Abstract
Fanconi anemia (FA) is a hereditary chromosomal instability disorder often displaying congenital abnormalities and characterized by a predisposition to progressive bone marrow failure (BMF) and cancer. Over the last 25 years since the discovery of the first linkage of genetic mutations to FA, its molecular genetic landscape has expanded tremendously as it became apparent that FA is a disease characterized by a defect in a specific DNA repair pathway responsible for the correction of covalent cross-links between the two complementary strands of the DNA double helix. This pathway has become increasingly complex, with the discovery of now over 20 FA-linked genes implicated in interstrand cross-link (ICL) repair. Moreover, gene products known to be involved in double-strand break (DSB) repair, mismatch repair (MMR), and nucleotide excision repair (NER) play roles in the ICL response and repair of associated DNA damage. While ICL repair is predominantly coupled with DNA replication, it also can occur in non-replicating cells. DNA damage accumulation and hematopoietic stem cell failure are thought to contribute to the increased inflammation and oxidative stress prevalent in FA. Adding to its confounding nature, certain FA gene products are also engaged in the response to replication stress, caused endogenously or by agents other than ICL-inducing drugs. In this review, we discuss the mechanistic aspects of the FA pathway and the molecular defects leading to elevated replication stress believed to underlie the cellular phenotypes and clinical features of FA.
1. FA Pathway and Models of Interstrand Cross-link Repair
Fanconi anemia (FA) is a hereditary disorder characterized by congenital abnormalities, progressive bone marrow failure (BMF), and predisposition to cancer, especially leukemia and aggressive head and neck squamous cell carcinoma (HNSCC) [1,2,3]. Head and neck squamous cell carcinoma is thought to arise within a geographical field of the mucosal epithelium, leading to tumors with chromosomal instability due to a deficiency in the FA pathway; contributions of human papilloma virus infection as well as genotoxic stress may contribute, but this is debated [3] Leukemia is thought to arise from chromosomal aberrations in the hematopoietic stem cells, leading to leukemic transformation by the rise of clonal populations [4] More research in this area is required to address the predominant origins of cancers observed in FA. Fanconi anemia patient-derived cells are extremely hypersensitive to agents that generate DNA interstrand cross-links (ICLs) that covalently bond the two strands of the DNA double helix to one another [5,6,7,8,9] There is a spectrum of structurally distorting and non-distorting DNA ICLs caused by endogenous sources that differ in their target sequences [10] Therefore, it is plausible that the physical structure at the ICL mediates the proteins that are involved in ICL unhooking and repair (discussed below). In addition, a variety of DNA lesions including ICLs can be induced by exogenous chemicals including cancer chemotherapy drugs (nitrogen mustards, cisplatin, mitomycin C (MMC), psoralen) that also differ in their target sequences and site-specific adducts [10] Among the agents leading to covalently cross-linked DNA and protein-DNA adducts is formaldehyde, which can arise endogenously. For a perspective on the chromosomal instability and hematopoietic stem cell attrition caused by formaldehyde, readers are referred to several recently published papers from the Patel lab and the references therein [11,12]. Emerging evidence suggests that the clinical and cellular features of FA are caused not only directly by ICL-induced chromosomal instability but also more globally by inflammation and defects in epigenetic regulation and telomere metabolism [13].
The connection of FA to accumulation of ICLs has prompted much interest in the source of ICLs and mechanism(s) for repair. Research in simple unicellular organisms such as yeast and bacteria has provided insight. Studies of E. coli dating back to the 1970s led researchers to believe that ICLs are repaired by both nucleotide excision repair (NER) and homologous recombination (HR) in sequential steps, but the details were unclear [14,15,16]. Later, it was proposed that ICL repair could also occur in nonreplicating bacteria by a non-recombinogenic mechanism requiring a translesion (TLS) DNA polymerase [17,18]. Moving to simple eukaryotes, research from a number of laboratories suggests that ICL repair in yeast is likely to be more complex, with a greater number of proteins from a more expansive list of classical repair pathways involved [19,20].
Since the discovery of the first FA gene FANCC over 25 years ago [21], mutations in a growing list of genes (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCN, FANCP, FANCQ, and FANCT) are linked to FA. In addition, mutations in three genes (FANCO, RAD51 (FANCR), and FANCS) are implicated in a FA-like syndrome in which patient cells are hypersensitive to ICL-inducing drugs, but the disease does not present with all the clinical features classically observed in FA [22,23]. Collectively, the FA gene products comprise a specialized pathway of ICL repair (Figure 1). In addition to these genes, FANCM is involved in ICL repair with other proteins in the FA pathway; however, it is considered an atypical FA gene because its linkage to FA has not been formally demonstrated [8,21]. Among the bona fide FA genes, FANCA, FANCC, and FANCG are most frequently inactivated by bi-allelic mutations linked to the hereditary disorder (Fanconi Anemia Database; http://www2.rockefeller.edu/fanconi/) [24]. Importantly, mono-allelic mutations in certain FA genes including FANCD1 (BRCA2), FANCS (BRCA1), FANCN (PALB2), FANCM, FANCJ (BRIP1), and FANCO (RAD51C) that are believed to operate downstream in the pathway and implicated in HR are associated with sporadic breast and ovarian cancer [24,25].
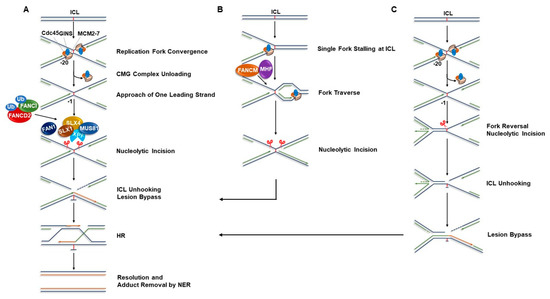
Figure 1.
Models of replication-coupled ICL repair. In replicating cells, ICLs are efficiently repaired by cooperative action of FA pathway proteins and the proteins involved in the NER, TLS, and HR pathways. Other than the classical ICL repair model of single fork convergence, there are different proposed mechanisms by which crosslinks can be repaired in a replication-dependent manner, enabling forks stalled at ICLs to resume DNA synthesis. (A) Dual fork convergence model. Crosslink repair is triggered when two replication forks converge at an ICL. The leading strands of two converging forks initially stall at ~20 nucleotides away from the lesion due to the steric hindrance imposed by the CMG complex (Cdc45/MCM2-7/GINS). Ubiquitin signaling promotes chromatin unloading of the CMG complex, which allows one of the leading strands on either side of the ICL to approach to within one nucleotide of the lesion. The FA pathway is simultaneously activated through the monoubiquitination of the FANCI‒FANCD2 complex. Ubiquitylated FANCI‒FANCD2 in turn promotes recruitment of SLX4 (FANCP), which acts as a molecular scaffold to recruit structure-specific endonucleases SLX1, XPF (FANCQ)-ERCC1, and MUS81 at the ICL site. Nucleolytic incision and subsequent unhooking of the ICL by the coordinated actions of these enzymes creates a DSB in one sister chromatid. FAN1, another endonuclease, is also implicated in the incision step. TLS polymerases, Pol ζ and REV1 subsequently extend the leading strand past the unhooked ICL and generate an intact DNA template suitable for HR repair. The DSB is finally repaired by HR and the unhooked ICL is removed by NER process. (B) Replication traverse model. In living cells, the majority (~60%) of replication forks encountering ICLs bypass the lesions without unhooking them in a manner dependent on the translocase activity of FANCM/MHF complex. The “X-shaped” DNA structures generated through the traverse process are subsequently processed by endonucleases followed by lesion bypass and post-replication repair as in dual fork convergence model. (C) Replication fork reversal at ICL. Following CMG complex unloading, one of the two converging forks undergoes reversal. The opposite fork is subsequently incised by the endonucleases leading to ICL unhooking. The lesion is then bypassed by the action of TLS polymerases and DSB intermediate generated during the incision process is finally repaired by HR. ICL: Interstrand cross-link; FA: Fanconi anemia; NER: Nucleotide excision repair; TLS: Translesion; HR: Homologous recombination.
Proteins implicated in the FA pathway primarily operate during the DNA synthesis (S)-phase of the cell cycle and cooperate with structure-specific endonucleases, DNA polymerases implicated in TLS, and HR proteins to fix the double-strand breaks (DSBs) that arise from processed ICLs. Although the dominant pathway of ICL repair in proliferating cells is replication-dependent, nondividing or slowly diving cells such as neurons and stem cells may employ a pathway of replication-independent repair (RIR) to correct these toxic lesions [26,27,28]. RIR operates through the coordination of TLS and NER, and requires DNA polymerase κ (Pol κ), DNA polymerase ζ (Pol ζ), and an ATP-dependent DNA translocase known as Cockayne syndrome Group B protein (CSB) to repair ICLs outside of S-phase [27,28,29]. The precise roles of FA gene products usually associated with the classical replication-dependent pathway in RIR remain to be thoroughly understood.
In the classical FA pathway of ICL repair, ICL-blocked replication forks are first recognized by a FANCM‒FAAP24‒MHF1‒MHF2 protein complex, which in turn activates the FA core complex to mono-ubiquitinate FANCD2-I heterodimer (ID-Ub); this represents a key signaling step in the FA pathway [8,30,31]. FANCD2-I mono-ubiquitination is followed by a series of events including ID-Ub-dependent incision and unhooking of the ICL by endonucleases, followed by TLS, and HR-mediated repair of the DSB intermediates. However, several models for processing of an ICL-stalled X-shaped replication fork have been proposed and these are being actively investigated (Figure 1). The simplest model depicts a single replication fork encounter of an ICL followed by unhooking, TLS, and fork restoration via HR [32,33]. Importantly, this model first delineated roles of FANCM (a DNA translocase) and FANCJ (a bona fide DNA helicase) in the FA pathway of ICL repair. FANCM was speculated to facilitate endonucleolytic cleavage by disrupting duplex DNA surrounding the ICL, whereas FANCJ was proposed to operate with other HR proteins to rejoin DSBs by a homologous DNA sequence-dependent pathway. However, this classical ICL-repair model lacked some mechanistic details regarding the precise roles of FA proteins and replisome proteins in the repair process.
More recently, a second model of ICL repair was proposed. Raschel et al. performed reconstitution experiments with Xenopus egg extract that was incubated with a plasmid carrying a single, site-specific ICL; they demonstrated that repair is triggered when two replication forks collide with the ICL [9]. In this dual fork convergence model of ICL repair (Figure 1A), the leading strands of two converging replication forks are initially stalled at ~20‒40 nucleotides (“-20 position”) away from the lesion due to steric hindrance imposed by the template bound CMG (CDC45, MCM2-7, and GINS) replicative helicase. Subsequent eviction of the CMG complex from the DNA allows the leading strands to approach further and extend up to one nucleotide away from the ICL (“-1 position”). The HR protein BRCA1 has been proposed to play a crucial role at this step by promoting unloading of the CMG complex, thereby paving the way for leading strand synthesis to extend towards the ICL [34]. Concurrent activation of the FA pathway via mono-ubiquitylation of the FANCD2-I complex in turn promotes the incision of one of the parental strands by XPF‒ERCC1 and another incision in the same strand by possibly another endonuclease(s), thereby “unhooking” the ICL and creating a DSB. TLS polymerases such as DNA polymerase ζ and REV1 facilitate lesion bypass on the opposite strand and recreate an intact duplex that serves as a template for subsequent HR-mediated repair of the DSB. The DSB is finally repaired by HR and the unhooked ICL remnant is removed by NER. This model proposes that the X-shaped structure formed when two replication forks converge at an ICL is the essential triggering factor for the repair process to begin. This mechanism was further supported by a subsequent study where a single replication fork stalled at an ICL was shown to be unable to promote ICL repair in a cell-free Xenopus egg extract [35]. Further studies are required to tease out the precise mechanistic details of the dual convergence model that operate in vivo. Moreover, it remains to be determined how precisely conserved are the molecular events of ICL repair in the Xenopus reconstituted system compared to mammalian cells.
Given the uncoupling of the CMG replicative helicase prior to ICL repair, the question has been raised how replication resumes without CMG loading following fork recovery unless another fork from the opposite direction arrives. This points towards the logical validity of the dual-fork convergence concept. However, given the long inter-origin distance (~100 kb) in eukaryotic cells, simultaneous arrival of two replication forks at an ICL seems unlikely and it is conceivable that one fork encounters the barrier before another fork arrives in vivo [36]. Therefore, both single fork collision and dual-fork convergence at ICLs are possible, but the relative frequencies might vary. Importantly, productive ICL repair was shown to occur in Xenopus egg extracts even when there was a significant time gap between the arrival of the two forks at an ICL [35]. In 2013, Seidman and colleagues proposed a new concept of ICL repair, which they termed “replication traverse” [37]. Using a specialized DNA fiber technique, they monitored DNA replication tracts at fluorescently labeled ICLs in living cells and showed that instead of ICL “unhooking”, the majority of the replication forks (~60%) bypass or “traverse” the ICLs in a manner dependent on the translocase activity of FANCM/MHF complex (Figure 1B). The remnant ICLs are likely to be removed through post-replication repair. ICL traverse of the replisome is essentially dependent on the functional interaction between FANCM and the Bloom’s syndrome helicase complex (BLM, Topoisomerase 3α, RMI1 and RMI2); together, the proteins operate in an epistatic manner in crosslink repair [38]. In a parallel study, Kisker’s group further showed that the replication traverse is dependent on the functional interaction of FANCM with the replicative polymerase co-factor PCNA [39]. Interestingly, FANCM plays apparently distinct roles in replication-transverse and the classical FA pathway of ICL repair. In addition to FANCM’s role as a DNA translocase to promote replication-traverse, FANCM also facilitates chromatin recruitment of the FA core complex and subsequent FANCD2-I mono-ubiquitination in response to ICL-induced DNA damage [40,41,42]. However, cells deficient of FANCM are less sensitive to ICLs compared to cells with an inactivated FA core complex, suggesting that replication traverse plays a less protective role than the conventional FA pathway when the replication machinery encounters an ICL [40,43].
Although conventional wisdom would argue that the replisome machinery cannot continue to synthesize DNA on the other side of a DNA ICL, experimental data supporting the Seidman model for replication traverse have recently been generated by the Lopes lab and provide further mechanistic insight [44]. Fork traverse requires global fork slowing mediated by fork reversal enacted by the double-stranded DNA translocase ZRANB3 and HR protein RAD51 as well as ATR checkpoint activation. Moreover, as reported by the Seidman group [37], FANCM was shown to promote efficient fork traverse thereby acting as a stimulatory component of the specialized fork restart pathway. Determination of the precise ATR targets supporting replisome progression and stability remains to be seen.
The molecular mechanism(s) of ICL repair are still being investigated and a direct comparison of the processing efficiency of single fork, dual fork, and traversed fork at ICLs would provide insight. Using Xenopus egg extracts, the Walter group showed that a classic psoralen-induced ICL or an abasic site ICL characterized by a nonnative N-glycosyl bond can be “unhooked” by NEIL3, a DNA glycosylase [45]. This distinct mechanism of replication coupled-ICL repair does not require FANCI-FANCD2-dependent incision, thereby preventing generation of DSB intermediates. In another interesting study, they further demonstrated that one of the two converging forks at an ICL undergoes reversal in a manner that is dependent on prior removal of the CMG from the template strand (Figure 1C) [46]. In this context, Lopes and co-workers recently reported that ICL induction causes replication fork slowing and reversal at crosslink sites as well as non-ICL sites [44]. Using DIG-TMP (Digoxygenin-tagged TMP)-based ICL immunolabeling combined with DNA fiber assay and electron microscopy, the authors provided experimental evidence that in response to ICLs, ATR-mediated global fork slowing rapidly recruits RAD51 to chromatin leading to fork reversal. This in turn facilitates single-fork ICL traverse and prevents chromosomal instability. This new information not only updated the existing dual fork convergence model of ICL repair but also presented a new avenue of research, namely fork protection in the context of ICL repair. Certain FA proteins (BRCA2, RAD51, FANCD2) and associated factors (DNA helicase-nuclease proteins DNA2 and WRN (the latter mutated in the premature aging disorder Werner syndrome [47]) protect forks from dysregulated degradation, which is critically important for genomic stability [48,49,50,51,52,53]. Therefore, it will be important to delineate the functions of WRN and DNA2 in fork protection at sites of ICLs.
2. Functional Involvement of FA Proteins with Players in Other DNA Repair Pathways
Functional integration of FA proteins with NER, TLS, and HR repair factors is indispensable for ICL repair. In addition, many proteins of the FA pathway collaborate with these repair factors to maintain genome integrity in response to non-ICL directed DNA damage.
2.1. Nucleotide Excision Repair
In eukaryotes, helix-distorting bulky DNA lesions induced by UV exposure or platinum-based cancer chemotherapeutics are primarily repaired by the NER pathway [54,55]. Bi-allelic mutations in NER genes cause Xeroderma pigmentosum (XP), an autosomal recessive disorder characterized by impaired tolerance to UV light and predisposition to skin cancer [56]. XP patient-derived cells are hypersensitive to ICL-generating drugs, suggesting a functional role of NER proteins in DNA cross-link repair [29,57]. Consistent with this, cells inactivated of structure-specific NER endonucleases XPF‒ERCC1 or MUS81‒EME1 are hypersensitive to DNA crosslinking agents [57,58,59,60,61]. Moreover, ERCC1-deficient mice are prone to develop a hematopoietic defect like that observed in FA, implicating a direct role of the heterodimeric XPF‒ERCC1 endonuclease complex in the FA pathway [62]. Importantly, FA patients with mutations in the ERCC4 (FANCQ) gene encoding XPF endonuclease have been reported [63] and ERCC4 restoration in FA patient-derived cells has been shown to complement MMC sensitivity [63,64]. At the mechanistic level, the ERCC1‒XPF nuclease complex acts in concert with FANCD2 and SLX4/FANCP to incise ICLs in the FA pathway [8,64,65,66]. Ubiquitylated FANCD2 facilitates the recruitment and activation of structure-specific nucleases including XPF‒ERCC1 and MUS81‒EME1 via the SLX4 scaffolding protein at the site of the crosslink and subsequently promotes ICL “unhooking,” thereby generating DSB intermediates. Consistent with the role of MUS81‒EME1 in the incision step, DSB induction is severely compromised in MUS81 -/- mouse embryonic stem (ES) cells upon MMC or cisplatin treatment [58]. Moreover, the in vitro purified MUS81‒EME1 complex has been shown to cause an incision at the 5´ side of psoralen-induced ICL [67]. Experimental evidence, however, also suggests involvement of the structure-specific endonucleases in the downstream HR-mediated repair of the ICL-induced DSB intermediates. In agreement with this notion, ERCC1-/- mouse embryonic fibroblast cells (MEFs) display accumulation of DSBs upon ICL induction [61]. Furthermore, cells depleted of SLX4 or SLX1 are defective in resolving cisplatin-induced DSBs, implying the role of these nucleases in the downstream HR repair step. Likewise, experimental evidence suggests molecular links between Mus81 and HR-mediated repair as MUS81 has been shown to physically interact with the HR factor RAD54 [58,68]. Overall, these pieces of evidence suggest that the observed ICL hypersensitivity of human cells lacking structure-specific endonucleases is attributed not only to the defects in the initial incision step but also to the inefficient downstream processing of the incision-induced DSB intermediates.
Certain FA proteins facilitate NER of bulky DNA adducts such as UV photoproducts. BRCA1 (FANCS)-deficient cells are hypersensitive to UV-C-induced damage, suggesting its role in photoproduct elimination [69]. BRCA1 promotes XPF‒ERCC1 recruitment to replication forks stalled at unrepaired UV lesions and facilitates lesion excision in a replication-dependent manner [54]. Two additional FA components, the ubiquitin-conjugating enzyme UBE2T and the DNA translocase FANCM, also influence NER [70]. Genetic dissection experiments in avian DT-40 cells revealed that these two FA proteins cooperatively promote NER of UV-induced DNA damage and are important for protecting cells from UV genotoxicity. However, FANCM-deficient chicken DT-40 cells were found to be as effective as wild-type cells in photolesion removal. This suggests that FANCM likely functions downstream of the lesion excision step in the NER repair pathway. In contrast, UBE2T possibly facilitates UV lesion excision as UBE2T null cells were found to be defective in lesion removal, but the mechanistic role of the ubiquitin-conjugating enzyme to confer UV resistance is still poorly understood. Nonetheless, the collective observations suggest that the interaction of protein players in the FA and NER pathways plays a pivotal role in repairing DNA cross-links and other bulky DNA lesions.
2.2. Double-Strand Break Repair
The functional link between FA and HR proteins in a common ICL repair pathway first became evident when FA patients with biallelic mutations in BRCA2, a key player in HR-mediated DSB repair, were reported [71]. This finding assigned BRCA2 as a bona fide FA gene, designated FANCD1. Subsequent clinical and mouse genetic studies revealed that BRCA2 loss gives rise to FA-like phenotypes including chromosomal abnormalities and robust hematological defects [72,73,74]. Genotype‒phenotype correlation studies in FA or FA-like patients further demonstrated functional roles for other HR factors including BRCA1 (FANCS), PALB2 (FANCN), RAD51 (FANCR), and FANCJ in ICL repair pathway [51,75,76,77,78,79,80,81,82,83]. BRCA1 in conjunction with PALB2, BRCA2, and RAD51 mediates the necessary downstream HR-dependent DSB repair events in the FA pathway [10,84,85,86,87]. However, BRCA1 has additional HR-independent functions upstream of the FANCD2-I monoubiquitylation event. Cell-based experiments provided evidence that BRCA1 is important for FANCD2 recruitment to ICL sites [88,89,90]. Moreover, BRCA1 promotes CMG helicase eviction from an ICL-stalled fork, an early step necessary for the repair process to begin [34]. FANCJ, like BRCA1, may also play an early role in the ICL response to facilitate chromatin loading of monoubiquitinated FANCD2 [91]. Zhang et al. presented data suggesting that cells expressing a FANCJ mutant specifically defective in its interaction with BRCA1 displayed impaired FANCD2 nuclear foci formation in unperturbed conditions as well as upon MMC treatment [92], consistent with coordinate involvement of FANCJ and its interacting partner BRCA1.
As opposed to its upstream role in the activation of the FA pathway, FANCD2 is also implicated in regulating HR-mediated repair of DSB intermediates formed during ICL repair, where it facilitates DNA end-resection through direct interactions with the CtIP and DNA2 nucleases [93,94,95]. In addition to its role in DSB processing, CtIP is involved in the initiation events of ICL repair in a BRCA1- and FANCM-dependent manner [89]. Thus, evidence suggests that HR proteins function both upstream and downstream of the FA core complex in the classical FA pathway of ICL repair. As the model of ICL repair is evolving, further research will provide more significant insights into the multi-level functional crosstalk between HR and FA proteins in repairing DNA crosslinks. Importantly, FA pathway components also contribute to repairing ICL-independent DSBs. FA proteins believed to operate both upstream (FANCA and FANCG) and downstream (FANCD2) in the pathway have been shown to promote HR-mediated repair of DSBs [96,97]. However, unlike BRCA1, BRCA2, and RAD51, mutations in the FA core complex or FANCD2 monoubiquitylation sites do not result in a severe HR defect, suggesting a non-essential role of upstream FA components in HR repair [97,98,99,100,101].
Although autosomal recessive mutations in certain RecQ helicase genes (WRN, BLM, RECQL4) are linked to accelerated aging diseases characterized by cancer predisposition, chromosomal instability, and a heightened sensitivity to DNA cross-linking agents and other genotoxic drugs, they are not genetically implicated in FA [102]. WRN, like BLM and RECQL4, plays functional roles in DSB repair that are likely to influence the downstream events of ICL repair [103,104]. Given its role in DNA end-resection in conjunction with DNA2, WRN might play an important role in generating 3´ ssDNA overhangs at DSB sites to initiate HR repair of the DSB intermediates [105]. On the other hand, WRN might also help in resolving HR products generated during the repair of DSB intermediates, as suggested by studies from the Monnat lab [104]. Indeed, WRN collaborates with BRCA1 in ICL repair [106], and it is conceivable that WRN might play a backup role to the FA pathway. In support of this idea, our laboratory showed that pharmacological suppression of WRN-catalyzed DNA unwinding by a WRN-specific helicase inhibitor (WRNi) compromises the cellular ICL response when the FA pathway is defective as evidenced by the hypersensitivity of FANCD2 or FANCA mutant cells to MMC upon WRNi treatment [107]. It is conceivable that WRN helps to resolve HR intermediates formed at the later stage of ICL repair and loss of WRN’s helicase function impairs homology-directed ICL repair leading to activation of nonhomologous end-joining (NHEJ) repair, particularly when a protein of the FA pathway (e.g., FANCD2, FANCA) is defective.
Aside from promoting HR-mediated DSB repair, the FA pathway likely suppresses the error-prone NHEJ mode of DNA repair [108,109,110]. In addition, data obtained from cell-based reporter assays suggest an important role of FA pathway components to promote alternative end-joining pathway of DSB repair [110]. In this context, Benitez et al. reported that FANCA plays a direct role in DSB repair by catalyzing single-strand annealing (SSA) and strand exchange (SE) in a FANCG-dependent manner [111]. Consistent with this, patient-derived FANCA mutants were found to be defective in both SSA and SE activities. Collectively, these observations suggest that FA pathway proteins ensure genome stability by promoting error-free pathways of DSB repair.
2.3. Mismatch Repair
Functional interplay between the FA and mismatch repair (MMR) pathways is important for a robust response to agents that induce ICLs. Components of MMR protein complexes, specifically MutSα and MutSβ, are involved in ICL recognition as well as unhooking and HR-mediated repair of the processed ICL-induced DSBs [112,113,114,115]. Furthermore, physical interactions of MMR proteins with FANCD2, BRCA1, FANCJ, and SLX4 (FANCP) have been reported [116,117,118,119,120,121,122]. Along with other DNA repair proteins, MMR factors are integral components of the BRCA1-associated genome surveillance complex (BASC), which is believed to act as a crucial sensor for cellular DNA damage [116]. Mass spectrometry analyses of BRCA1 immunoprecipitated proteins from HeLa nuclear extracts revealed physical interactions between BRCA1 and the MMR proteins MSH2, MSH6, and MLH1 [116]. As a part of the BASC complex, the MSH2‒MSH6 heterodimer may recognize ICL-induced distorted helix structures and trigger DNA damage response signaling. Perhaps BRCA1 regulates DNA damage sensing by the MMR proteins or conversely these MMR factors might help BRCA1 to execute its repair functions in the FA pathway. High-throughput proteomic studies provided evidence for molecular interactions of MMR proteins with the structure-specific endonuclease FAN1 and scaffolding protein SLX4 implicated in FA [99,117,123,124]. LC-MS/MS-based proteomic analyses of HA-tagged SLX4 immune complex isolated from 293TREX cells showed that MSH2 and MSH3 reside in a large multimeric protein complex formed by SLX4 [117]. Moreover, protein‒protein interaction studies based on a tandem affinity purification (TAP) technique demonstrated that FAN1 interacts with MLH1 and PMS2 [123,124].
FANCJ directly interacts with MLH1, a component of the MutLα complex, through its helicase domain [122]. This interaction is essential for ICL repair as an impaired FANCJ‒MLH1 interaction renders cells sensitive to DNA cross-linking agents [122]. The MMR protein MSH2 is important in activating the FA pathway following ICL generation [121]. Although interactions of MSH2 and MLH1 with FANCD2 are both induced by DNA crosslinks, only MSH2 is important for FA pathway activation [121]. These findings suggest that while MSH2 functions upstream of FANCD2 monoubiquitination and triggers FA pathway activation, MLH1 functions downstream in the FA pathway and facilitates the repair of DSB intermediates. Consistent with this, either MSH2-deficient or MLH1-deficient cells are sensitive to ICL-inducing agents. Interestingly, MSH2 depletion in cells lacking a functional FANCJ‒MLH1 interaction suppresses ICL sensitivity and promotes replication restart [125]. Therefore, the MSH2-dependent toxic DNA damage response renders FA-defective cells sensitive to DNA cross-linking agents. Thus, an accurate functional coordination between FA and MMR pathways is critical to suppress a deleterious DNA damage response to ICLs.
3. Involvement of the FA Pathway in Replication Fork Stabilization
When replication forks encounter DNA damage, an unusual DNA structure (e.g., G-quadruplex), or a transcribing RNA polymerase complex, the fork can be hindered in its progression. Stalled forks can also arise when cells are exposed to a DNA polymerase inhibitor (e.g., aphidicolin (APH)), an agent that causes nucleotide pool depletion (e.g., HU), or oncogene activation. Different structures form at stalled replication forks, depending on the nature of the block. In these situations, certain proteins are recruited to the site of the stalled fork to help manage the cellular response to the distress. Perhaps the myriad of DNA structures (e.g., leading strand gap, lagging strand gap, non-canonical DNA structure stabilized by Hoogsteen hydrogen bonds, regressed four-stranded Holliday Junction-like structure, etc.) helps to explain the diversity and redundancy of factors involved in replication fork protection. The metabolism of the stalled fork is growing to be much more complex than previously thought, but in simplest terms, a stalled fork center is dealt with by proteins and events that are focused on preserving the fork’s integrity while enforcing appropriate cell cycle checkpoints and allowing DNA synthesis to resume as efficiently as possible. ATP-dependent DNA translocases or fork remodeling enzymes, DNA binding proteins, DNA nucleases, and helicases are among the key players called upon to deal with stalled forks. For further reading on the more global topic of fork management, we recommend some recently published reviews on the topic [126,127,128,129,130].
Replication fork stabilization under conditions of stress (e.g., nucleotide deprivation or genotoxin exposure that induces replication-blocking lesions) allows for fork protection from deleterious nucleolytic resection and timely restart of stalled forks [126,127,128]. Besides their canonical ICL repair functions, proteins of the FA pathway play a central role in stabilizing stressed replication forks, thereby ensuring genome integrity. The FA pathway is rapidly activated (as evidenced by FANCD2 mono-ubiquitination and FANCD2 nuclear foci formation) upon cellular exposure to HU or APH that causes replication fork stalling but does not generate ICLs [131,132]. Moreover, increased chromosomal instability observed in FA-deficient cells experiencing replication stress is attributed to impaired stabilization of the stressed replication forks [49,131,133,134].
A direct role of the FA/HR pathway in protecting stalled forks entered the spotlight through two breakthrough discoveries from the Jasin lab. Using the single-molecule DNA fiber assay, Schlacher et al. showed that BRCA2 (FANCD1) protects stalled replication forks from MRE11-mediated over-resection by stabilizing Rad51 nucleofilament [48]. A similar fork protective function of FANCD2 and BRCA1 (FANCS) was subsequently described in their second study [49]. Like BRCA2, stalled fork protection by FANCD2 also requires RAD51 filament stabilization and is epistatic with RAD51 [49]. Furthermore, a non-epistatic interaction between FANCD2 and BRCA2 has been delineated [135,136]. In the absence of BRCA2, FANCD2 protects stalled forks from MRE11-dependent degradation [135]. Although blocking MRE11 function completely rescues the fork degradation phenotype in cells deficient of either BRCA2 or FANCD2, only partial recovery of the phenotype is observed in cells lacking both factors [135]. Thus, an additional nuclease(s) other than MRE11 plays a role in fork resection governed by BRCA2 and FANCD2.
Although fork de-protection leads to increased chromosomal instability in FA-defective cells, they are not overtly sensitive to replication stalling agents such as HU, camptothecin, or gemcitabine [49], suggesting redundant pathways ensure cell viability but perhaps at the cost of chromosomal instability. This implies a crucial molecular link between FA pathway loss and tumorigenesis under conditions of replication stress when chromosomal aberrations/mutations arise due to severe fork instability. FANCA loss or a ubiquitination defect in FANCD2 also results in stalled fork degradation upon HU treatment [49]. This suggests that factors functioning upstream of FANCD2 mono-ubiquitination in the FA pathway are also involved in stalled fork protection [49]. Consistent with this notion, the FA core complex protein FANCB was shown to operate in collaboration with RECQL5 helicase to prevent nascent strand degradation upon HU treatment [137]. Very recently a fork protective role of FANCJ has also emerged (see FANCJ section) [138]. Furthermore, a novel protein designated Biorientation Defect 1-like (BODL1), associated with nascent DNA and sharing homology in its N-terminal region with the mitotic regulator BOD1, was discovered [139]. A role of BODL1 in the DNA damage response is suggested by the presence of multiple ATM/ATR phosphorylation sites. It was proposed that BODL1, like FANCJ (another downstream component of the FA/HR pathway) is involved in RAD51-dependent stalled fork protection. In the absence of BODL1, BLM/FBH1-mediated destabilization of RAD51 nucleofilament causes nascent DNA strand degradation behind the stalled fork and promotes genomic instability. However, BODL1, unlike BRCA2 and FANCD2, protects stalled forks from DNA2 nucleolytic degradation instead of MRE11 nuclease attack. Thus, different FA pathway proteins prevent stalled fork degradation by counteracting distinct nucleases and possibly act in concert to provide stability to the stressed forks. Intriguingly, although they counteract the function of two distinct nucleases at stalled fork, BODL1 depletion does not further exacerbate fork degradation in BRCA2-deficient cells [139]. This observation suggests that when both BRCA2 and BODL1 functions are lost, either one of the nucleolytic resection pathways dominates over the other or both pathways act in concert in a controlled manner at stalled forks. In this context, Liao et al. recently proposed that a negative feedback mechanism restrains elevated resection to prevent deleterious single-stranded DNA accumulation under conditions when both fork protection factors are absent [128]. However, further research to identify additional nucleases functioning at stressed replication forks in defined genetic backgrounds is required to obtain a better understanding of the functional interplay among different FA pathway proteins in protecting stalled replication forks.
Besides its role in preventing deleterious nucleolytic resection of stalled forks, proteins of the FA pathway have additional functions to counteract replication stress. FANCD2 cooperates with BLM to facilitate stalled fork restart and suppress new origin firing [140]. Importantly, while FANCD2 prevents stalled fork degradation independently of BLM, BLM promotes fork progression following restart in a FANCD2-independent manner [49,140]. Thus, FANCD2 and BLM have cooperative as well as independent roles in the context of stalled fork recovery. Furthermore, upon replication fork stalling the FA-associated nuclease FAN1 interacts directly with the chromatin-bound FANCD2‒BLM complex and cooperatively promotes replication fork restart [141]. Here, FAN1 chromatin access is tightly regulated by FANCD2, which prevents FAN1 nuclease-mediated over-resection of the stalled fork [141]. In the absence of FANCD2, uncontrolled nucleolytic activity of FAN1 along with MRE11 results in nascent DNA strand degradation behind the fork [141]. Lachaud et al. further showed that ubiquitinated FANCD2 promotes stalled fork processing by directly recruiting FAN1 nuclease to stalled replication forks [134]. This in turn restrains DNA replication under stressed conditions and prevents chromosomal instability [134]. Apart from FAN1, CtIP nuclease is also implicated in the FANCD2-driven recovery of stalled replication forks [142]. Importantly, FANCD2-mediated stalled fork recovery is completely independent of the FA core complex, suggesting the process operates with downstream FA pathway proteins such as BRCA2 and FANCJ [143].
In response to replication stalling, experimental evidence suggests that FA proteins directly interact with the replisome and regulate DNA synthesis. Data obtained from experiments utilizing a technique known as isolation of proteins on nascent DNA (iPOND) followed by mass spectrometry (MS) analysis revealed that in HU-treated cells FANCD2 and FANCI are recruited to newly synthesized DNA in response to nucleotide depletion [144]. Moreover, in response to HU treatment, FANCD2 binds to the MCM2-7 replicative helicase complex and regulates its function to restrain DNA synthesis at stalled forks thereby preventing accumulation of single-stranded DNA gaps and genomic instability [144]. While FANCD2 restricts DNA synthesis, FANCI induces MCM2-7 helicase activity to promote dormant origin firing to help rescue stalled replication forks in response to mild replication stress [145]. Although not mechanistically clear, this function of FANCI in promoting dormant origin firing is inhibited by FANCD2 [145]. However, under conditions of severe stress (e.g., > 2 mM HU) exposure or prolonged (>18 h) HU incubation), activated ATR signaling phosphorylates FANCI to suppress new origin firing and provide cells with enough time to repair the damage and resume replication [145]. DNA synthesis by the translesion DNA polymerase kappa in response to HU-induced fork stalling is regulated by mono-ubiquitinated FANCD2 (as well as PCNA polyubiquitylation), suggesting that FA pathway activation is required for replication stress recovery [146]. Thus, the FA pathway functions at three distinct levels to recover stalled replication forks: i) fork protection from nucleolytic degradation, ii) stalled fork restart, and iii) suppression of new origin firing. Future studies will provide more mechanistic insights into these important non-canonical functions of the FA pathway to deal with replication stress.
4. New Developments in Understanding FANCJ’s Role in the Replication Stress Response
Although much insight to the roles of RecQ helicases in managing fork stress has been garnered over the years [102], up to only very recently have there been advances in the characterization of other auxiliary helicases such as those from the iron‒sulfur (Fe‒S) family, which includes FANCJ, RTEL1, DDX11, and XPD [147]. Among these, FANCJ is implicated in both hereditary disease and cancer and thought to have a specialized role(s) during DNA replication [148]. Since the discovery of FANCJ mutations linked to FA, there have been numerous studies on the molecular and cellular functions of the Fe‒S DNA helicase that address its broader roles in genome metabolism. Beyond its involvement in ICL repair through a still poorly understood series of downstream events to mediate HR repair of the ICL-induced DSB, it is now evident that FANCJ is a key player in the replication stress response. In this section, we will summarize the latest developments on FANCJ and mechanistic aspects of how the helicase is proposed to operate at stalled replication forks. The reader is referred to earlier reviews on FANCJ to provide a historical backdrop for this perspective [149,150,151].
Recently, we addressed the catalytic requirements of FANCJ necessary for its function in DNA repair versus response to replication stress [152]. By characterizing patient-derived FANCJ missense mutations residing in the helicase core domain both biochemically and genetically, we determined that a minimal threshold of FANCJ helicase activity is required for the cellular response to the DNA polymerase inhibitor APH, G-quadruplex ligand telomestatin (TMS), or the DNA strand-breaking agent bleomycin (BLEO). A catalytically inactive FANCJ-H396D mutant failed to rescue FANCJ null cells for any of these agents, whereas a partially active FANCJ-R707C mutant characterized by a dimerization defect and reduced processivity was competent to confer significant cellular resistance to APH, TMS, or BLEO. Interestingly, expression of either FANCJ-H396D or FANCJ-R707C completely failed to restore resistance to the DNA cross-linking agents MMC or cisplatin, indicating that optimal FANCJ helicase activity is required for ICL repair. This observation is consistent with a previous study in which it was shown that a patient-derived FANCJ missense mutation in the Fe-S domain that cripples helicase activity but leaves ATPase and single-stranded DNA translocase functions intact abolishes its ability to restore resistance of FANCJ null cells to an ICL-inducing agent [153]. Furthermore, these findings reinforce the belief that sensitivity of FANCJ patient cells to ICL-inducing agents is an excellent biomarker for FA. Further studies in this area should address if other missense mutations in FANCJ (linked to FA or associated with breast or ovarian cancer) that specifically interfere with key protein interactions of FANCJ but leave the catalytic function intact are able to differentially function in ICL repair versus response to agents that impede but do not entirely arrest cellular DNA replication. In terms of the replication fork, we surmise that it makes sense that the role of FANCJ in fork reconstruction via HR repair of ICL-induced damage would require maximal catalytic DNA unwinding (or possibly branch-migration activity), whereas the role of the helicase to help remodel or protect stalled forks during replication stress would only require partially active FANCJ helicase. New studies from the Cantor lab (discussed below) may begin to shed light on FANCJ’s role at stalled forks.
The Cantor lab took a novel pair-wise approach to address FANCJ’s role in replication [138]. They examined the effects of FANCJ deficiency on fork protection as measured by the popular DNA fiber analysis and determined the identity of unique proteins associated with replication forks by iPOND in CRISPR (Cas9) FANCJ knockout (KO) cells compared to the same cells endogenously expressing wild-type FANCJ. The DNA fiber results showed that nascently synthesized DNA is degraded in FANCJ-KO U2OS cells exposed to 4 mM HU in a MRE11 nuclease-dependent manner. The fork degradation was also observed in FA-J patient fibroblasts; suppression of fork degradation was dependent on expression of helicase-active FANCJ. In terms of the iPOND results, the abundance of several proteins in unstressed FANCJ-deficient cells was decreased, including ones involved in chromatin remodeling (e.g., SMARCAL1) and HR promotion (KIF4A). Notably, the replication fork remodeling enzyme HLTF was enriched at the fork and found to be preferentially associated with chromatin in FANCJ-KO cells. Moreover, DNA fiber analysis showed that HTLF depletion in HU-treated FANCJ-KO cells decreased fork degradation; however, it was not determined if ATP hydrolysis-dependent catalytic function of HLTF was involved. Further study demonstrated that HLTF helps FANCJ-KO cells recover from stress imposed by HU. Finally, Peng et al. provided experimental evidence that the associated unrestrained replication and single-stranded DNA gaps characteristic of HLTF-depleted cells were prevented by FANCJ deficiency. This led the authors to propose a model in which the FANCJ DNA helicase and HLTF fork remodeler keep each other in check to balance fork degradation and DNA synthesis elongation under conditions of replication stress.
There are several important questions to be addressed regarding the significance of the FANCJ-HLTF fork protection model in vivo and from a biomedical perspective: 1) Does the model apply under conditions of replication stress imposed by non-B-form DNA structures such as endogenous G-quadruplexes (in which FANCJ is already implicated [154,155,156,157,158,159]) or covalent DNA damage imposed endogenously (e.g., formaldehyde by-products [12]) or exogenously by compounds such as those used in chemotherapy [7]? 2) Are chromosomal fragile sites contributing to genomic instability prevalent in cancer and other disease states and aggravated by replication stress found in greater abundance when FANCJ and HLTF activities are not balanced properly? 3) Do defects in the coordinate action of FANCJ and HLTF contribute to FA, and are there other chromosomal instability disorders that by analogy arise due to deficiencies in balancing catalytic activities of specific molecular motors at the stalled replication fork? 4) Do FANCJ and HLTF represent druggable targets to enhance anticancer treatments, as proposed for other DNA damage response proteins [160]. Broadly speaking, an improved understanding of fork protection and its importance for human disease and cancer will help researchers and clinicians to bridge gaps in our knowledge bringing enlightenment for better diagnosis, treatment, and cures.
Finally, while small molecule inhibitors of RecQ helicases WRN [107,161,162] and BLM [163] have attracted attention as an approach to enhance anticancer therapies, we propose that FANCJ may also be a useful target for pharmacological inhibition. Given its importance in ICL repair induced by chemotherapy drugs (e.g., MMC, cisplatin), the emerging role of FANCJ to help cells deal with stalled forks suggests that the helicase may be a good target for inhibition to enhance the killing effects of drugs that perturb fork progression in rapidly dividing cancer cells. Here it may be advantageous to identify FANCJ-specific DNA helicase inhibitors using molecular docking interfaced with a computational approach; however, as we await a solved FANCJ structure, the approach would have to rely on a homology model of the FANCJ helicase core domain based on the sequence-related Fe‒S helicase XPD [164].
In addition to its fork protection role, FANCJ is important for fixing broken replication forks via HR repair of DSBs during the S and G2 phases of the cell cycle. This idea builds upon the original observation that FANCJ physically binds to the tumor suppressor BRCA1, which is also implicated in HR repair [80]. Most recently, the Nagaraju lab acquired experimental evidence that FANCJ regulates sister chromatid recombination in a manner that is dependent on its helicase activity and interaction with BRCA1 [165]. Flipping the coin, FANCJ may play a role in preventing fork stalling in the first place by resolving alternate DNA structures such as G-quadruplexes, as mentioned above. Recently, two laboratories independently showed that FANCJ prevents microsatellite instability during replication to suppress chromosome breakage and translocations, presumably by resolving non-B DNA structures in a manner that is independent of the classic FA pathway used to repair ICLs [166,167]. Thus, FANCJ operates at four distinct levels during replication stress or DNA repair to maintain genomic stability (Figure 2).
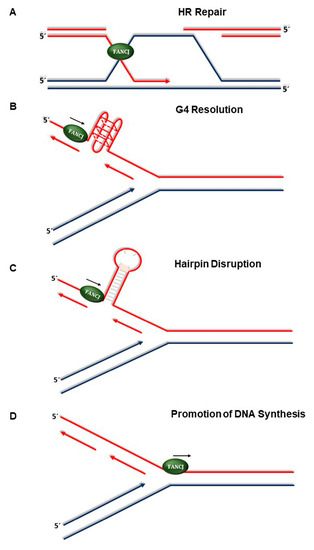
Figure 2.
Roles of FANCJ in the replication stress response. (A) FANCJ facilitates homology-directed repair of DSBs or ICL-induced broken replication forks. (B) FANCJ resolves G-quadruplex structures formed at transient single-stranded DNA regions during replication or possibly transcription. (C) FANCJ unwinds hairpin duplexes formed in single-stranded DNA templates during replication fork progression through repetitive DNA sequences of the genome thereby suppressing microsatellite instability. (D) FANCJ promotes DNA synthesis at stalled replication forks to minimize HLTF-mediated fork regression and prevent fork degradation.
5. Involvement of FA Proteins in the Response to R-Loop-Induced Replication Stress
Recent studies highlighted a crucial non-canonical role of the FA pathway in regulating R-loop-driven genomic instability [168,169] (Figure 3). R-loops are transcription-associated three-stranded nucleic acid structures that form when nascent RNA anneals with the template DNA strand (DNA: RNA hybrid) while displacing the other strand of the duplex DNA. Although programmed R-loop formation is beneficial for certain biological processes including control of gene expression, abnormal accumulation of these hybrid structures causes DNA damage and genomic instability [170,171,172]. Moreover, persistent R-loops impair transcription and obstruct replication fork movement, thereby generating replication stress [170,172,173,174,175,176]. However, cells have multiple ways to either prevent the unscheduled formation of the co-transcriptional R-loops or efficiently remove them from the genome to minimize replication‒transcription conflicts [177].
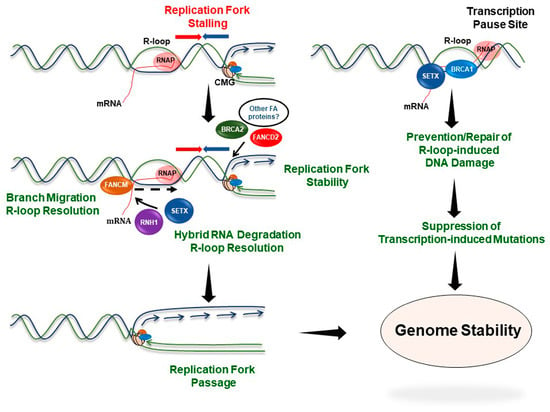
Figure 3.
The Fanconi anemia pathway suppresses R-loop-associated genome instability. A replication fork encountering a R-loop is stabilized by the coordinated functions of proteins implicated in FA. BRCA2, FANCD2, and possibly other FA proteins are recruited to the replication fork encountering a DNA‒RNA hybrid to prevent fork collapse. The FA proteins may facilitate R-loop resolving enzymes such as RNH1 or SETX. On the other hand, FANCM directly resolves R-loop structures to facilitate resumption of DNA synthesis. Thus, proteins involved in the FA pathway mitigate R-loop-associated transcription‒replication conflicts and ensure genome stability. BRCA1 in association with SETX is recruited to the R-loop sites formed at transcriptional termination regions of highly transcribed genes and acts to repair/prevent R-loop-driven DNA damage at these genomic loci.
Among these mechanisms, BRCA2 (FANCD1) was the first FA protein implicated in the suppression of R-loop-associated genome instability [178]. Using the procedure of proximity ligation assay (PLA) with a fluorescently tagged hybrid-interacting RNaseH1 peptide and a hybrid-specific antibody, Bhatia et al. showed that BRCA2 functionally interacts with a TREX-2 mRNA export factor PCID2 and prevents R-loop accumulation in vivo. Furthermore, results from DNA-RNA immunoprecipitation (DRIP) experiments revealed hybrid enrichment at actively transcribed genes in BRCA2-deficient cells [178]. The authors proposed a model whereby BRCA2 and other FA proteins directly act on the replication fork ahead of a R-loop and thereby stabilize the fork, prevent R-loop extension, promote R-loop dissolution and possibly facilitate fork restart in replicating cells [178]. On non-replicating chromatin, BRCA2 might get recruited or stabilized in the vicinity of transcribed regions by TREX-2 complex, where it promotes R-loop dissolution by providing greater access of R-loop resolving enzymes to the hybrid sites [178]. Like BRCA2, the downstream FA pathway protein BRCA1 (FANCS) is also implicated in R-loop metabolism [179]. Together with DNA/RNA helicase senataxin (SETX), BRCA1 prevents/repairs R-loop-induced DNA damage at transcription pause sites of actively transcribed genes. This is consistent with insertions/deletions found particularly at the hybrid containing transcription termination regions in breast tumors carrying germline BRCA1 mutations [179]. BRCA1/SETX knockdown results in a single-stranded DNA break and R-loop accumulation at transcription termination loci. Because recruitment of BRCA1/SETX complex to R-loop sites was shown to be BRCA1-dependent, it is plausible that BRCA1 prevents R-loop accumulation by facilitating hybrid sites access to SETX. It should be noted, however, that BRCA1/SETX depletion also causes DSBs but they are not R-loop-associated.
Schwab et al. further demonstrated that under normal physiological conditions, components of the FA pathway are required to mitigate transcription‒replication conflict and limit unscheduled R-loop accumulation [169]. Results obtained from PLA experiments revealed that FANCD2 colocalizes with elongating RNA polymerase II in vivo in a transcription-dependent manner. Furthermore, inhibition of transcription alleviated replication fork progression defects and replication stress observed in FANCD2-deficient cells. These observations suggest that under normal growth conditions, transcription complexes are natural impediments to DNA replication and that the FA pathway plays a critical role to overcome these barriers. Apart from this, experimental evidence suggests that the FA pathway is directly involved in preventing unusual accumulation of co-transcriptional R-loops [169]. Overexpression of the R-loop resolving nuclease RNase H1 suppressed FANCD2 nuclear foci formation and mono-ubiquitination in replicating cells. Moreover, RNase H1 overexpression prevented R-loop accumulation and suppressed replication fork arrest and chromosomal instability in FANCD2 knockdown cells. Consistent with these results and suggesting a broader role of the FA pathway in R-loop metabolism, FA patient-derived FANCD2-/- or FANCA-/- cells display elevated hybrid accumulation and associated chromosomal aberrations [168].
In addition to suppressing R-loop formation, the FANCM DNA translocase directly resolves DNA:RNA hybrids arising from transcription [169]. FANCM-deficient cells exhibit increased immunofluorescence staining by a R-loop-specific antibody. Moreover, results from in vitro unwinding assays showed that purified FANCM efficiently resolves DNA: RNA hybrid substrates [169], further supported by a very recent report demonstrating that FANCM has a R-loop metabolizing function in vitro and that FANCM-deficient cells are highly sensitive to R-loop stabilizing drugs [180]. Apart from R-loop resolution, a functional FA pathway might essentially provide stability to the replication forks stalled at hybrid sites, thereby preventing replication fork collapse and DNA breaks [169]. Importantly, the role of FANCD2 in facilitating replication at common fragile sites (CFS) is also attributed to its R-loop removal function [133]; however, the precise role of FANCD2 is not well understood. Overall, the experimental data suggest that R-loop-associated replication‒transcription conflicts that persist when the FA pathway is defective contributes to the elevated chromosomal instability observed in cells from FA patients.
R-loop-induced genomic instability is implicated in various human diseases including cancer [181]. Given the role of cancer predisposition factors such as BRCA1/BRCA2 and FA pathway in mitigating R-loops, it is reasonable that persistent R-loops are major contributing factors to cancer-associated genomic instability. Indeed, as mentioned above BRCA1-deficient breast tumors are highly enriched for mutations at R-loop sites located at transcriptional termination regions in the genome [179]. This suggests that unresolved R-loops might be the chief source of mutagenesis in cancer when tumor suppressors like BRCA1 are lost. Additional whole-genome sequencing of human cancers harboring driver mutations in major cancer susceptibility genes would be helpful to further dissect the molecular link between R-loop biology and cancer-associated genome instability. Oncogene-induced replication stress is considered as an early driver of genomic instability in cancer [182]. In this scenario, global upregulation of transcription activity and increased R-loop accumulation have been shown to be major driving factors of replication stress upon oncogene activation [183]. Thus, suppressing unscheduled R-loop formation by crucial cellular pathways such as FA under conditions of stress is vital to prevent neoplastic transformation. Better understanding of the molecular relationships between the FA pathway and R-loop biology in the context of FA pathogenesis and cancer will be an exciting field of research.
6. Conclusions
Although FA is a complex genetic disorder with many genes involved and an array of cellular phenotypes and clinical symptoms, the underlying cause at the cellular level seems to revolve around molecular defects contributing to a compromised DNA damage response and DNA repair. While evidence suggests that ICLs are the major culprit, there are other drivers of replication stress and genomic instability in FA. In this review, we have discussed the roles of various FA gene products and associated DNA repair factors in the metabolism of not only ICLs but also other bulky lesions, unusual replication fork structures caused by replisome-stalling agents, and R-loops. Continued research in this area will help to elucidate potential strategies to ameliorate the molecular and cellular defects of FA. Furthermore, fundamental advances in understanding the basis for the chromosomal instability leading to various cancers associated with mutations in genes (particularly downstream members) of the FA pathway and their interacting partners will help to identify new biomarkers and therapeutic strategies.
Author Contributions
A.D. and R.M.B.J. each wrote original sections of the manuscript draft, and together edited it to generate the final paper.
Funding
This work was supported in full by the Intramural Research Program of the National Institutes of Health, NIA.
Acknowledgments
We wish to thank Michael Seidman (National Institute on Aging, NIH) for his comments and helpful discussion.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kee, Y.; D’Andrea, A.D. Molecular pathogenesis and clinical management of Fanconi Anemia. J. Clin. Investig. 2012, 122, 3799–3806. [Google Scholar] [CrossRef] [PubMed]
- Romick-Rosendale, L.E.; Hoskins, E.E.; Privette Vinnedge, L.M.; Foglesong, G.D.; Brusadelli, M.G.; Potter, S.S.; Komurov, K.; Brugmann, S.A.; Lambert, P.F.; Kimple, R.J.; et al. Defects in the Fanconi Anemia pathway in head and neck cancer cells stimulate tumor cell invasion through DNA-PK and Rac1 signaling. Clin. Cancer Res. 2016, 22, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- Romick-Rosendale, L.E.; Lui, V.W.; Grandis, J.R.; Wells, S.I. The Fanconi anemia pathway: Repairing the link between DNA damage and squamous cell carcinoma. Mutat. Res. 2013, 743–744, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Bochtler, T.; Frohling, S.; Kramer, A. Role of chromosomal aberrations in clonal diversity and progression of acute myeloid leukemia. Leukemia 2015, 29, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Kottemann, M.C.; Smogorzewska, A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 2013, 493, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; D’Andrea, A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012, 26, 1393–1408. [Google Scholar] [CrossRef] [PubMed]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Knipscheer, P.; Raschle, M.; Smogorzewska, A.; Enoiu, M.; Ho, T.V.; Scharer, O.D.; Elledge, S.J.; Walter, J.C. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 2009, 326, 1698–1701. [Google Scholar] [CrossRef] [PubMed]
- Raschle, M.; Knipscheer, P.; Enoiu, M.; Angelov, T.; Sun, J.; Griffith, J.D.; Ellenberger, T.E.; Scharer, O.D.; Walter, J.C. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 2008, 134, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Clauson, C.; Scharer, O.D.; Niedernhofer, L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspect. Biol. 2013, 5, a012732. [Google Scholar] [CrossRef] [PubMed]
- Garaycoechea, J.I.; Crossan, G.P.; Langevin, F.; Mulderrig, L.; Louzada, S.; Yang, F.; Guilbaud, G.; Park, N.; Roerink, S.; Nik-Zainal, S.; et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 2018, 553, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Pontel, L.B.; Rosado, I.V.; Burgos-Barragan, G.; Garaycoechea, J.I.; Yu, R.; Arends, M.J.; Chandrasekaran, G.; Broecker, V.; Wei, W.; Liu, L.; et al. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Mol. Cell 2015, 60, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr.; Bellani, M.; Liu, Y.; Seidman, M.M. Fanconi Anemia: A DNA repair disorder characterized by accelerated decline of the hematopoietic stem cell compartment and other features of aging. Ageing Res. Rev. 2017, 33, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Cole, R.S.; Sinden, R.R. Repair of cross-linked DNA in Escherichia coli. Basic Life Sci. 1975, 5b, 487–495. [Google Scholar] [PubMed]
- Lin, P.F.; Bardwell, E.; Howard-Flanders, P. Initiation of genetic exchanges in lambda phage—Prophage crosses. Proc. Natl. Acad. Sci. USA 1977, 74, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Sinden, R.R.; Cole, R.S. Repair of cross-linked DNA and survival of Escherichia coli treated with psoralen and light: Effects of mutations influencing genetic recombination and DNA metabolism. J. Bacteriol. 1978, 136, 538–547. [Google Scholar] [PubMed]
- Berardini, M.; Foster, P.L.; Loechler, E.L. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 1999, 181, 2878–2882. [Google Scholar] [PubMed]
- Berardini, M.; Mackay, W.; Loechler, E.L. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry 1997, 36, 3506–3513. [Google Scholar] [CrossRef] [PubMed]
- Daee, D.L.; Myung, K. Fanconi-like crosslink repair in yeast. Genome Integr. 2012, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- McHugh, P.J.; Ward, T.A.; Chovanec, M. A prototypical Fanconi anemia pathway in lower eukaryotes? Cell Cycle 2012, 11, 3739–3744. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, C.A.; Gavish, H.; Shannon, W.R.; Buchwald, M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature 1992, 356, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Bogliolo, M.; Surralles, J. Fanconi anemia: A model disease for studies on human genetics and advanced therapeutics. Curr. Opin. Genet. Dev. 2015, 33, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Gueiderikh, A.; Rosselli, F.; Neto, J.B.C. A never-ending story: The steadily growing family of the FA and FA-like genes. Genet. Mol. Biol. 2017, 40, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, S.; Wu, Z. Fanconi anemia pathway defects in inherited and sporadic cancers. Transl. Pediatr. 2014, 3, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Kawasoe, Y.; Williams, H.; Coates, E.; Roy, U.; Shi, Y.; Beese, L.S.; Scharer, O.D.; Yan, H.; Gottesman, M.E.; et al. Sensing and Processing of DNA Interstrand Crosslinks by the Mismatch Repair Pathway. Cell Rep. 2017, 21, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Davies, A.A.; Ulrich, H.D.; McHugh, P.J. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006, 25, 1285–1294. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.L.; Gottesman, M.E.; Gautier, J. Replication-independent repair of DNA interstrand crosslinks. Mol. Cell 2012, 47, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Enoiu, M.; Jiricny, J.; Scharer, O.D. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012, 40, 8953–8964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Walter, J.C. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair 2014, 19, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Niedernhofer, L.J.; Lalai, A.S.; Hoeijmakers, J.H. Fanconi anemia (cross)linked to DNA repair. Cell 2005, 123, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Wang, W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007, 8, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Long, D.T.; Joukov, V.; Budzowska, M.; Walter, J.C. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol. Cell 2014, 56, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dewar, J.M.; Budzowska, M.; Motnenko, A.; Cohn, M.A.; Walter, J.C. DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 2015, 22, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Berezney, R.; Dubey, D.D.; Huberman, J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 2000, 108, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, S.; Bellani, M.A.; Thazhathveetil, A.K.; Ling, C.; de Winter, J.P.; Wang, Y.; Wang, W.; Seidman, M.M. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol. Cell 2013, 52, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Huang, J.; Yan, Z.; Li, Y.; Ohzeki, M.; Ishiai, M.; Xu, D.; Takata, M.; Seidman, M.; Wang, W. Bloom syndrome complex promotes FANCM recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell Discov. 2016, 2, 16047. [Google Scholar] [CrossRef] [PubMed]
- Rohleder, F.; Huang, J.; Xue, Y.; Kuper, J.; Round, A.; Seidman, M.; Wang, W.; Kisker, C. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res. 2016, 44, 3219–3232. [Google Scholar] [CrossRef] [PubMed]
- Mosedale, G.; Niedzwiedz, W.; Alpi, A.; Perrina, F.; Pereira-Leal, J.B.; Johnson, M.; Langevin, F.; Pace, P.; Patel, K.J. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005, 12, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Meetei, A.R.; Medhurst, A.L.; Ling, C.; Xue, Y.; Singh, T.R.; Bier, P.; Steltenpool, J.; Stone, S.; Dokal, I.; Mathew, C.G.; et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005, 37, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Kee, Y.; Gurtan, A.; D’Andrea, A.D. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 2008, 111, 5215–5222. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leung, J.W.; Jiang, Y.; Lowery, M.G.; Do, H.; Vasquez, K.M.; Chen, J.; Wang, W.; Li, L. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell 2013, 49, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Mutreja, K.; Krietsch, J.; Hess, J.; Ursich, S.; Berti, M.; Roessler, F.K.; Zellweger, R.; Patra, M.; Gasser, G.; Lopes, M. ATR-mediated global fork slowing and reversal assist fork traverse and prevent chromosomal breakage at DNA interstrand cross-links. Cell Rep. 2018, 24, 2629–2642.e25. [Google Scholar] [CrossRef] [PubMed]
- Semlow, D.R.; Zhang, J.; Budzowska, M.; Drohat, A.C.; Walter, J.C. Replication-dependent unhooking of DNA interstrand cross-Links by the NEIL3 glycosylase. Cell 2016, 167, 498–511.e14. [Google Scholar] [CrossRef] [PubMed]
- Amunugama, R.; Willcox, S.; Wu, R.A.; Abdullah, U.B.; El-Sagheer, A.H.; Brown, T.; McHugh, P.J.; Griffith, J.D.; Walter, J.C. Replication fork reversal during DNA interstrand crosslink repair requires CMG unloading. Cell Rep. 2018, 23, 3419–3428. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.E.; Oshima, J.; Fu, Y.H.; Wijsman, E.M.; Hisama, F.; Alisch, R.; Matthews, S.; Nakura, J.; Miki, T.; Ouais, S.; et al. Positional cloning of the Werner’s syndrome gene. Science 1996, 272, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Schlacher, K.; Christ, N.; Siaud, N.; Egashira, A.; Wu, H.; Jasin, M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011, 145, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Schlacher, K.; Wu, H.; Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Berti, M.; Vindigni, A. Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 2016, 23, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.T.; Kim, T.; Wagner, J.E.; Conti, B.A.; Lach, F.P.; Huang, A.L.; Molina, H.; Sanborn, E.M.; Zierhut, H.; Cornes, B.K.; et al. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell 2015, 59, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Mukherjee, S.; Yang, Y.; Mori, E.; Bhattacharya, S.; Kobayashi, J.; Yannone, S.M.; Chen, D.J.; Asaithamby, A. Nonenzymatic role for WRN in preserving nascent DNA strands after replication stress. Cell Rep. 2014, 9, 1387–1401. [Google Scholar] [CrossRef] [PubMed]
- Iannascoli, C.; Palermo, V.; Murfuni, I.; Franchitto, A.; Pichierri, P. The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic Acids Res. 2015, 43, 9788–9803. [Google Scholar] [CrossRef] [PubMed]
- Scharer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harbor Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef] [PubMed]
- DiGiovanna, J.J.; Kraemer, K.H. Shining a light on xeroderma pigmentosum. J. Investig. Dermatol. 2012, 132, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Saffi, J.; Agnoletto, M.H.; Guecheva, T.N.; Batista, L.F.; Carvalho, H.; Henriques, J.A.; Stary, A.; Menck, C.F.; Sarasin, A. Effect of the anti-neoplastic drug doxorubicin on XPD-mutated DNA repair-deficient human cells. DNA Repair 2010, 9, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Budzowska, M.; Modesti, M.; Maas, A.; Wyman, C.; Essers, J.; Kanaar, R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006, 25, 4921–4932. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, N.G.; Raams, A.; Silengo, M.C.; Wijgers, N.; Niedernhofer, L.J.; Robinson, A.R.; Giglia-Mari, G.; Hoogstraten, D.; Kleijer, W.J.; Hoeijmakers, J.H.; et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 2007, 80, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Kobertz, W.R.; Ariza, R.R.; Biggerstaff, M.; Essigmann, J.M.; Wood, R.D. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 2000, 275, 26632–26636. [Google Scholar] [CrossRef] [PubMed]
- Niedernhofer, L.J.; Odijk, H.; Budzowska, M.; van Drunen, E.; Maas, A.; Theil, A.F.; de Wit, J.; Jaspers, N.G.; Beverloo, H.B.; Hoeijmakers, J.H.; et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 2004, 24, 5776–5787. [Google Scholar] [CrossRef] [PubMed]
- Prasher, J.M.; Lalai, A.S.; Heijmans-Antonissen, C.; Ploemacher, R.E.; Hoeijmakers, J.H.; Touw, I.P.; Niedernhofer, L.J. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1-/- mice. EMBO J. 2005, 24, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Bogliolo, M.; Schuster, B.; Stoepker, C.; Derkunt, B.; Su, Y.; Raams, A.; Trujillo, J.P.; Minguillon, J.; Ramirez, M.J.; Pujol, R.; et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 2013, 92, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Klein Douwel, D.; Boonen, R.A.; Long, D.T.; Szypowska, A.A.; Raschle, M.; Walter, J.C.; Knipscheer, P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 2014, 54, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.N.; Kobayashi, S.; Tsuda, M.; Kurumizaka, H.; Takata, M.; Kono, K.; Jiricny, J.; Takeda, S.; Hirota, K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 6492–6496. [Google Scholar] [CrossRef] [PubMed]
- Hodskinson, M.R.; Silhan, J.; Crossan, G.P.; Garaycoechea, J.I.; Mukherjee, S.; Johnson, C.M.; Scharer, O.D.; Patel, K.J. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol. Cell 2014, 54, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Benitez, A.; Yuan, F.; Nakajima, S.; Wei, L.; Qian, L.; Myers, R.; Hu, J.J.; Lan, L.; Zhang, Y. Damage-dependent regulation of MUS81-EME1 by Fanconi anemia complementation group A protein. Nucleic Acids Res. 2014, 42, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Interthal, H.; Heyer, W.D. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000, 263, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Nguyen, J.; Hill, S.J.; Scully, R.; Adelmant, G.O.; Marto, J.A.; Feunteun, J.; Livingston, D.M. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell 2011, 44, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Kelsall, I.R.; Langenick, J.; MacKay, C.; Patel, K.J.; Alpi, A.F. The Fanconi anaemia components UBE2T and FANCM are functionally linked to nucleotide excision repair. PLoS ONE 2012, 7, e36970. [Google Scholar] [CrossRef] [PubMed]
- Howlett, N.G.; Taniguchi, T.; Olson, S.; Cox, B.; Waisfisz, Q.; De Die-Smulders, C.; Persky, N.; Grompe, M.; Joenje, H.; Pals, G.; et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 2002, 297, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Alter, B.P. The association between FANCD1/BRCA2 mutations and leukaemia. Br. J. Haematol 2006, 133, 446–448; author reply 448. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.E.; Tolar, J.; Levran, O.; Scholl, T.; Deffenbaugh, A.; Satagopan, J.; Ben-Porat, L.; Mah, K.; Batish, S.D.; Kutler, D.I.; et al. Germline mutations in BRCA2: Shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood 2004, 103, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.; Meza, N.W.; Quintana-Bustamante, O.; Casado, J.A.; Jacome, A.; McAllister, K.; Puerto, S.; Surralles, J.; Segovia, J.C.; Bueren, J.A. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol. Ther. 2006, 14, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Dorsman, J.C.; Ameziane, N.; de Vries, Y.; Rooimans, M.A.; Sheng, Q.; Pals, G.; Errami, A.; Gluckman, E.; Llera, J.; et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 2007, 39, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.; Schindler, D.; Hanenberg, H.; Barker, K.; Hanks, S.; Kalb, R.; Neveling, K.; Kelly, P.; Seal, S.; Freund, M.; et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 2007, 39, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Ameziane, N.; May, P.; Haitjema, A.; van de Vrugt, H.J.; van Rossum-Fikkert, S.E.; Ristic, D.; Williams, G.J.; Balk, J.; Rockx, D.; Li, H.; et al. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat. Commun. 2015, 6, 8829. [Google Scholar] [CrossRef] [PubMed]
- Domchek, S.M.; Tang, J.; Stopfer, J.; Lilli, D.R.; Hamel, N.; Tischkowitz, M.; Monteiro, A.N.; Messick, T.E.; Powers, J.; Yonker, A.; et al. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 2013, 3, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.T.; Smogorzewska, A. SnapShot: Fanconi anemia and associated proteins. Cell 2015, 160, 354–354.e1. [Google Scholar] [CrossRef] [PubMed]
- Cantor, S.B.; Bell, D.W.; Ganesan, S.; Kass, E.M.; Drapkin, R.; Grossman, S.; Wahrer, D.C.; Sgroi, D.C.; Lane, W.S.; Haber, D.A.; et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 2001, 105, 149–160. [Google Scholar] [CrossRef]
- Levitus, M.; Waisfisz, Q.; Godthelp, B.C.; de Vries, Y.; Hussain, S.; Wiegant, W.W.; Elghalbzouri-Maghrani, E.; Steltenpool, J.; Rooimans, M.A.; Pals, G.; et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J. Nat. Genet. 2005, 37, 934–935. [Google Scholar] [CrossRef] [PubMed]
- Levran, O.; Attwooll, C.; Henry, R.T.; Milton, K.L.; Neveling, K.; Rio, P.; Batish, S.D.; Kalb, R.; Velleuer, E.; Barral, S.; et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 2005, 37, 931–933. [Google Scholar] [CrossRef] [PubMed]
- Litman, R.; Peng, M.; Jin, Z.; Zhang, F.; Zhang, J.; Powell, S.; Andreassen, P.R.; Cantor, S.B. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 2005, 8, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. The role of BRCA1 in homologous recombination repair in response to replication stress: Significance in tumorigenesis and cancer therapy. Cell Biosci. 2013, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Fan, Q.; Ren, K.; Andreassen, P.R. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 2009, 7, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Doty, T.; Gibson, B.; Heyer, W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010, 17, 1260–1262. [Google Scholar] [CrossRef] [PubMed]
- McHugh, P.J.; Spanswick, V.J.; Hartley, J.A. Repair of DNA interstrand crosslinks: Molecular mechanisms and clinical relevance. Lancet Oncol. 2001, 2, 483–490. [Google Scholar] [CrossRef]
- Bunting, S.F.; Callen, E.; Kozak, M.L.; Kim, J.M.; Wong, N.; Lopez-Contreras, A.J.; Ludwig, T.; Baer, R.; Faryabi, R.B.; Malhowski, A.; et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell 2012, 46, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Duquette, M.L.; Zhu, Q.; Taylor, E.R.; Tsay, A.J.; Shi, L.Z.; Berns, M.W.; McGowan, C.H. CtIP is required to initiate replication-dependent interstrand crosslink repair. PLoS Genet. 2012, 8, e1003050. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Higuera, I.; Taniguchi, T.; Ganesan, S.; Meyn, M.S.; Timmers, C.; Hejna, J.; Grompe, M.; D’Andrea, A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 2001, 7, 249–262. [Google Scholar] [CrossRef]
- Chen, X.; Wilson, J.B.; McChesney, P.; Williams, S.A.; Kwon, Y.; Longerich, S.; Marriott, A.S.; Sung, P.; Jones, N.J.; Kupfer, G.M. The Fanconi anemia proteins FANCD2 and FANCJ interact and regulate each other’s chromatin localization. J. Biol. Chem. 2014, 289, 25774–25782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Fan, Q.; Ren, K.; Auerbach, A.D.; Andreassen, P.R. FANCJ/BRIP1 recruitment and regulation of FANCD2 in DNA damage responses. Chromosoma 2010, 119, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Murina, O.; von Aesch, C.; Karakus, U.; Ferretti, L.P.; Bolck, H.A.; Hanggi, K.; Sartori, A.A. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014, 7, 1030–1038. [Google Scholar] [CrossRef] [PubMed]
- Unno, J.; Itaya, A.; Taoka, M.; Sato, K.; Tomida, J.; Sakai, W.; Sugasawa, K.; Ishiai, M.; Ikura, T.; Isobe, T.; et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014, 7, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Karanja, K.K.; Cox, S.W.; Duxin, J.P.; Stewart, S.A.; Campbell, J.L. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 2012, 11, 3983–3996. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Herceg, Z.; Nakanishi, K.; Demuth, I.; Piccoli, C.; Michelon, J.; Hildebrand, G.; Jasin, M.; Digweed, M.; Wang, Z.Q. The Fanconi anemia group A protein modulates homologous repair of DNA double-strand breaks in mammalian cells. Carcinogenesis 2005, 26, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Yang, Y.G.; Pierce, A.J.; Taniguchi, T.; Digweed, M.; D’Andrea, A.D.; Wang, Z.Q.; Jasin, M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA 2005, 102, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K.; Cavallo, F.; Perrouault, L.; Giovannangeli, C.; Moynahan, M.E.; Barchi, M.; Brunet, E.; Jasin, M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat. Struct. Mol. Biol. 2011, 18, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Westermark, U.K.; Reyngold, M.; Olshen, A.B.; Baer, R.; Jasin, M.; Moynahan, M.E. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 2003, 23, 7926–7936. [Google Scholar] [CrossRef] [PubMed]
- Moynahan, M.E.; Pierce, A.J.; Jasin, M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 2001, 7, 263–272. [Google Scholar] [CrossRef]
- Stark, J.M.; Pierce, A.J.; Oh, J.; Pastink, A.; Jasin, M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004, 24, 9305–9316. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 2013, 13, 542–558. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, S.; Lee, L.; Davalos, A.; Schiestl, R.H.; Campisi, J.; Oshima, J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell 2003, 2, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Saintigny, Y.; Makienko, K.; Swanson, C.; Emond, M.J.; Monnat, R.J., Jr. Homologous recombination resolution defect in Werner syndrome. Mol. Cell. Biol. 2002, 22, 6971–6978. [Google Scholar] [CrossRef] [PubMed]
- Sturzenegger, A.; Burdova, K.; Kanagaraj, R.; Levikova, M.; Pinto, C.; Cejka, P.; Janscak, P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 2014, 289, 27314–27326. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Kusumoto, R.; Opresko, P.L.; Sui, X.; Huang, S.; Nicolette, M.L.; Paull, T.T.; Campisi, J.; Seidman, M.; Bohr, V.A. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006, 34, 2751–2760. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Banerjee, T.; Sommers, J.A.; Iannascoli, C.; Pichierri, P.; Shoemaker, R.H.; Brosh, R.M., Jr. Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Res. 2013, 73, 5497–5507. [Google Scholar] [CrossRef] [PubMed]
- Adamo, A.; Collis, S.J.; Adelman, C.A.; Silva, N.; Horejsi, Z.; Ward, J.D.; Martinez-Perez, E.; Boulton, S.J.; La Volpe, A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell 2010, 39, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Pace, P.; Mosedale, G.; Hodskinson, M.R.; Rosado, I.V.; Sivasubramaniam, M.; Patel, K.J. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 2010, 329, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.M.; Yanez, D.A.; Stark, J.M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015, 11, e1004943. [Google Scholar] [CrossRef] [PubMed]
- Benitez, A.; Liu, W.; Palovcak, A.; Wang, G.; Moon, J.; An, K.; Kim, A.; Zheng, K.; Zhang, Y.; Bai, F.; et al. FANCA promotes DNA double-strand break repair by catalyzing single-strand annealing and strand exchange. Mol. Cell 2018, 71, 621–628.e4. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; O’Regan, E.; Brown, R.; Karran, P. Selective recognition of a cisplatin-DNA adduct by human mismatch repair proteins. Nucleic Acids Res. 1997, 25, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Lu, X.; Zhang, X.; Peterson, C.A.; Legerski, R.J. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell. Biol. 2002, 22, 2388–2397. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jain, A.; Iyer, R.R.; Modrich, P.L.; Vasquez, K.M. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic Acids Res. 2009, 37, 4420–4429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Liu, X.; Li, L.; Legerski, R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair 2007, 6, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cortez, D.; Yazdi, P.; Neff, N.; Elledge, S.J.; Qin, J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000, 14, 927–939. [Google Scholar] [PubMed]
- Svendsen, J.M.; Smogorzewska, A.; Sowa, M.E.; O’Connell, B.C.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 2009, 138, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ghosal, G.; Yuan, J.; Chen, J.; Huang, J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 2010, 329, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Smogorzewska, A.; Desetty, R.; Saito, T.T.; Schlabach, M.; Lach, F.P.; Sowa, M.E.; Clark, A.B.; Kunkel, T.A.; Harper, J.W.; Colaiacovo, M.P.; et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 2010, 39, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Kennedy, R.; Ali, A.M.; Moreau, L.A.; Meetei, A.R.; D’Andrea, A.D.; Chen, C.C. Human MutS and FANCM complexes function as redundant DNA damage sensors in the Fanconi Anemia pathway. DNA Repair 2011, 10, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Wilson, J.B.; Clark, A.P.; Mitson-Salazar, A.; Tomashevski, A.; Ananth, S.; Glazer, P.M.; Semmes, O.J.; Bale, A.E.; Jones, N.J.; et al. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum. Mol. Genet. 2011, 20, 4395–4410. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Litman, R.; Xie, J.; Sharma, S.; Brosh, R.M., Jr.; Cantor, S.B. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007, 26, 3238–3249. [Google Scholar] [CrossRef] [PubMed]
- Cannavo, E.; Gerrits, B.; Marra, G.; Schlapbach, R.; Jiricny, J. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J. Biol. Chem. 2007, 282, 2976–2986. [Google Scholar] [CrossRef] [PubMed]
- Kratz, K.; Schopf, B.; Kaden, S.; Sendoel, A.; Eberhard, R.; Lademann, C.; Cannavo, E.; Sartori, A.A.; Hengartner, M.O.; Jiricny, J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 2010, 142, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Xie, J.; Ucher, A.; Stavnezer, J.; Cantor, S.B. Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. EMBO J. 2014, 33, 1698–1712. [Google Scholar] [CrossRef] [PubMed]
- Quinet, A.; Lemacon, D.; Vindigni, A. Replication Fork Reversal: Players and Guardians. Mol. Cell 2017, 68, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Sidorova, J. A game of substrates: Replication fork remodeling and its roles in genome stability and chemo-resistance. Cell Stress 2017, 1, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Ji, F.; Helleday, T.; Ying, S. Mechanisms for stalled replication fork stabilization: New targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Ait Saada, A.; Lambert, S.A.E.; Carr, A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 2018. [Google Scholar] [CrossRef] [PubMed]
- Pasero, P.; Vindigni, A. Nucleases acting at stalled forks: How to reboot the replication program with a few shortcuts. Annu. Rev. Genet. 2017, 51, 477–499. [Google Scholar] [CrossRef] [PubMed]
- Howlett, N.G.; Taniguchi, T.; Durkin, S.G.; D’Andrea, A.D.; Glover, T.W. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005, 14, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Wilson, J.B.; Medhurst, A.L.; Hejna, J.; Witt, E.; Ananth, S.; Davies, A.; Masson, J.Y.; Moses, R.; West, S.C.; et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 2004, 13, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Madireddy, A.; Kosiyatrakul, S.T.; Boisvert, R.A.; Herrera-Moyano, E.; Garcia-Rubio, M.L.; Gerhardt, J.; Vuono, E.A.; Owen, N.; Yan, Z.; Olson, S.; et al. FANCD2 facilitates replication through common fragile sites. Mol. Cell 2016, 64, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Lachaud, C.; Moreno, A.; Marchesi, F.; Toth, R.; Blow, J.J.; Rouse, J. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 2016, 351, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Michl, J.; Zimmer, J.; Buffa, F.M.; McDermott, U.; Tarsounas, M. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat. Struct. Mol. Biol. 2016, 23, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Kais, Z.; Rondinelli, B.; Holmes, A.; O’Leary, C.; Kozono, D.; D’Andrea, A.D.; Ceccaldi, R. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA Repair. Cell Rep. 2016, 15, 2488–2499. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.M.; Son, M.Y.; Dodds, S.; Hu, L.; Luo, G.; Hasty, P. RECQL5 and BLM exhibit divergent functions in cells defective for the Fanconi anemia pathway. Nucleic Acids Res. 2015, 43, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Cong, K.; Panzarino, N.J.; Nayak, S.; Calvo, J.; Deng, B.; Zhu, L.J.; Morocz, M.; Hegedus, L.; Haracska, L.; et al. Opposing roles of FANCJ and HLTF protect forks and restrain replication during stress. Cell Rep. 2018, 24, 3251–3261. [Google Scholar] [CrossRef] [PubMed]
- Higgs, M.R.; Reynolds, J.J.; Winczura, A.; Blackford, A.N.; Borel, V.; Miller, E.S.; Zlatanou, A.; Nieminuszczy, J.; Ryan, E.L.; Davies, N.J.; et al. BOD1L Is required to suppress deleterious resection of stressed replication forks. Mol. Cell 2015, 59, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, I.; Sareen, A.; Raghunandan, M.; Sobeck, A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013, 41, 6444–6459. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, I.; Stroik, D.R.; Sobeck, A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 2014, 34, 3939–3954. [Google Scholar] [CrossRef] [PubMed]
- Yeo, J.E.; Lee, E.H.; Hendrickson, E.A.; Sobeck, A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 2014, 23, 3695–3705. [Google Scholar] [CrossRef] [PubMed]
- Raghunandan, M.; Chaudhury, I.; Kelich, S.L.; Hanenberg, H.; Sobeck, A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle 2015, 14, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Lossaint, G.; Larroque, M.; Ribeyre, C.; Bec, N.; Larroque, C.; Decaillet, C.; Gari, K.; Constantinou, A. FANCD2 binds MCM proteins and controls replisome function upon activation of S phase checkpoint signaling. Mol. Cell 2013, 51, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Jones, M.J.; Yin, Y.; Crist, S.B.; Colnaghi, L.; Sims, R.J., 3rd; Rothenberg, E.; Jallepalli, P.V.; Huang, T.T. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 2015, 58, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Tonzi, P.; Yin, Y.; Lee, C.W.T.; Rothenberg, E.; Huang, T.T. Translesion polymerase kappa-dependent DNA synthesis underlies replication fork recovery. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Estep, K.N.; Brosh, R.M., Jr. RecQ and Fe-S helicases have unique roles in DNA metabolism dictated by their unwinding directionality, substrate specificity, and protein interactions. Biochem. Soc. Trans. 2018, 46, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Cantor, S.B.; Guillemette, S. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011, 7, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Cantor, S.B.; Nayak, S. FANCJ at the FORK. Mutat. Res. 2016, 788, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr.; Cantor, S.B. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi Anemia. Front. Genet. 2014, 5, 372. [Google Scholar] [CrossRef] [PubMed]
- Bharti, S.K.; Awate, S.; Banerjee, T.; Brosh, R.M. Getting ready for the dance: FANCJ irons out DNA wrinkles. Genes 2016, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Bharti, S.K.; Sommers, J.A.; Awate, S.; Bellani, M.A.; Khan, I.; Bradley, L.; King, G.A.; Seol, Y.; Vidhyasagar, V.; Wu, Y.; et al. A minimal threshold of FANCJ helicase activity is required for its response to replication stress or double-strand break repair. Nucleic Acids Res. 2018, 46, 6238–6256. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sommers, J.A.; Suhasini, A.N.; Leonard, T.; Deakyne, J.S.; Mazin, A.V.; Shin-Ya, K.; Kitao, H.; Brosh, R.M., Jr. Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood 2010, 116, 3780–3791. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shin-ya, K.; Brosh, R.M., Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef] [PubMed]
- London, T.B.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Murat, P.; Phillips, L.G.; Patel, K.J.; Balasubramanian, S.; Sale, J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012, 40, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Bharti, S.K.; Sommers, J.A.; George, F.; Kuper, J.; Hamon, F.; Shin-ya, K.; Teulade-Fichou, M.P.; Kisker, C.; Brosh, R.M., Jr. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 2013, 288, 28217–28229. [Google Scholar] [CrossRef] [PubMed]
- Castillo Bosch, P.; Segura-Bayona, S.; Koole, W.; van Heteren, J.T.; Dewar, J.M.; Tijsterman, M.; Knipscheer, P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014, 33, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.G.; Spies, M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016, 44, 8742–8753. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Helleday, T. DNA REPAIR. Drugging DNA repair. Science 2016, 352, 1178–1179. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Sommers, J.A.; Shoemaker, R.H.; Brosh, R.M., Jr. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1525–1530. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Banerjee, T.; Sommers, J.A.; Brosh, R.M., Jr. Targeting an Achilles’ heel of cancer with a WRN helicase inhibitor. Cell Cycle 2013, 12, 3329–3335. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.H.; Dexheimer, T.S.; Rosenthal, A.S.; Chu, W.K.; Singh, D.K.; Mosedale, G.; Bachrati, C.Z.; Schultz, L.; Sakurai, M.; Savitsky, P.; et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 2013, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Brosh, R.M., Jr. New insights into DNA helicases as druggable targets for cancer therapy. Front. Mol. Biosci. 2018, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Somyajit, K.; Mishra, A.; Scully, R.; Nagaraju, G. FANCJ helicase controls the balance between short- and long-tract gene conversions between sister chromatids. Nucleic Acids Res. 2017, 45, 8886–8900. [Google Scholar] [CrossRef] [PubMed]
- Barthelemy, J.; Hanenberg, H.; Leffak, M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2017, 45, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Borel, V.; Adelman, C.A.; Schindler, D.; Boulton, S.J. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi Anemia pathway. Genes Dev. 2015, 29, 2532–2546. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, M.L.; Perez-Calero, C.; Barroso, S.I.; Tumini, E.; Herrera-Moyano, E.; Rosado, I.V.; Aguilera, A. The Fanconi Anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015, 11, e1005674. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.A.; Nieminuszczy, J.; Shah, F.; Langton, J.; Lopez Martinez, D.; Liang, C.C.; Cohn, M.A.; Gibbons, R.J.; Deans, A.J.; Niedzwiedz, W. The Fanconi Anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell 2015, 60, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Gomez-Gonzalez, B. DNA-RNA hybrids: The risks of DNA breakage during transcription. Nat. Struct. Mol. Biol. 2017, 24, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Garcia-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A. The connection between transcription and genomic instability. EMBO J. 2002, 21, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P.; Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 2011, 44, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.Y.; Stirling, P.C. Replication fork protection factors controlling R-loop bypass and suppression. Genes 2017, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Barroso, S.I.; Garcia-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Hatchi, E.; Skourti-Stathaki, K.; Ventz, S.; Pinello, L.; Yen, A.; Kamieniarz-Gdula, K.; Dimitrov, S.; Pathania, S.; McKinney, K.M.; Eaton, M.L.; et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 2015, 57, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Hodson, C.; O’Rourke, J.J.; van Twest, S.; Murphy, V.J.; Dunn, E.; Deans, A.J. FANCM-family branchpoint translocases remove co-transcriptional R-loops. bioRxiv 2018. [Google Scholar] [CrossRef]
- Richard, P.; Manley, J.L. R loops and links to human disease. J. Mol. Biol. 2017, 429, 3168–3180. [Google Scholar] [CrossRef] [PubMed]
- Hills, S.A.; Diffley, J.F. DNA replication and oncogene-induced replicative stress. Curr. Biol. 2014, 24, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Kotsantis, P.; Silva, L.M.; Irmscher, S.; Jones, R.M.; Folkes, L.; Gromak, N.; Petermann, E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016, 7, 13087. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).