Abstract
The Drosophila grainyhead (grh) and vertebrate Grainyhead-like (Grhl) transcription factors are among the most critical genes for epithelial development, maintenance and homeostasis, and are remarkably well conserved from fungi to humans. Mutations affecting grh/Grhl function lead to a myriad of developmental and adult onset epithelial disease, such as aberrant skin barrier formation, facial/palatal clefting, impaired neural tube closure, age-related hearing loss, ectodermal dysplasia, and importantly, cancers of epithelial origin. Recently, mutations in the family member GRHL3 have been shown to lead to both syndromic and non-syndromic facial and palatal clefting in humans, particularly the genetic disorder Van Der Woude Syndrome (VWS), as well as spina bifida, whereas mutations in mammalian Grhl2 lead to exencephaly and facial clefting. As transcription factors, Grhl proteins bind to and activate (or repress) a substantial number of target genes that regulate and drive a cascade of transcriptional networks. A multitude of large-scale datasets have been generated to explore the grh/Grhl-dependent transcriptome, following ablation or mis-regulation of grh/Grhl-function. Here, we have performed a meta-analysis of all 41 currently published grh and Grhl RNA-SEQ, and microarray datasets, in order to identify and characterise the transcriptional networks controlled by grh/Grhl genes across disparate biological contexts. Moreover, we have also cross-referenced our results with published ChIP and ChIP-SEQ datasets, in order to determine which of the critical effector genes are likely to be direct grh/Grhl targets, based on genomic occupancy by grh/Grhl genes. Lastly, to interrogate the predictive strength of our approach, we experimentally validated the expression of the top 10 candidate grhl target genes in epithelial development, in a zebrafish model lacking grhl3, and found that orthologues of seven of these (cldn23, ppl, prom2, ocln, slc6a19, aldh1a3, and sod3) were significantly down-regulated at 48 hours post-fertilisation. Therefore, our study provides a strong predictive resource for the identification of putative grh/grhl effector target genes.
1. Introduction
Among the earliest tissues formed during embryonic development are epithelia, the thin, cellular layers that line the surfaces of all organs and body cavities. More than just semi-permeable “barriers”, epithelia are, in fact, highly dynamic tissues, actively regulating embryonic growth and expansion, as well as providing essential biochemical and mechanical signal-transduction cues. Understanding the nature of the genetic networks that regulate normal epithelial development is therefore a cornerstone of understanding birth defects due to epithelial aberrations. These not only include significant congenital abnormalities, (e.g., neural tube defects, cleft lip/palate, intestinal dysfunction), but, importantly, also underpin numerous epithelial disorders in later life (e.g., psoriasis, eczema, ectodermal dysplasia and, critically, cancers). Therefore, identifying the genes that drive epithelial formation and maintenance is a necessary first step to developing therapies that may ultimately ameliorate both the severity and incidence of congenital and adult onset epithelial pathologies.
Among the most critical genes for epithelial development and homeostasis are the highly conserved Grainyhead-like (Grhl) transcription factors. First described in Drosophila [1] as grainyhead (grh), this gene family is found in all organisms within the animal kingdom, and is remarkable both for its extremely high evolutionary conservation across eukaryotes [2,3], and for the extensive and disparate functions that are controlled by its various orthologues, in the context of both embryonic development and adult onset disease. In Drosophila, grh regulates key processes of epithelial development, such as Planar Cell Polarity (PCP), formation of the chitinous head “skeleton”, dorsal hole closure, and cuticle integrity and maintenance [1], whereas fungal Grhl is essential for cell wall remodelling and asexual spore (conidial) separation [4]. In vertebrates, Grainyhead-like orthologues display a conserved functional homology of regulating processes, required for epithelial development. Studies in mice [2,5], Xenopus [6] and zebrafish [7] have determined remarkable conserved functional homology to Drosophila grh, apparent in the morphogenic development of the epidermal barrier [8,9,10,11], closure of the neural tube [12,13,14,15], maintenance of neural cell survival [16], and the growth and development of the craniofacial skeleton [17,18]. Together, these findings point to a critical role for Grhl function in the development of all organisms within the animal kingdom.
Recent work has also shown that Grhl function is highly conserved in humans, and that these genes regulate both development and adult disease. Critical recent work has demonstrated that loss of GRHL3 leads to the craniofacial defect Van der Woude Syndrome (VWS), characterised by clefting of the secondary palate. This anomaly is due to aberrant maintenance of the oral periderm, a thin, extra-epithelial tissue that lines the developing palatal shelves [19]. Additionally, GRHL3 mutations are also associated with non-syndromic palatal clefting [20] and spina bifida [21]. As substantial, disparate craniofacial defects are also observed in Drosophila, zebrafish and murine models lacking grh/Grhl2/Grhl3 function, and, as these factors are expressed almost exclusively in epithelia, it becomes clear that both clefting and other facial, jaw and skull deformities observed in these models are secondary to a primary epithelial defect, following abrogation of grh/Grhl function.
After development, Grainyhead-like (Grhl) plays an important a role in regulating genes required for the maintenance of the epithelial barrier during growth and injury. Studies of human Grainyhead-like (Grhl) orthologues have begun to uncover the role that loss of function or gene variants have in adult-onset diseases, such as hearing loss/deafness [22,23], defects of epidermal integrity and, particularly, numerous forms of cancer [9,24,25,26]. The Grhl-genes are also implicated in the aetiology of a number of human diseases, including cleft palate [27,28], spina bifida [5,14,29], cancer [30], ectodermal dysplasia [28] and asthma [31]. This incredible functional diversity is mediated, at least in part, through a grh/Grhl transcription factor binding to a promoter or enhancer element and regulating the subsequent transactivation or repression of target effector genes through recruitment of transcriptional co-factors.
The grh/Grhl binding site is exceptionally well-conserved through evolution, broadly taking the form AACCGGTT (with the first “C” and second “G” largely invariant in experimentally validated assays). Although several upstream regulatory factors, such as branchless/FGF8 and Erk [32,33], and interacting co-factors, such as LMO4 [10,34], have been described, the delineation of grh/Grhl-dependent transcriptional networks has largely focussed on the identification and characterisation of these direct downstream target genes. Many candidate genes have been experimentally validated in the literature, both through targeted approaches (e.g., Tgm1/5 [35], ARHGEF19 [36], Dsg1 [37], eng2a [16], spec1 [16], edn1 [17] and PTEN [30], and, more recently, through large-scale screening approaches, such as RNA-SEQ and ChIP-SEQ. However, the precise mechanisms by which the grh/Grhl-dependent transcriptome regulates numerous disparate cellular and developmental functional processes are still unclear. Nevertheless, it is clear that the functional interaction between grainyhead-signalling and target gene regulation is dependent on tissue, and developmental/disease stage and organism, and the incredible heterogeneity of these regulations suggest that delineation of these grh/Grhl transcriptional networks will shed significant light on both embryonic development and disease.
Currently, 41 Microarray/RNA-SEQ datasets probing Grhl function have been reported in the literature, from both Drosophila and mouse tissues, as well as several human cancer/short-hairpin RNA (shRNA) cell lines (Table 1). In order to delineate the grh/Grhl transcriptional network, we have interrogated these datasets to bring greater clarity to the Grainyhead-dependent transcriptome, and to begin to understand the critical effector genes that operate broadly across disparate contexts, as well as identifying those that have more restricted roles in specific processes, particularly epithelial development.

Table 1.
Published datasets used for meta-analysis. Microarray and RNA-SEQ datasets including tissue of origin were classified as being derived from either “primary epithelia”, “cell line—cancer”, “cell line—non-cancer”, “other mammalian” or “other non-mammalian”, for subsequent analyses.
Lastly, using our recently described grhl3-/- zebrafish that present with epithelial defects of the enveloping layer, EVL [7], we performed Q-RT-PCR to determine the expression of zebrafish orthologues of the ten most differentially regulated epithelial genes, thereby functionally testing the predictive strength of our meta-analysis approach in a novel animal model. Together, this study collates multiple “big-data” approaches and provides an experimental pipeline for determining and testing novel grh/Grhl-dependent functional interactions.
2. Methods
2.1. RNA-SEQ and Microarray Meta-Analysis and Database Construction
The datasets from individual studies were converted to a common file format (.csv) and imported into MySQL (www.mysql.com), a relational database management system that enables the identification of relationships between datasets. Inside the database, the data were combined into species-specific tables. Using information from Ensembl v.91, gene symbols from the original human data files were mapped to the human Ensembl gene_stable_ids; gene symbols from other species were mapped to human gene symbols and gene_stable_ids of their orthologous human genes. After mapping, all the human-mapped data were joined together into a single table, searchable by the original gene symbol in every species, as well as the gene symbol and gene_stable_id of the human orthologue, publication name, dataset name, type of Grhl regulator, and direction of Grhl regulation (positive or negative), and were designated as being derived from either “cell-line” or “tissue”. For interactive visualization and exploring, the data were exported from the MySQL database, re-structured and uploaded into the Ordino tool (https://ordino.caleydoapp.org). To achieve a common gene ranking, the fold change rank-based scores from individual datasets were aggregated into the sum of scores for each gene, separately for positively and negatively regulated genes. For datasets where p-values/false discovery rates (FDRs) were available, another set of scores, based on p-value/FDR ranks, and computed independently for positively and negatively regulated genes, are also available. For singular genes, and the direction of regulation, p-values/FDRs from multiple datasets were combined into a single p-value, using the standard Fisher’s method. The ranks were then transformed into normalised scores that permitted aggregation of rank-based information from datasets comprising different numbers of genes. In order to do this, we used a scoring function f(r) =1/r^k, where k was an arbitrary parameter, allowing us to determine the significance of a genes’ rank across datasets. The constant of k = 3 was chosen, as it balanced the number of datasets that a given gene appeared in with instances in which this gene was highly ranked within an individual dataset. Therefore, the top ranked gene (Rank #1) within a given dataset would achieve a normalised score of 1 (1/1^3), whereas, if a gene did not appear within a particular dataset, it would be given a score of 0.
2.2. Zebrafish Experimentation
All zebrafish experiments were conducted in accordance with La Trobe University guidelines for the care and housing of zebrafish, under Animal Ethics Committee approval number AEC16-91. The grhl3-/- fish line was reported previously [7].
2.3. RNA Extraction and cDNA Synthesis
RNA was isolated from grhl3-/- and wild type embryos (at 48 hours post-fertilisation; hpf) as described previously [38]. cDNA was synthesized (from three biological replicates, using WT and phenotypic grhl3-/- embryos at 48hpf, utilising 50 embryos per group per replicate) using the iScript™ cDNA (Life Science Research, Bio-Rad, Hercules, CA, USA) synthesis kit, according to the manufacturer’s instructions. Quantitative Reverse Transcription Polymerase Chain Reaction (Q-RT-PCR) using SsoFast™ EvaGreen® Supermix (Bio-Rad) and a BioRad CFX96 Real-Time System/C1000 Thermal Cycler was employed to quantify expression levels of 10 genes (comprising 17 fish orthologues) that were predicted to be differentially regulated in epithelial establishment following loss of grhl3, using methodology described previously [38]; expression levels were normalised to the EF1α housekeeping gene, as described previously [7]. Oligonucleotide sequences used for Q-RT-PCR are shown in Table S1.
3. Results
3.1. Meta-Analysis Reveals the Most Consistently Differentially-Regulated Genes Following grh/Grhl Misexpression
In order to analyse the grh/Grhl transcriptome across multiple disparate biological contexts, we standardised all published grh/Grhl Microarray/RNA-SEQ datasets into a common format that allowed for direct comparison of differing methodologies and results, together with the corresponding published fold-changes and p-values (Figure 1). Differences in methodologies and data reporting in published studies created the need to integrate lists of regulated genes that were of different lengths, selected using different (study-specific) criteria. As studies varyingly examined the effects of either loss or gain of function of grh/Grhl, we divided the data based on whether a gene was positively or negatively regulated under normal homeostatic conditions. That is, if a gene was found to be downregulated in an experimental paradigm following loss of grh/Grhl, we deemed it to be positively regulated. Next, the positively and the negatively regulated genes were separately ranked, on the basis of the reported fold-change within each dataset (Figure 1).

Figure 1.
Meta-analysis pipeline methodology All published grh/Grhl Microarray/RNA-SEQ datasets were converted to a common file format (.csv, A) and imported into MySQL (B). Next, we determined whether genes within each dataset were positively or negatively regulated following functional modulation of grh/Grhl (C), and genes across the datasets were ranked according to their regulation within each dataset. To achieve a common gene ranking, the fold change rank-based scores from individual datasets were aggregated into the sum of scores for each gene, separately for positively and negatively regulated genes. (D). Each gene symbol within all datasets was mapped to the appropriate gene using Ensembl. After mapping, a table of all the human-mapped data was generated, searchable by the original gene symbol in every species (E). For interactive visualization and exploring, the data were exported from the MySQL database, re-structured and uploaded into Ordino (F).
Our analysis allowed us to determine the genes that are both most frequently and most substantially differentially regulated, following gain/loss of grh/Grhl function. From these comparisons, we determined which genes exhibited the greatest degree of regulation following disruption of grh/Grhl signalling. We determined this by: (1) frequency of altered expression (the number of datasets a given gene appeared in as “regulated”); and (2) degree of regulation, measured by the score, based on the rank given to a gene in a particular dataset, with respect to other differentially regulated genes. These analyses allowed us to reveal the genes that appeared within at least one dataset and determine the top differentially regulated genes following grh/Grhl de-regulation or misexpression, across all biological contexts thus far tested and described in the literature (Figure 2).
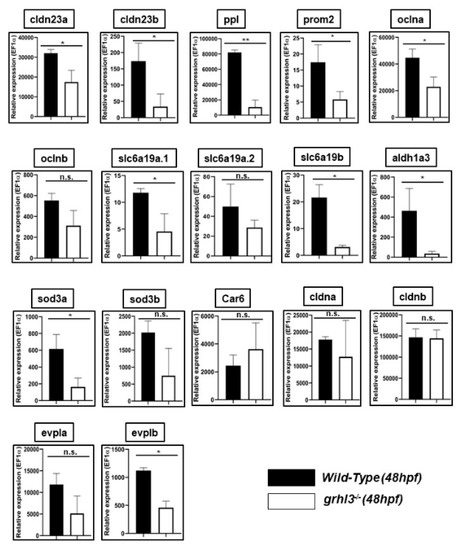
Figure 2.
Q-RT-PCR analysis of zebrafish orthologue expression in 48 hours post-fertilisation (hpf) WT and grhl3-/- embryos. Gene expression was normalised to expression of housekeeping gene EF1a. Of the 18 fish orthologues selected, 10 showed differential expression in WT (black bars) vs grhl3-/- (white bars) embryos. * p < 0.05, ** p < 0.01, n.s., non-significant.
From these analyses, we compiled a list of the top 50 most-highly differentially regulated genes (following grh/Grhl modulation) across various tissues and cell lines, and at different developmental time points (Table 2). As grh/Grhl transcription factors bind to the promoter of target genes in order to drive transcription, we determined whether any of these genes had previously been identified as being bound by grh/Grhl factors in ChIP or ChIP-SEQ datasets. In total, 15 of the top 50 genes in our list (30%: tmem54, prom2, cldn4, ppl, cdh1, rab15, lad1, rab25, Epcam, Tacstd2, st14, Esrp1, Prss22, spint1 and prss8) are known to be bound to grh/Grhl factors, supporting the robustness of our analysis algorithm. Additionally, seven genes in our list—cldn4, rab15, rab25, epcam, cdh1, tslp, and spint1—had been previously characterised as direct Grhl-target genes through biological validation experiments, further highlighting the predictive strength of our approach.

Table 2.
The top 50 most differentially regulated genes in all biological contexts following modulation of grh/Grhl function. The overall top 50 differentially regulated (both positively and negatively) genes of grh/grhl as identified by analysis of all published Microarray and RNA-SEQ datasets. 15 out of the 50 predicted target genes had previously been validated in literature through either large-scale ChIP-SEQ genome mining experiments, or through experimental validation (highlighted in red).
3.2. Biological Pathways Regulated by grhl in Epithelia as Identified by GO Analysis
In order to determine the overall biological processes regulated by the grh/Grhl pathway, we performed GO (Gene Ontology) pathway analysis, using gProfiler’s GOStat (Gene Group Functional Profiling), with the default background set of all human genes. Mindful of obvious limitations, such as tissue origin, disease state and temporal/developmental time point, we nonetheless analysed all the disparate datasets to determine an “overall” snapshot of grhl-target gene biology (Table 3). We found that this analysis largely confirmed previous known roles of the Grhl family. The major biological functions ascribed to genes regulated by the Grhl family were “cell–cell adhesion” ”tissue (epithelia/epidermis) development”, “animal organ/skin development”, “water homeostasis” and “epithelial cell morphogenesis”, consistent with known grh/Grhl-dependent functions in epithelial morphogenesis and maintenance, as well as preventing trans-epidermal water loss and maintaining epithelial integrity through suppressing the epithelial–mesenchymal transition (EMT). Somewhat surprisingly, the GO term “protein cross-linking” was also highlighted in our grh/Grhl transcriptome analysis. grh/Grhl factors are thought to be constitutively bound to the promoters/enhancers of target genes, achieving transcriptional specificity through the recruitment of partner protein co-activators as development proceeds [39]. These co-factors may include members of the Polycomb and Trithorax groups in Drosophila [39,40], and the vertebrate transcription factor LMO4 [34] in mice, although, as few such co-factors are known, enrichment of this GO term may suggest a novel, hitherto unsuspected grh/Grhl role in the assembly, formation or maintenance of the core transcriptional apparatus.

Table 3.
Gene Ontology (GO) analysis of the most differentially regulated cellular processes in all biological contexts, following modulation of grh/Grhl function. Functional Gene Ontology (GO) annotations are over-represented among the 200 genes positively regulated by grh/grhl, with the highest evidence of regulation across all species (highest fold change score aggregated over all the datasets).
3.3. Grhl-Dependent Target Genes are Well-Conserved between Mouse and Human; Less so between Mouse and Drosophila
In order to determine how well conserved the grh/Grhl-dependent transcriptional pathways were between mouse, Drosophila and human, we generated separate lists for each organism, using only species-specific, large-scale microarray/RNA-SEQ datasets. This analysis comprised 20 datasets from mouse, six datasets from Drosophila, and 12 datasets from human (Table 1). Using the top 50-ranked mouse genes as our root list for comparison, we determined which of these genes were ranked in the top 10% differentially-regulated genes in both Drosophila (in the top 1300 of ~13,000 genes in the Drosophila genome) and human (top 2500 out of ~25,000 total human genes), reasoning that genes which fell below this ranking threshold were less likely to be biologically-relevant in the context of grh/Grhl transcription (Table 4). Our analyses showed that 32 of the top 50 mouse genes had an identifiable Drosophila orthologue, with 4/32 orthologues of these mouse genes—Grhl2 (grh), Car6 (CAH7), Car2 (CAH1) and unc93a (GC4928)—also being differentially-regulated in Drosophila. Unsurprisingly, the degree of conservation was greater in human, with 47/50 human orthologues identified, and 17/47 of these mouse targets appearing in the top 10% differentially regulated human genes. Moreover, 13 of these orthologues—PROM2, CLDN4, PPL, GRHL2, CDH1, LAD1, RAB25, TMPRSS13, AP1M2, KLK6, SFN, CLDN1 and RPTN—appeared within the top 500 (top ~2%) most differentially regulated genes in humans, suggesting that these genes may substantially underpin Grhl-dependent transcriptional networks in mammalian species. Analysis of these 13 genes shows that eight of these—Prom2, Cldn4, Ppl, Cdh1, Lad1, Tmprss13, Cldn1 and RPTN—are involved in maintaining structural integrity within epithelia, particularly the epidermis. Of the others, there is a member of the Ras superfamily of small GTPases, involved in vesicular trafficking to the cell membrane (Rab25), a transcription factor (Grhl2 itself), a serine protease (KLK6), a clathrin-associated adaptor in the Golgi/endosome network (Ap1M2) and a cell-cycle checkpoint protein (SFN). This suggests that no other biological processes are as well conserved among Grhl-differentially regulated transcripts across multiple species as epithelial and epidermal integrity. These data are particularly striking, given the large differences in the source material used to generate each of the datasets, comprising different organisms, disease states, developmental timepoints and manipulating different grh/grhl orthologues.

Table 4.
Conservation of differentially regulated genes between mouse, Drosophila and human across all biological contexts, following modulation of grh/Grhl function.
3.4. Refining Meta-Analyses to Encompass Only Large-Scale Datasets Generated from Epithelial Tissue or Cancer Cell Lines Reveals grh/Grhl-dependent Transcriptome Specificity
As the grh/Grhl family are key regulators of epithelial establishment, development, maintenance and homeostasis, we refined our meta-analyses to only include datasets generated from primary epithelial tissues (Table 1). These data encompassed 18 datasets from seven separate publications, and restricted our data to Grhl-dependent transcriptomes, derived from epithelia from mouse embryonic skin [10], bladder [41], non-neural ectoderm [42] and lung epithelium [43], human neonatal epidermal keratinocytes [44], and undifferentiated primary human bronchial epithelial cells [31], as well as adult mouse and human psoriatic skin [45], and allowed us to identify the most-differentially regulated genes in epithelial development, the top 50 of which are shown in Table 5. This list includes numerous well characterized genes, known to be expressed in epithelia, which are involved in the maintenance of epithelial integrity and structural stability, such as Claudins 1/4/23, Periplakin, Envoplakin, Occludin and Prominin 2 (Table 5). Interestingly, the top 50 genes include five “predicted” genes—Gm19601, Gm3579, 231000H18Rik, Gm20305 (possible pseudogene) and Gm10639—that are yet to be ascribed a function. Next, we determined which of these top 50 “epithelial” grh/Grhl-regulated genes may also be de-regulated in Grhl-influenced cancerous cells (Table 6). We generated a dataset of differentially regulated genes in those RNA-SEQ/Microarray datasets that were designated as being derived from “cancer cell-lines” (Table 6). These experiments typically included control cancer cells, compared to those in which Grhl activity was modulated through short-hairpin (shRNA) knockdown, and included 4T1 breast tumor cells [46], human mammary epithelial (HMLE-Twist-ER) cells [47], MSP (mesenchymal sub-population) cells obtained from HMLE cells [48], OVCA429 (ovarian cystadenocarcinoma; intermediate epithelial (IE) phenotype) cells [49] and LNCaP (human prostate carcinoma) cells [50]. Surprisingly, only two genes appeared in both the “epithelial targets” and “cancer targets” tables—Tmem54 and Claudin-4, a previously experimentally validated Grhl-target. We did note several instances of different family members expressed across both sets (e.g., Sod3, Krt6b/78, Tmprss4/11bnl and Cldn1/23/b8 in epithelia, and Sod1/2, Krt7/19, Tmprss13 and Cldn7 in cancer), suggesting that loss of Grhl-mediated transcription may lead to similar biological consequences, albeit mediated via different (possibly secondary, or indirect) target pathways. However, these results indicate that the Grhl-dependent transcriptome is largely context and tissue-type dependent, with little evidence of conserved mechanisms operating in epithelial homeostasis and epithelial cancer.

Table 5.
The top 50 most differentially regulated genes in primary epithelia following modulation of grh/Grhl function. Orthologues of the genes highlighted in red were screened and validated in subsequent Q-RT-PCR experiments in grhl3-/- zebrafish.

Table 6.
The top 50 most differentially regulated genes in cancer cell lines following modulation of grh/Grhl function.
3.5. Expression of Differentially-Regulated Epithelial Genes in a Novel Vertebrate Model
In order to determine the predictive power of our meta-analysis approach, to identify novel grhl-dependent putative target genes (both direct and indirect), we analysed the expression of our top-ranked genes that were differentially regulated in epithelia, by performing Q-RT-PCR on cDNA from grhl3-/- mutant zebrafish. This model was utilised to test our meta-analyses, as none of the large-scale datasets were generated from zebrafish tissue, hence, this model would speak to the genetic conservation of grhl-dependent pathways across vertebrates.
From the genes that were differentially regulated in epithelia (Table 5), we identified the top 10 genes with an identifiable zebrafish orthologue, or orthologues. The roles of these ten mammalian genes—Cldn23, Ppl, Prom2, Ocln, Slc6a19, Aldh1a3, Sod3, Car6, Cldn4 and Evpl—were sub-functionalised into a total of 17 zebrafish orthologues, based on sequence conservation homology (Table S2). Next, we performed Q-RT-PCR on fifty, 48 hours post-fertilisation (hpf) WT and grhl3-/- embryos (experiment performed in triplicate, see methods) to determine the relative expression of each of these fish orthologues. Of the 17 zebrafish orthologues analysed, we found that 10—cldn23a, cldn23b, ppl, prom2, oclna, slc6a19a.1, slc6a19b, aldh1a3, sod3a and evplb—showed statistically significant differences in expression between WT and grhl3-/- fish (primarily down-regulation). We utilised whole embryos for our analyses, not just epithelial tissue, and we only performed analyses at a single developmental timepoint. Nonetheless, even within these experimental constraints, the fact that 10/17 zebrafish orthologues were significantly differentially regulated highlights the remarkable conservation of grhl-dependent pathways across the evolutionary spectrum, and the strong predictive power of our approach for identifying novel, biologically relevant transcriptional networks downstream of grh/Grhl function.
4. Discussion
In this study, we interrogated large-scale transcriptomic data to uncover novel genetic mechanisms that operate downstream of activation or repression by grh/Grhl genes. We collected all currently published large-scale RNA-SEQ and microarray datasets from multiple disparate animal models, developmental timepoints, tissues and cancer cell lines, to delineate conserved grh/Grhl-dependent genetic networks. Using a novel, ranking-based algorithm approach to draw synergies between disparate methodologies, we identified the top-differentially regulated genes following gain or loss of grh/Grhl function in all datasets, as well as refining our analyses to the top-differentially regulated genes in primary epithelia and in cancer. We also examined the conservation of mechanisms in mouse, Drosophila and human genes, and, lastly, we tested the predictive strength of our meta-analysis approach to determine which of the top-ranked genes were differentially regulated in a novel vertebrate grhl-loss-of-function model, the zebrafish. Taken together, our study brings cohesion and clarity to the existing “big-data” approaches to tackle grh/Grhl-dependent transcription, and opens up new avenues (and identifies novel putative targets) to further experimentally delineate grh/Grhl-dependent transcriptional genetic regulatory networks.
Across the 41 disparate Microarray/RNA-SEQ datasets, of the top 50 genes we identified, 15 had previous connections to Grhl transcription factors, either through ChIP/ChIP-SEQ experiments to identify regions of genome occupancy by Grhl proteins, or through targeted biological experiments to empirically determine novel functional relationships. The genes to which Grhl-factors bind, and drive/repress transcription can be considered true “direct” target genes. A number of our top 50 genes fall into this category, namely, Cldn4, Cdh1 (E-Cadherin), Rab25, Epcam, Esrp1 and Spint1, with a further nine genes—Tmem54, Prom2, Ppl, Lad1, Tacstd2, St14, Prss22, and Prss8—showing instances of promoter-binding by Grhl-factors in ChIP-analyses, but currently lacking empirical experimental validation to determine true biological interactions. As such, these nine genes would appear to be the prime candidates for future studies into identifying novel Grhl-target genes through established analyses, such as in vitro luciferase activation assays, genetic complementarity in animal models (e.g., through generating Grhl/gene-of-interest doubly heterozygous mice) or models to rescue Grhl loss-of function phenotypes, as we have shown previously by rescuing aberrant developmental phenotypes in grhl2b or grhl3-deficient zebrafish embryos, with eng2a [16] and edn1 [17] mRNA micro-injection, respectively.
The Grhl-family are well-known in both embryogenesis and cancer. Multiple studies have highlighted substantial developmental abnormalities in animal models lacking Grhl factors. These include numerous skin formation, maintenance and healing defects [2,11,28,36,51], impaired neural tube closure [5], facial and palatal clefting [19,52], regulation of skull bone apposition [18] and impaired formation of the lower jaw [17]. These latter (craniofacial) defects are interesting, owing to the fact that they are secondary consequences of disrupted epithelial formation—either the overlying cranial skin not allowing cranial bones to grow and expand [18], or through decreased endodermal edn1-signalling from the endodermal epithelium to cranial neural crest cells [17]. These studies parallel recent findings in human craniofacial disorders, namely Van der Woude Syndrome (VWS), a congenital syndrome characterised by palatal clefting. Disruption of the thin epithelial layer covering the developing palatal shelves—the periderm—leads to clefting in both mice and humans due to adhesions forming between the palatal shelves and tongue in utero [19]. Critically, VWS is also known to be caused by mutations in a critical Grhl-co-factor, IRF6 [53] in both mice and humans. These studies show that understanding the Grhl-dependent transcriptome is likely to uncover further novel biological pathways in the aetiology of craniofacial (and other epithelial) disorders caused by a primary epithelial defect.
In the context of cancer, Grhl-factors perform tumor-suppression roles in the epithelia of both the skin [30] and oesophagus [54], as well as maintaining epithelial identity through suppression of the epithelial–mesenchymal transition (EMT) in cancers of epithelial origin [47]. Strikingly, and somewhat surprisingly, our study found virtually no correlation between the top 50 differentially regulated genes in normal epithelia as compared to cancerous cells, with only two genes—Tmem54 and Claudin4—appearing in both lists. This could suggest that, in general terms, the normal grh/Grhl-dependent transcriptional pathways, that operate in epithelial development, homeostasis and maintenance, are not the most highly dysregulated in epithelial cancers. Caveats exist in this interpretation, chief among them being the lack of a direct comparison between normal and cancerous epithelia derived from the same source, as well as the substantial differences that exist between cells and tissues of origin utilised for the individual RNA-SEQ/Microarray datasets. Additionally, the published microarray/RNA-SEQ datasets defining the role of Grhl in cancers follow the experimental paradigm of inhibiting or over-expressing Grhl-factors in already tumorigenic cell lines, which is a separate biological question to analysis of the transcriptome of cancers that arise directly due to aberrant Grhl-function. Transcriptomic analyses of tumors arising in Grhl3-deficient mice, relative to non-cancerous adjoining tissue, such as the skin and esophagus [30,54], would add important data points to our current meta-analyses.
Overall, this study demonstrates that meta-analysis of previously published RNA-Seq datasets is able to identify novel targets of grhl transcription. The depth and robustness of this approach is clearly demonstrated by the fact that many of the genes identified via this method have been previously validated in the literature. Moreover, the experimental validation of altered expression of our identified target genes, using a vertebrate model that was not included in any of the published data sets, highlights the power of this type of analysis to predict conserved gene targets across different model organisms. Our experimental paradigm naturally has certain caveats and limitations, which any analysis must keep in mind, such as the degree and direction of target regulation in fish compared to mammals, the analysis of gene expression over separate developmental and, perhaps, adult timepoints, and specific analyses through ISH/IHC of mRNA/protein distribution, specifically in epithelial tissues, e.g., developing EVL and the skin. Moreover, grh (and presumably also Grhl) proteins are known to be post-translationally modified through processes such as phosphorylation [32,55], which further confounds the precise nature of grh/Grhl-dependent regulation (or co-regulation) of transcriptional targets. Nonetheless our experimental approach in fish successfully identified 10 gene orthologues that are significantly differentially regulated, as predicted by the meta-analysis database construction algorithms we have developed and utilised here. Lastly, our analyses did not determine which of these genes may be true direct targets, based on promoter occupancy of the target gene promoter by grh/Grhl, although such direct binding and activation/repression approaches would be a logical extension of our meta-analyses. Thus, our database is an excellent resource for researchers in the field of grhl function, helping to identify and categorise novel molecular pathways underlying various developmental and disease processes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/11/876/s1, Table S1: Oligonucleotide sequences used in the present study; Table S2: Zebrafish orthologues of differentially regulated mammalian genes. The top 10 differentially regulated genes following Grhl-dependent modulation in primary epithelia that had identifiable zebrafish orthologues, showing the function of each gene in mammals. Zebrafish orthologues of these ten mammalian genes comprise a total of 18 separate genes.
Author Contributions
Conceptualization, S.D.; Data curation, N.M., L.B.M., T.W., M.C.K., A.C., M.D. and S.D.; Formal analysis, N.M., T.W., A.C., M.D. and S.D.; Funding acquisition, S.D.; Investigation, N.M., L.B.M., P.J.A., M.C.K., A.C., M.D. and S.D.; Methodology, N.M., M.C.K., A.C. and M.D.; Project administration, S.D.; Resources, P.J.A., B.L.G., S.J.M., A.C. and M.D.; Software, A.C. and M.D.; Supervision, L.B.M., B.L.G., M.D. and S.D.; Writing—original draft, N.M. and S.D.; Writing—review and editing, L.B.M., P.J.A., T.W., B.L.G., S.J.M., M.C.K., M.D. and S.D.
Funding
This project was funded by La Trobe University Research Focus Area Grants GR2000003053 and GR2000002381 and Capital Equipment Grant 4.0150.35.31 to S.D.
Conflicts of Interest
The authors declare no conflict of interest
References
- Bray, S.J.; Kafatos, F.C. Developmental function of Elf-1: An essential transcription factor during embryogenesis in Drosophila. Genes Dev. 1991, 5, 1672–1683. [Google Scholar] [CrossRef] [PubMed]
- Ting, S.B.; Caddy, J.; Hislop, N.; Wilanowski, T.; Auden, A.; Zhao, L.L.; Ellis, S.; Kaur, P.; Uchida, Y.; Holleran, W.M.; et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 2005, 308, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, S.; Jane, S.M.; Darido, C. The planar cell polarity pathway in vertebrate epidermal development, homeostasis and repair. Organogenesis 2011, 7, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Pare, A.; Kim, M.; Juarez, M.T.; Brody, S.; McGinnis, W. The functions of grainy head-like proteins in animals and fungi and the evolution of apical extracellular barriers. PLoS ONE 2012, 7, e36254. [Google Scholar] [CrossRef]
- Ting, S.B.; Wilanowski, T.; Auden, A.; Hall, M.; Voss, A.K.; Thomas, T.; Parekh, V.; Cunningham, J.M.; Jane, S.M. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat. Med. 2003, 9, 1513–1519. [Google Scholar] [CrossRef]
- Tao, J.; Kuliyev, E.; Wang, X.; Li, X.; Wilanowski, T.; Jane, S.M.; Mead, P.E.; Cunningham, J.M. BMP4-dependent expression of Xenopus Grainyhead-like 1 is essential for epidermal differentiation. Development 2005, 132, 1021–1034. [Google Scholar] [CrossRef]
- Miles, L.B.; Darido, C.; Kaslin, J.; Heath, J.K.; Jane, S.M.; Dworkin, S. Mis-expression of grainyhead-like transcription factors in zebrafish leads to defects in enveloping layer (EVL) integrity, cellular morphogenesis and axial extension. Sci. Rep. 2017, 7, 17607. [Google Scholar] [CrossRef]
- Wang, S.; Samakovlis, C. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr. Top. Dev. Biol. 2012, 98, 35–63. [Google Scholar]
- Mlacki, M.; Darido, C.; Jane, S.M.; Wilanowski, T. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS ONE 2014, 9, e89247. [Google Scholar] [CrossRef]
- Yu, Z.; Lin, K.K.; Bhandari, A.; Spencer, J.A.; Xu, X.; Wang, N.; Lu, Z.; Gill, G.N.; Roop, D.R.; Wertz, P.; et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 2006, 299, 122–136. [Google Scholar] [CrossRef]
- Goldie, S.J.; Cottle, D.L.; Tan, F.H.; Roslan, S.; Srivastava, S.; Brady, R.; Partridge, D.D.; Auden, A.; Smyth, I.M.; Jane, S.M.; et al. Loss of GRHL3 leads to TARC/CCL17-mediated keratinocyte proliferation in the epidermis. Cell Death Dis. 2018, 9, 1072. [Google Scholar] [CrossRef]
- Rifat, Y.; Parekh, V.; Wilanowski, T.; Hislop, N.R.; Auden, A.; Ting, S.B.; Cunningham, J.M.; Jane, S.M. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 2010, 345, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Kimura-Yoshida, C.; Mochida, K.; Ellwanger, K.; Niehrs, C.; Matsuo, I. Fate specification of neural plate border by canonical wnt signaling and Grhl3 is crucial for neural tube closure. EBioMedicine 2015, 2, 513–527. [Google Scholar] [CrossRef] [PubMed]
- De Castro, S.C.P.; Gustavsson, P.; Marshall, A.R.; Gordon, W.M.; Galea, G.; Nikolopoulou, E.; Savery, D.; Rolo, A.; Stanier, P.; Andersen, B.; et al. Overexpression of Grainyhead-like 3 causes spina bifida and interacts genetically with mutant alleles of Grhl2 and Vangl2 in mice. Hum. Mol. Genet. 2018, 27, 4218–4230. [Google Scholar] [CrossRef] [PubMed]
- Kousa, Y.A.; Zhu, H.; Fakhouri, W.D.; Lei, Y.; Kinoshita, A.; Roushangar, R.R.; Patel, N.K.; Agopian, A.J.; Yang, W.; Leslie, E.J.; et al. The TFAP2A-IRF6-GRHL3 genetic pathway is conserved in neurulation. Hum. Mol. Genet. 2019, 28, 1726–1737. [Google Scholar] [CrossRef]
- Dworkin, S.; Darido, C.; Georgy, S.R.; Wilanowski, T.; Srivastava, S.; Ellett, F.; Pase, L.; Han, Y.; Meng, A.; Heath, J.K.; et al. Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development 2012, 139, 525–536. [Google Scholar] [CrossRef]
- Dworkin, S.; Simkin, J.; Darido, C.; Partridge, D.D.; Georgy, S.R.; Caddy, J.; Wilanowski, T.; Lieschke, G.J.; Doggett, K.; Heath, J.K.; et al. Grainyhead-like 3 regulation of endothelin-1 in the pharyngeal endoderm is critical for growth and development of the craniofacial skeleton. Mech. Dev. 2014, 133, 77–90. [Google Scholar] [CrossRef]
- Goldie, S.J.; Arhatari, B.D.; Anderson, P.; Auden, A.; Partridge, D.D.; Jane, S.M.; Dworkin, S. Mice lacking the conserved transcription factor Grainyhead-like 3 (Grhl3) display increased apposition of the frontal and parietal bones during embryonic development. BMC Dev. Biol. 2016, 16, 37. [Google Scholar] [CrossRef]
- Peyrard-Janvid, M.; Leslie, E.J.; Kousa, Y.A.; Smith, T.L.; Dunnwald, M.; Magnusson, M.; Lentz, B.A.; Unneberg, P.; Fransson, I.; Koillinen, H.K. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet. 2014, 94, 23–32. [Google Scholar] [CrossRef]
- Mangold, E.; Bohmer, A.C.; Ishorst, N.; Hoebel, A.K.; Gultepe, P.; Schuenke, H.; Klamt, J.; Hofmann, A.; Golz, L.; Raff, R.; et al. Sequencing the GRHL3 Coding Region Reveals Rare Truncating Mutations and a Common Susceptibility Variant for Nonsyndromic Cleft Palate. Am. J. Hum. Genet. 2016, 98, 755–762. [Google Scholar] [CrossRef]
- Lemay, P.; De Marco, P.; Emond, A.; Spiegelman, D.; Dionne-Laporte, A.; Laurent, S.; Merello, E.; Accogli, A.; Rouleau, G.A.; Capra, V.; et al. Rare deleterious variants in GRHL3 are associated with human spina bifida. Hum. Mutat. 2017, 38, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, M.; Fujioka, M.; Ogawa, K.; Okano, H. Distinct expression patterns of causative genes responsible for hereditary progressive hearing loss in non-human primate cochlea. Sci. Rep. 2016, 6, 22250. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Mu, Y.; Li, X.; Xu, P.; Tong, J.; Liu, Z.; Ma, T.; Zeng, G.; Yang, S.; Du, J.; et al. Grhl2 deficiency impairs otic development and hearing ability in a zebrafish model of the progressive dominant hearing loss DFNA28. Hum. Mol. Genet. 2011, 20, 3213–3226. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, C.; Zhao, Z.; Gao, N.; Chen, G.; Wang, Y.; Cui, J. Clinical implications of GRHL3 protein expression in breast cancer. Tumour. Biol. 2014, 35, 1827–1831. [Google Scholar] [CrossRef]
- Xiang, J.; Fu, X.; Ran, W.; Chen, X.; Hang, Z.; Mao, H.; Wang, Z. Expression and role of grainyhead-like 2 in gastric cancer. Med. Oncol. 2013, 30, 714. [Google Scholar] [CrossRef]
- Chen, W.; Yi, J.K.; Shimane, T.; Mehrazarin, S.; Lin, Y.-L.; Shin, K.-H.; Kim, R.H.; Park, N.-H.; Kang, M.K. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis 2016, 37, 500–510. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Huang, Y.; Pan, Y.; Jia, Z.; Ma, L.; Ma, L.; Lan, F.; Zhou, Y.; Shi, J.; et al. Association study between Van der Woude Syndrome causative gene GRHL3 and nonsyndromic cleft lip with or without cleft palate in a Chinese cohort. Gene 2016, 588, 69–73. [Google Scholar] [CrossRef]
- Petrof, G.; Nanda, A.; Howden, J.; Takeichi, T.; McMillan, J.R.; Aristodemou, S.; Ozoemena, L.; Liu, L.; South, A.P.; Pourreyron, C.; et al. Mutations in GRHL2 result in an autosomal-recessive ectodermal Dysplasia syndrome. Am. J. Hum. Genet. 2014, 95, 308–314. [Google Scholar] [CrossRef]
- Brouns, M.R.; De Castro, S.C.; Terwindt-Rouwenhorst, E.A.; Massa, V.; Hekking, J.W.; Hirst, C.S.; Savery, D.; Munts, C.; Partridge, D.; Lamers, W.; et al. Over-expression of Grhl2 causes spina bifida in the Axial defects mutant mouse. Hum. Mol. Genet. 2011, 20, 1536–1546. [Google Scholar] [CrossRef]
- Darido, C.; Georgy, S.R.; Wilanowski, T.; Dworkin, S.; Auden, A.; Zhao, Q.; Rank, G.; Srivastava, S.; Finlay, M.J.; Papenfuss, A.T.; et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 2011, 20, 635–648. [Google Scholar] [CrossRef]
- Gao, X.; Vockley, C.M.; Pauli, F.; Newberry, K.M.; Xue, Y.; Randell, S.H.; Reddy, T.E.; Hogan, B.L. Evidence for multiple roles for grainyheadlike 2 in the establishment and maintenance of human mucociliary airway epithelium. Proc. Natl. Acad. Sci. USA 2013, 110, 9356–9361. [Google Scholar] [CrossRef] [PubMed]
- Hemphala, J.; Uv, A.; Cantera, R.; Bray, S.; Samakovlis, C. Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development 2003, 130, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Mace, K.A.; Pearson, J.C.; McGinnis, W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 2005, 308, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Hislop, N.R.; Caddy, J.; Ting, S.B.; Auden, A.; Vasudevan, S.; King, S.L.; Lindeman, G.J.; Visvader, J.E.; Cunningham, J.M.; Jane, S.M. Grhl3 and Lmo4 play coordinate roles in epidermal migration. Dev. Biol. 2008, 321, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Cangkrama, M.; Darido, C.; Georgy, S.R.; Partridge, D.; Auden, A.; Srivastava, S.; Wilanowski, T.; Jane, S.M. Two ancient gene families are critical for maintenance of the mammalian skin barrier in postnatal life. J. Investig. Dermatol. 2016, 136, 1438–1448. [Google Scholar] [CrossRef]
- Caddy, J.; Wilanowski, T.; Darido, C.; Dworkin, S.; Ting, S.B.; Zhao, Q.; Rank, G.; Auden, A.; Srivastava, S.; Papenfuss, T.A.; et al. Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev. Cell 2010, 19, 138–147. [Google Scholar] [CrossRef]
- Wilanowski, T.; Caddy, J.; Ting, S.B.; Hislop, N.R.; Cerruti, L.; Auden, A.; Zhao, L.L.; Asquith, S.; Ellis, S.; Sinclair, R.; et al. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 2008, 27, 886–897. [Google Scholar] [CrossRef]
- Dworkin, S.; Heath, J.K.; de Jong-Curtain, T.A.; Hogan, B.M.; Lieschke, G.J.; Malaterre, J.; Ramsay, R.G.; Mantamadiotis, T. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev. Biol. 2007, 307, 127–141. [Google Scholar] [CrossRef]
- Nevil, M.; Bondra, E.R.; Schulz, K.N.; Kaplan, T.; Harrison, M.M. Stable binding of the conserved transcription factor grainy head to its target genes throughout drosophila melanogaster development. Genetics 2017, 205, 605–620. [Google Scholar] [CrossRef]
- Tuckfield, A.; Clouston, D.R.; Wilanowski, T.M.; Zhao, L.L.; Cunningham, J.M.; Jane, S.M. Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol. Cell. Biol. 2002, 22, 1936–1946. [Google Scholar] [CrossRef]
- Yu, Z.; Mannik, J.; Soto, A.; Lin, K.K.; Andersen, B. The epidermal differentiation-associated Grainyhead gene Get1/Grhl3 also regulates urothelial differentiation. EMBO J. 2009, 28, 1890–1903. [Google Scholar] [CrossRef] [PubMed]
- Ray, J.H.; Niswander, L.A. Grainyhead-like 2 downstream targets act to suppress epithelial-to-mesenchymal transition during neural tube closure. Development 2016, 143, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Kersbergen, A.; Best, S.A.; Dworkin, S.; Ah-Cann, C.; de Vries, M.E.; Asselin-Labat, M.L.; Ritchie, M.E.; Jane, S.M.; Sutherland, K.D. Lung morphogenesis is orchestrated through Grainyhead-like 2 (Grhl2) transcriptional programs. Dev. Biol. 2018, 443, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hopkin, A.S.; Gordon, W.; Klein, R.H.; Espitia, F.; Daily, K.; Zeller, M.; Baldi, P.; Andersen, B. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012, 8, e1002829. [Google Scholar] [CrossRef] [PubMed]
- Gordon, W.M.; Zeller, M.D.; Klein, R.H.; Swindell, W.R.; Ho, H.; Espetia, F.; Gudjonsson, J.E.; Baldi, P.F.; Andersen, B. A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. J. Clin. Investig. 2014, 124, 5205–5218. [Google Scholar] [CrossRef]
- Xiang, X.; Deng, Z.; Zhuang, X.; Ju, S.; Mu, J.; Jiang, H.; Zhang, L.; Yan, J.; Miller, D.; Zhang, H.G. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS ONE 2012, 7, e50781. [Google Scholar] [CrossRef]
- Cieply, B.; Riley, P.; Pifer, P.M.; Widmeyer, J.; Addison, J.B.; Ivanov, A.V.; Denvir, J.; Frisch, S.M. Suppression of the Epithelial-Mesenchymal Transition by Grainyhead-Like-2. Cancer Res. 2012, 72, 2440–2453. [Google Scholar] [CrossRef]
- Farris, J.C.; Pifer, P.M.; Zheng, L.; Gottlieb, E.; Denvir, J.; Frisch, S.M. Grainyhead-like 2 reverses the metabolic changes induced by the oncogenic epithelial-mesenchymal transition: Effects on anoikis. Mol. Cancer Res. 2016, 14, 528–538. [Google Scholar] [CrossRef]
- Chung, V.Y.; Tan, T.Z.; Tan, M.; Wong, M.K.; Kuay, K.T.; Yang, Z.; Ye, J.; Muller, J.; Koh, C.M.; Guccione, E.; et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 2016, 6, 19943. [Google Scholar] [CrossRef]
- Paltoglou, S.; Das, R.; Townley, S.L.; Hickey, T.E.; Tarulli, G.A.; Coutinho, I.; Fernandes, R.; Hanson, A.R.; Denis, I.; Carroll, J.S.; et al. Novel androgen receptor coregulator GRHL2 exerts both oncogenic and antimetastatic functions in prostate cancer. Cancer Res. 2017, 77, 3417–3430. [Google Scholar] [CrossRef]
- Boglev, Y.; Wilanowski, T.; Caddy, J.; Parekh, V.; Auden, A.; Darido, C.; Hislop, N.R.; Cangkrama, M.; Ting, S.B.; Jane, S.M. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev. Biol. 2011, 349, 512–522. [Google Scholar] [CrossRef]
- Pyrgaki, C.; Liu, A.; Niswander, L. Grainyhead-like 2 regulates neural tube closure and adhesion molecule expression during neural fold fusion. Dev. Biol. 2011, 353, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, C.R.; Kinoshita, A.; Kondo, S.; Yang, B.; Sajan, S.; Trout, K.J.; Malik, M.I.; Dunnwald, M.; Goudy, S.L.; Lovett, M.; et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet. 2006, 38, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Georgy, S.R.; Cangkrama, M.; Srivastava, S.; Partridge, D.; Auden, A.; Dworkin, S.; McLean, C.A.; Jane, S.M.; Darido, C. Identification of a novel proto-oncogenic network in head and neck squamous cell carcinoma. J. Natl. Cancer Inst. 2015, 107, pii: djv152. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; McGinnis, W. Phosphorylation of Grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc. Natl. Acad. Sci. USA 2011, 108, 650–655. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).