westmoreland, J.; Mieczkowski, P.A.; Gawel, M.; Petes, T.D.; Resnick, M.A. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 2008, 105, 11845–11850. [Google Scholar] [CrossRef]
- Bilinski, P.; Albert, P.S.; Berg, J.J.; Birchler, J.A.; Grote, M.N.; Lorant, A.; Quezada, J.; Swarts, K.; Yang, J.; Ross-Ibarra, J. Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLoS Genet. 2018, 14, e1007162. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Díez, C.M.; Gaut, B.S.; Meca, E.; Scheinvar, E.; Montes-Hernandez, S.; Eguiarte, L.E.; Tenaillon, M.I. Genome size variation in wild and cultivated maize along altitudinal gradients. New Phytol. 2013, 199, 264–276. [Google Scholar] [CrossRef]
- Rayburn, A.L.; Dudley, J.W.; Biradar, D.P. Selection for early flowering results in simultaneous selection for reduced nuclear DNA content in maize. Plant Breeding 1994, 112, 318–322. [Google Scholar] [CrossRef]
- Higgins, D.M.; Lowry, E.G.; Kanizay, L.B.; Becraft, P.W.; Hall, D.W.; Dawe, R.K. Fitness costs and variation in transmission distortion associated with the abnormal chromosome 10 meiotic drive system in maize. Genetics 2018, 208, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Kupiec, M.; Petes, T.D. Allelic and ectopic recombination between Ty elements in yeast. Genetics 1988, 119, 549–559. [Google Scholar] [PubMed]
- Han, K.; Lee, J.; Meyer, T.J.; Remedios, P.; Goodwin, L.; Batzer, M.A. L1 recombination-associated deletions generate human genomic variation. Proc. Natl. Acad. Sci. USA 2008, 105, 19366–19371. [Google Scholar] [CrossRef]
- Michael, T.P. Plant genome size variation: Bloating and purging DNA. Brief. Funct. Genom. 2014, 13, 308–317. [Google Scholar] [CrossRef]
- Grover, C.E.; Wendel, J.F. Recent insights into mechanisms of genome size change in plants. J. Bot. 2010, 2010, 382732. [Google Scholar] [CrossRef]
- Hawkins, J.S.; Proulx, S.R.; Rapp, R.A.; Wendel, J.F. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 17811–17816. [Google Scholar] [CrossRef]
- Devos, K.M.; Brown, J.K.; Bennetzen, J.L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002, 12, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Devos, K.M.; Bennetzen, J.L. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004, 14, 860–869. [Google Scholar] [CrossRef]
- Vitte, C.; Panaud, O.; Quesneville, H. LTR retrotransposons in rice (Oryza sativa L.): Recent burst amplifications followed by rapid DNA loss. BMC Genom. 2007, 8, 218. [Google Scholar] [CrossRef] [PubMed]
- Vitte, C.; Panaud, O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol. Biol. Evol. 2003, 20, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; McCarthy, E.M.; Ganko, E.W.; McDonald, J.F. Evolutionary history of Oryza sativa LTR retrotransposons: A preliminary survey of the rice genome sequences. BMC Genom. 2004, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Kim, H.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006, 16, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, Q.; Gao, D.; Wang, J.; Lang, Y.; Liu, T.; Li, B.; Bai, Z.; Luis Goicoechea, J.; Liang, C.; et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nature Commun. 2013, 4, 1595. [Google Scholar] [CrossRef]
- Schubert, I.; Vu, G.T.H. Genome stability and evolution: Attempting a holistic view. Trends Plant Sci. 2016, 21, 749–757. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Rouzé, P.; Van de Peer, Y. Higher intron loss rate in Arabidopsis thaliana than A. lyrata is consistent with stronger selection for a smaller genome. Mol. Biol. Evol. 2012, 29, 849–859. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Birol, I.; Bousquet, J.; Ingvarsson, P.K.; Jansson, S.; Jones, S.J.M.; Keeling, C.I.; MacKay, J.; Nilsson, O.; Ritland, K.; et al. Insights into conifer giga-genomes. Plant Physiol. 2014, 166, 1724–1732. [Google Scholar] [CrossRef]
- Kovach, A.; Wegrzyn, J.L.; Parra, G.; Holt, C.; Bruening, G.E.; Loopstra, C.A.; Hartigan, J.; Yandell, M.; Langley, C.H.; Korf, I.; et al. The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genom. 2010, 11, 420. [Google Scholar] [CrossRef]
- Hamberger, B.; Hall, D.; Yuen, M.; Oddy, C.; Keeling, C.I.; Ritland, C.; Ritland, K.; Bohlmann, J. Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol. 2009, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Ling, J.J.; Xiao, Y.; Zhang, H.G.; Ouyang, F.Q.; Wang, J.H. ConTEdb: A comprehensive database of transposable elements in conifers. Database J. Biol. Databases Curation 2018. [Google Scholar] [CrossRef]
- Morse, A.M.; Peterson, D.G.; Islam-Faridi, M.N.; Smith, K.E.; Magbanua, Z.; Garcia, S.A.; Kubisiak, T.L.; Amerson, H.V.; Carlson, J.E.; Nelson, C.D.; et al. Evolution of genome size and complexity in Pinus. PLoS ONE 2009, 4, 1–11. [Google Scholar] [CrossRef]
- Wegrzyn, J.L.; Lin, B.Y.; Zieve, J.J.; Dougherty, W.M.; Martínez-García, P.J.; Koriabine, M.; Holtz-Morris, A.; de Jong, P.; Crepeau, M.; Langley, C.H.; et al. Insights into the loblolly pine genome: Characterization of BAC and fosmid sequences. PLoS ONE 2013, 8, e72439. [Google Scholar] [CrossRef] [PubMed]
- Cossu, R.M.; Casola, C.; Giacomello, S.; Vidalis, A.; Scofield, D.G.; Zuccolo, A. LTR retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol. Evol. 2017, 9, 3449–3462. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo-Correa, J.P.; Verdú, M.; González-Martínez, S.C. The contribution of recombination to heterozygosity differs among plant evolutionary lineages and life-forms. BMC Evol. Biol. 2010, 10, 22. [Google Scholar] [CrossRef]
- Stapley, J.; Feulner, P.G.D.; Johnston, S.E.; Santure, A.W.; Smadja, C.M. Recombination: The good, the bad and the variable. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20170279. [Google Scholar] [CrossRef]
- Leitch, I.J.; Leitch, A.R. Genome size diversity and evolution in land plants. In Diversity of Plant Genomes Volume 2: Physical Structure, Behaviour and Evolution of Plant Genomes; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer-Verlag: New York, NY, USA, 2012. [Google Scholar]
- Fawcett, J.A.; Van de Peer, Y.; Maere, S. Significance and Biological Consequences of Polyploidization in Land Plants; Springer-Verlag: Wien, Austria, 2013. [Google Scholar]
- Williams, C.G. Conifer Reproductive Biology; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Brown, G.R.; Gill, G.P.; Kuntz, R.J.; Langley, C.H.; Neale, D.B. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc. Natl. Acad. Sci. USA 2004, 101, 15255–15260. [Google Scholar] [CrossRef] [PubMed]
- Pavy, N.; Namroud, M.C.; Gagnon, F.; Isabel, N.; Bousquet, J. The heterogeneous levels of linkage disequilibrium in white spruce genes and comparative analysis with other conifers. Heredity 2012, 108, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Namroud, M.C.; Guillet-Claude, C.; Mackay, J.; Isabel, N.; Bousquet, J. Molecular evolution of regulatory genes in spruces from different species and continents: Heterogeneous patterns of linkage disequilibrium and selection but correlated recent demographic changes. J. Mol. Evol. 2010, 70, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Savolainen, O.; Pyhäjärvi, T.; Knürr, T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 595–619. [Google Scholar] [CrossRef]
- Prunier, J.; Verta, J.P.; MacKay, J.J. Conifer genomics and adaptation: At the crossroads of genetic diversity and genome function. New Phytol. 2016, 209, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Sena, J.S.; Giguère, I.; Boyle, B.; Rigault, P.; Birol, I.; Zuccolo, A.; Ritland, K.; Ritland, C.; Bohlmann, J.; Jones, S.; et al. Evolution of gene structure in the conifer Picea glauca: A comparative analysis of the impact of intron size. BMC Plant Biol. 2014, 14, 95. [Google Scholar] [CrossRef]
- Castillo-Davis, C.I.; Mekhedov, S.L.; Hartl, D.L.; Koonin, E.V.; Kondrashov, F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002, 31, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Gregory, T.R. The case for junk DNA. PLoS Genet. 2014, 10, e1004351. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Soltis, P.S.; Bennett, M.D.; Leitch, I.J. Evolution of genome size in the angiosperms. Am. J. Bot. 2003, 90, 1596–1603. [Google Scholar] [CrossRef]
- Nosaka, M.; Itoh, J.I.; Nagato, Y.; Ono, A.; Ishiwata, A.; Sato, Y. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet. 2012, 8, e1002953. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.; Allshire, R.C. RNA silencing and genome regulation. Trends Cell Biol. 2005, 15, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Fahlgren, N.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Carrington, J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007, 5, e57. [Google Scholar] [CrossRef]
- Zhang, X.; Henderson, I.R.; Lu, C.; Green, P.J.; Jacobsen, S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 2007, 104, 4536–4541. [Google Scholar] [CrossRef]
- Takuno, S.; Ran, J.H.; Gaut, B.S. Evolutionary patterns of genic DNA methylation vary across land plants. Nature Plants 2016, 2, 15222. [Google Scholar] [CrossRef]
- Zhuang, J.L.; Wang, J.; Theurkauf, W.; Weng, Z.P. TEMP: A computational method for analyzing transposable element polymorphism in populations. Nucleic Acids Res. 2014, 42, 6826–6838. [Google Scholar] [CrossRef]
- Platzer, A.; Nizhynska, V.; Long, Q. TE-Locate: A tool to locate and group transposable element occurrences using paired-end next-generation sequencing data. Biology (Basel) 2012, 1, 395–410. [Google Scholar] [CrossRef]
- Steinbiss, S.; Willhoeft, U.; Gremme, G.; Kurtz, S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009, 37, 7002–7013. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008, 9, 411–412. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).