DNA Damage: From Threat to Treatment
Abstract
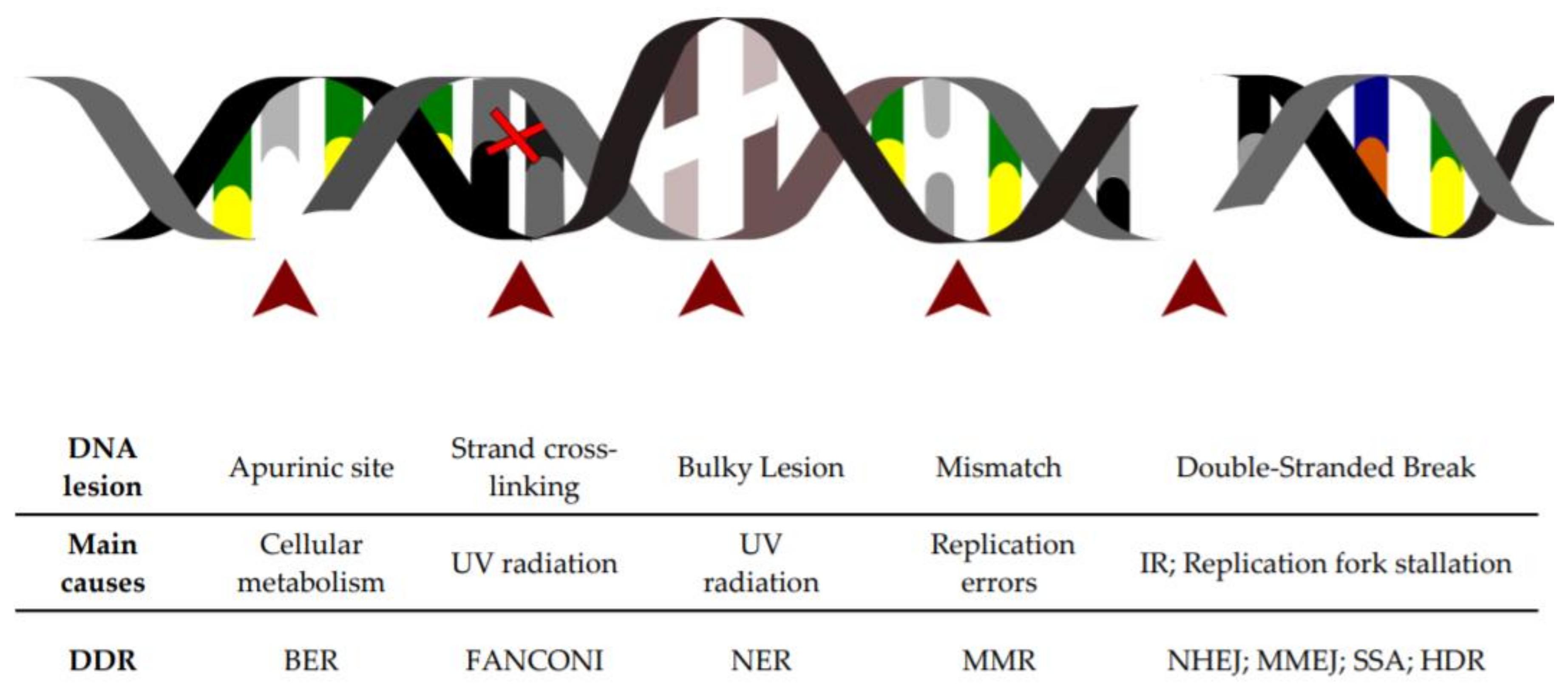
1. DNA Lesions as a Constant Threat to the Cell
2. The DNA Damage Response Is Activated upon Sensing of DNA Lesions
3. DNA Damage and Cell Cycle Checkpoints
4. DNA Repair Pathways
4.1. Fanconi Anemia (FA) Pathway
4.2. Nucleotide Excision Repair (NER)
4.3. Base Excision Repair (BER)
4.4. Mismatch Repair (MMR)
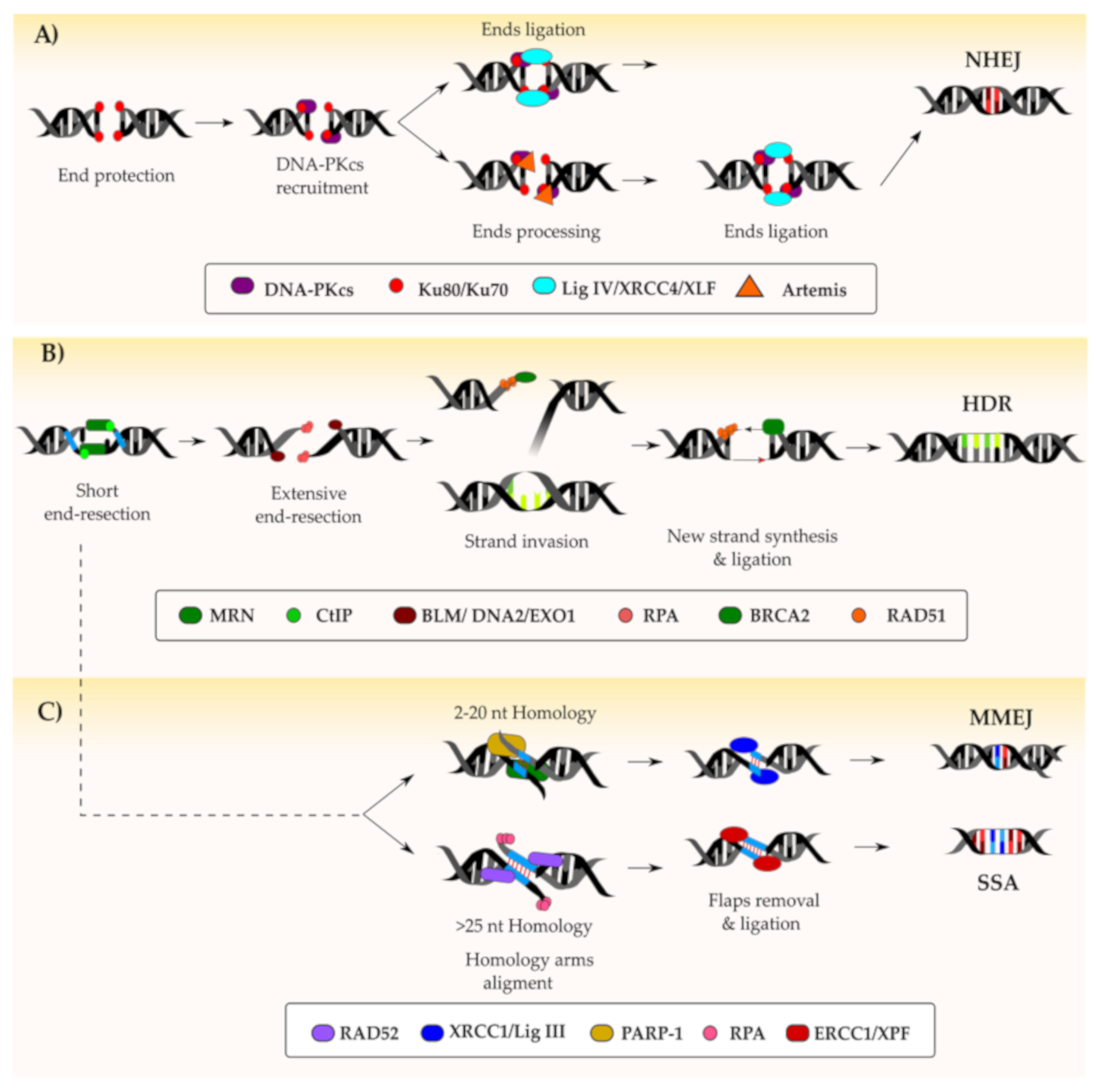
4.5. DNA Double-Stranded Break Repair Pathways
4.5.1. Non-Homologous End-Joining
4.5.2. Homology-Directed Repair
4.5.3. Alternative DSB Repair Pathways
5. DNA Repair Defects
5.1. Linking DDR Failure to Cell Disorders and Cancer
5.2. Exploiting Defects in DNA Repair to Treat Cancer
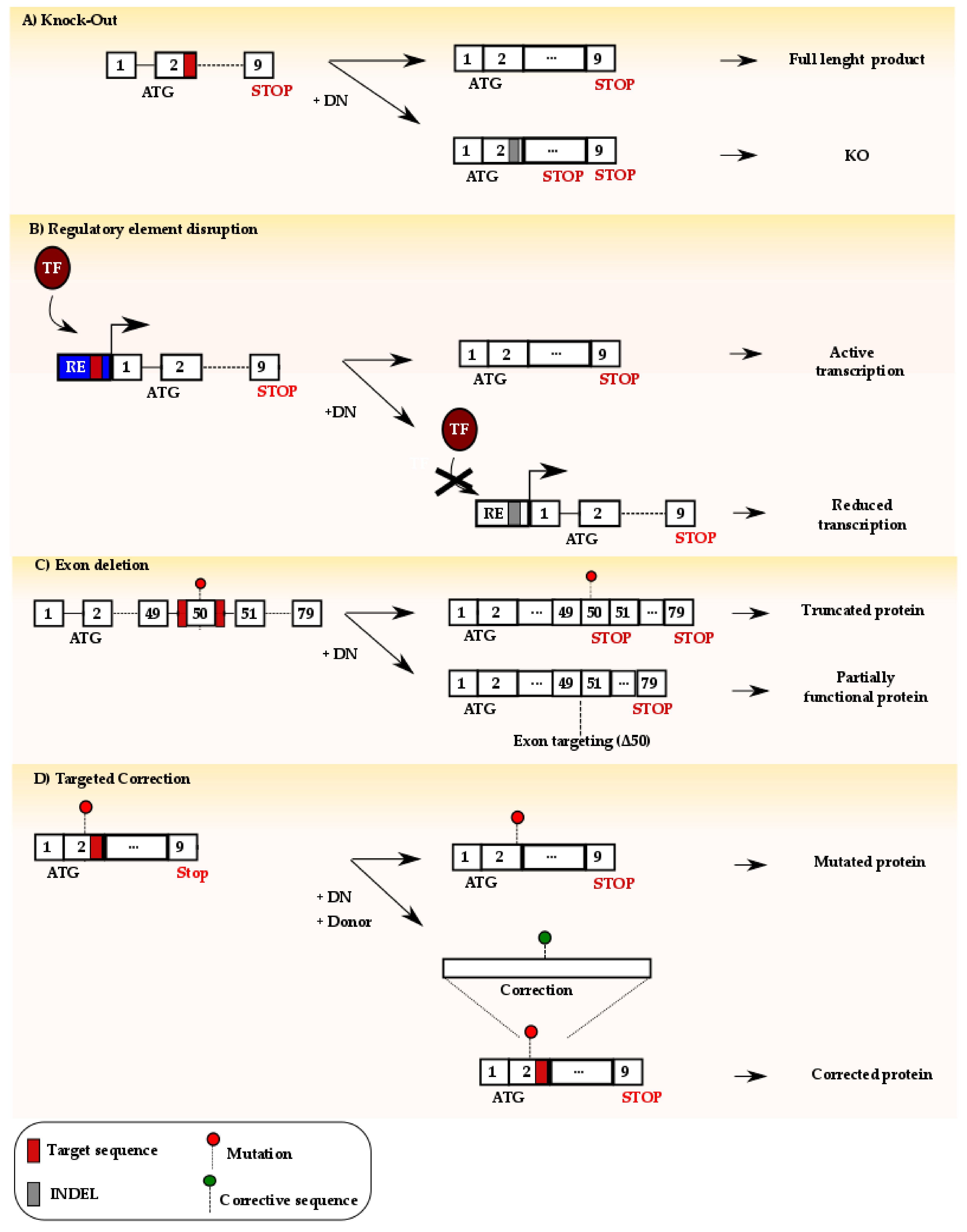
6. Exploiting DSB Repair to Develop Innovative Therapeutic Strategies
6.1. Use of NHEJ-Mediated Repair for Therapy
6.2. Use of HDR-Mediated Repair for Therapy
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Barnes, D. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Boil. 2000, 65, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Ganai, R.A.; Johansson, E. DNA Replication—A Matter of Fidelity. Mol. Cell 2016, 62, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Wagner, J.R. DNA Base Damage by Reactive Oxygen Species, Oxidizing Agents, and UV Radiation. Cold Spring Harb. Perspect. Boil. 2013, 5, a012559. [Google Scholar] [CrossRef]
- Van Houten, B.; Santa-Gonzalez, G.A.; Camargo, M. DNA repair after oxidative stress: Current challenges. Curr. Opin. Toxicol. 2017, 7, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Lin, Y.; Yan, S. Single-Strand Break End Resection in Genome Integrity: Mechanism and Regulation by APE2. Int. J. Mol. Sci. 2018, 19, 2389. [Google Scholar] [CrossRef]
- Abbotts, R.; Wilson, D.M. Coordination of DNA single strand break repair. Free. Radic. Boil. Med. 2017, 107, 228–244. [Google Scholar] [CrossRef]
- Callen, E.; Zong, D.; Wu, W.; Wong, N.; Stanlie, A.; Ishikawa, M.; Pavani, R.; Dumitrache, L.C.; Byrum, A.K.; Mendez-Dorantes, C.; et al. 53BP1 Enforces Distinct Pre- and Post-resection Blocks on Homologous Recombination. Mol. Cell 2020, 77, 26–38.e7. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pearlman, A.H.; Hsieh, P. DNA mismatch repair and the DNA damage response. DNA Repair 2016, 38, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, L.; Zhang, Y.; Gao, D. Crosstalk between signaling pathways and DNA damage response. Genome Instab. Dis. 2019, 1, 81–91. [Google Scholar] [CrossRef]
- Martinez, D.L.; Liang, C.-C.; Cohn, M.A. Cellular response to DNA interstrand crosslinks: The Fanconi anemia pathway. Cell. Mol. Life Sci. 2016, 73, 3097–3114. [Google Scholar] [CrossRef] [PubMed]
- Motnenko, A.; Liang, C.-C.; Yang, D.; Martinez, D.L.; Yoshikawa, Y.; Zhan, B.; Ward, K.E.; Tian, J.; Haas, W.; Spingardi, P.; et al. Identification of UHRF2 as a novel DNA interstrand crosslink sensor protein. PLoS Genet. 2018, 14, e1007643. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.; Wu, W.-F.; Langelier, M.-F.; Yang, J.-C.; Easton, L.E.; Riccio, A.A.; Pascal, J.M.; Neuhaus, D. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell 2015, 60, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Alexander, J.L.; Orr-Weaver, T.L. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 2016, 30, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Reginato, G.; Cejka, P. The MRE11 complex: A versatile toolkit for the repair of broken DNA. DNA Repair 2020, 102869. [Google Scholar] [CrossRef]
- Polo, S.; Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, P.; Foiani, M.; Kumar, A. ATM and ATR signaling at a glance. J. Cell Sci. 2015, 128, 4255–4262. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, I.A.; Krenning, L.; Bruinsma, W.; Medema, R. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 2015, 128, 607–620. [Google Scholar] [CrossRef]
- Houtgraaf, J.H.; Versmissen, J.; Van Der Giessen, W.J. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revascularization Med. 2006, 7, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, J.M.; Lees, J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Boil. 2002, 3, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Dean, D.C. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000, 14, 2393–2409. [Google Scholar] [CrossRef]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [PubMed]
- Petr, M.A.; Tulika, T.; Carmona-Marin, L.M.; Scheibye-Knudsen, M. Protecting the Aging Genome. Trends Cell Boil. 2020, 30, 117–132. [Google Scholar] [CrossRef]
- Wang, J.Y.J. Cell Death Response to DNA Damage. Yalej. Biol. Med. 2019, 92, 771–779. [Google Scholar]
- Fischer, M.; Quaas, M.; Nickel, A.; Engeland, K. Indirect p53-dependent transcriptional repression of Survivin, CDC25C, and PLK1 genes requires the cyclin-dependent kinase inhibitor p21/CDKN1A and CDE/CHR promoter sites binding the DREAM complex. Oncotarget 2015, 6, 41402–41417. [Google Scholar] [CrossRef] [PubMed]
- Thornton, T.M. Non-Classical P38 Map Kinase Functions: Cell Cycle Checkpoints and Survival. Int. J. Boil. Sci. 2009, 5, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Earnest, S.; Zhang, K.; Zhao, Y.; Cobb, M.H. TAO kinases mediate activation of p38 in response to DNA damage. Embo J. 2007, 26, 2005–2014. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, H.C.; Aslanian, A.S.; Lees, J.A.; Yaffe, M.B. p53-Deficient Cells Rely on ATM- and ATR-Mediated Checkpoint Signaling through the p38MAPK/MK2 Pathway for Survival after DNA Damage. Cancer Cell 2007, 11, 175–189. [Google Scholar] [CrossRef]
- Kim, G.-Y.; Mercer, S.E.; Ewton, D.Z.; Yan, Z.; Jin, K.; Friedman, E. The Stress-activated Protein Kinases p38α and JNK1 Stabilize p21Cip1 by Phosphorylation. J. Boil. Chem. 2002, 277, 29792–29802. [Google Scholar] [CrossRef] [PubMed]
- Barr, A.R.; Cooper, S.; Heldt, F.S.; Butera, F.; Stoy, H.; Mansfeld, J.; Novak, B.; Bakal, C. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 2017, 8, 14728. [Google Scholar] [CrossRef] [PubMed]
- Rhind, N.; Russell, P. Checkpoints: It takes more than time to heal some wounds. Curr. Boil. 2001, 10, R908–R911. [Google Scholar] [CrossRef]
- Bartek, J.; Lukas, C.; Lukas, J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Boil. 2004, 5, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Yekezare, M.; Gómez-González, B.; Diffley, J.F.X. Controlling DNA replication origins in response to DNA damage–inhibit globally, activate locally. J. Cell Sci. 2013, 126, 1297–1306. [Google Scholar] [CrossRef]
- Gire, V.; Dulić, V. Senescence from G2 arrest, revisited. Cell Cycle 2015, 14, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.; Papamichos-Chronakis, E.; Peterson, C.L. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 2013, 4, 2084. [Google Scholar] [CrossRef] [PubMed]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Douwel, D.K.; Boonen, R.A.; Long, D.; Szypowska, A.A.; Räschle, M.; Walter, J.C.; Knipscheer, P. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell 2014, 54, 460–471. [Google Scholar] [CrossRef]
- Puumalainen, M.-R.; Rüthemann, P.; Min, J.-H.; Naegeli, H. Xeroderma pigmentosum group C sensor: Unprecedented recognition strategy and tight spatiotemporal regulation. Cell. Mol. Life Sci. 2015, 73, 547–566. [Google Scholar] [CrossRef] [PubMed]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D. Nucleotide Excision Repair in Eukaryotes. Cold Spring Harb. Perspect. Boil. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjørås, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Freudenthal, B.D. Base excision repair of oxidative DNA damage from mechanism to disease. Front. Biosci. 2017, 22, 1493–1522. [Google Scholar] [CrossRef]
- Erie, D.A.; Weninger, K.R. Single molecule studies of DNA mismatch repair. DNA Repair 2014, 20, 71–81. [Google Scholar] [CrossRef]
- Hsieh, P.; Zhang, Y. The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2017, 114, 3552–3554. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Boil. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Rawal, C.C.; Liberi, G.; Pellicioli, A. Regulation of DNA Double Strand Breaks Processing: Focus on Barriers. Front. Mol. Biosci. 2019, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Boil. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Ahnesorg, P.; Smith, P.; Jackson, S.P. XLF Interacts with the XRCC4-DNA Ligase IV Complex to Promote DNA Nonhomologous End-Joining. Cell 2006, 124, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, I.; Sitters, G.; Candelli, A.; Heerema, S.J.; Heller, I.; De, A.J.M.; Zhang, H.; Normanno, D.; Modesti, M.; Peterman, E.J.; et al. Sliding sleeves of XRCC4–XLF bridge DNA and connect fragments of broken DNA. Nature 2016, 535, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q. Structural mechanism of DNA-end synapsis in the non-homologous end joining pathway for repairing double-strand breaks: Bridge over troubled ends. Biochem. Soc. Trans. 2019, 47, 1609–1619. [Google Scholar] [CrossRef]
- Goodarzi, A.A.; Yu, Y.; Riballo, E.; Douglas, P.; Walker, S.; Ye, R.; Härer, C.; Marchetti, C.; Morrice, N.; Jeggo, P.A.; et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006, 25, 3880–3889. [Google Scholar] [CrossRef] [PubMed]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Weinfeld, M.; Mani, R.S.; Abdou, I.; Aceytuno, R.D.; Glover, J.M.; Acetuno, R.D. Tidying up loose ends: The role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011, 36, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Watanabe, G.; Gerodimos, C.A.; Ochi, T.; Blundell, T.L.; Jackson, S.P.; Lieber, M.R. Different DNA End Configurations Dictate Which NHEJ Components Are Most Important for Joining Efficiency. J. Boil. Chem. 2016, 291, 24377–24389. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yu, Y.; Gupta, S.; Cho, Y.-M.; Lees-Miller, S.P.; Meek, K. Autophosphorylation of DNA-Dependent Protein Kinase Regulates DNA End Processing and May Also Alter Double-Strand Break Repair Pathway Choice. Mol. Cell. Boil. 2005, 25, 10842–10852. [Google Scholar] [CrossRef]
- Fumagalli, M.; Rossiello, F.; Mondello, C.; Di Fagagna, F.D. Stable Cellular Senescence Is Associated with Persistent DDR Activation. PLoS ONE 2014, 9, e110969. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2015, 16, 20–33. [Google Scholar] [CrossRef]
- Syed, A.; A Tainer, J. The MRE11–RAD50–NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem. 2018, 87, 263–294. [Google Scholar] [CrossRef]
- Myler, L.R.; Gallardo, I.F.; Soniat, M.M.; Deshpande, R.A.; Gonzalez, X.B.; Kim, Y.; Paull, T.T.; Finkelstein, I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell 2017, 67, 891–898.e4. [Google Scholar] [CrossRef] [PubMed]
- Jasin, M.; Rothstein, R. Repair of Strand Breaks by Homologous Recombination. Cold Spring Harb. Perspect. Boil. 2013, 5, a012740. [Google Scholar] [CrossRef]
- Hopfner, K.-P.; Craig, L.; Moncalián, G.; Zinkel, R.A.; Usui, T.; Owen, B.A.L.; Karcher, A.; Henderson, B.; Bodmer, J.-L.; McMurray, C.T.; et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 2002, 418, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Phelps, S.E.L.; Gray, S.; Neale, M.J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479, 241–244. [Google Scholar] [CrossRef]
- Langerak, P.; Mejia-Ramirez, E.; Limbo, O.; Russell, P. Release of Ku and MRN from DNA Ends by Mre11 Nuclease Activity and Ctp1 Is Required for Homologous Recombination Repair of Double-Strand Breaks. PLoS Genet. 2011, 7, e1002271. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, L.Z.; Wong, C.C.; Han, X.; Hwang, P.Y.-H.; Truong, L.N.; Zhu, Q.; Shao, Z.; Chen, D.J.; Berns, M.W.; et al. The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair. PLoS Genet. 2013, 9, e1003277. [Google Scholar] [CrossRef]
- Daley, J.M.; Jimenez-Sainz, J.; Wang, W.; Miller, A.S.; Xue, X.; Nguyen, K.A.; Jensen, R.B.; Sung, P. Enhancement of BLM-DNA2-Mediated Long-Range DNA End Resection by CtIP. Cell Rep. 2017, 21, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Guirouilh-Barbat, J.; Gelot, C.; Xie, A.; Dardillac, E.; Scully, R.; Lopez, B.S. 53BP1 Protects against CtIP-Dependent Capture of Ectopic Chromosomal Sequences at the Junction of Distant Double-Strand Breaks. PLoS Genet. 2016, 12, e1006230. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Jones, O.A.; Chan, K.-L. 53BP1 can limit sister-chromatid rupture and rearrangements driven by a distinct ultrafine DNA bridging-breakage process. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Misenko, S.M.; Patel, D.S.; Her, J.; Bunting, S.F. DNA repair and cell cycle checkpoint defects in a mouse model of ‘BRCAness’ are partially rescued by 53BP1 deletion. Cell Cycle 2018, 17, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Yates, L.; Aramayo, R.J.; Pokhrel, N.; Caldwell, C.; Kaplan, J.A.; Perera, R.; Spies, M.; Antony, E.; Zhang, X. A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Nat. Commun. 2018, 9, 5447. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.K.; Chen, H.; Symington, L.S. Replication protein A prevents promiscuous annealing between short sequence homologies: Implications for genome integrity. BioEssays 2014, 37, 305–313. [Google Scholar] [CrossRef]
- Ma, C.J.; Gibb, B.; Kwon, Y.; Sung, P.; Greene, E.C. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res. 2016, 45, 749–761. [Google Scholar] [CrossRef]
- Godin, S.K.; Sullivan, M.R.; Bernstein, K.A. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem. Cell Boil. 2016, 94, 407–418. [Google Scholar] [CrossRef]
- Van Der Heijden, T.; Modesti, M.; Hage, S.; Kanaar, R.; Wyman, C.; Dekker, C. Homologous Recombination in Real Time: DNA Strand Exchange by RecA. Mol. Cell 2008, 30, 530–538. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Boil. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- Sallmyr, A.; Tomkinson, A.E. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Boil. Chem. 2018, 293, 10536–10546. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Boil. 2015, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Pon, J.R.; Marra, M.A. Driver and Passenger Mutations in Cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 25–50. [Google Scholar] [CrossRef] [PubMed]
- Rogozin, I.B.; Goncearenco, A.; Lada, A.G.; De, S.; Yurchenko, V.; Nudelman, G.; Panchenko, A.R.; Cooper, D.N.; Pavlov, Y.V. DNA polymerase η mutational signatures are found in a variety of different types of cancer. Cell Cycle 2018, 17, 348–355. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.H.; Sample, K.M.; Pilarski, R.; Walsh, T.; Grosel, T.; Kinnamon, D.; Boru, G.; Massengill, J.B.; Schoenfield, L.; Kelly, B.; et al. Whole Exome Sequencing Identifies Candidate Genes Associated with Hereditary Predisposition to Uveal Melanoma. Ophthalmology 2020, 127, 668–678. [Google Scholar] [CrossRef]
- Tiwari, V.; Wilson, D.M. DNA Damage and Associated DNA Repair Defects in Disease and Premature Aging. Am. J. Hum. Genet. 2019, 105, 237–257. [Google Scholar] [CrossRef]
- Woodbine, L.; Gennery, A.R.; Jeggo, P.A. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair 2014, 16, 84–96. [Google Scholar] [CrossRef]
- Chi, X.; Li, Y.; Qiu, X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: Mechanism and regulation. Immunology 2020. [Google Scholar] [CrossRef]
- Mansilla-Soto, J.; Cortes, P. VDJ recombination: Artemis and its in vivo role in hairpin opening. J. Exp. Med. 2003, 197, 543–547. [Google Scholar] [CrossRef]
- Justiz Vaillant, A.A.; Mohseni, M. Severe Combined Immunodeficiency; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Mersch, J.; Jackson, M.A.; Park, M.; Nebgen, D.R.; Peterson, S.K.; Singletary, C.N.; Arun, B.K.; Litton, J.K. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 2014, 121, 269–275. [Google Scholar] [CrossRef]
- Noh, J.M.; Choi, D.H.; Baek, H.; Nam, S.J.; Lee, J.E.; Kim, J.W.; Ki, C.-S.; Park, W.; Huh, S.J. Associations betweenBRCAMutations in High-Risk Breast Cancer Patients and Familial Cancers Other than Breast or Ovary. J. Breast Cancer 2012, 15, 283–287. [Google Scholar] [CrossRef]
- Buisson, R.; Niraj, J.; Rodrigue, A.; Ho, C.K.; Kreuzer, J.; Foo, T.K.; Hardy, E.J.-L.; Dellaire, G.; Haas, W.; Xia, B.; et al. Coupling of Homologous Recombination and the Checkpoint by ATR. Mol. Cell 2017, 65, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Macedo, G.S.; Alemar, B.; Ashton-Prolla, P. Reviewing the characteristics of BRCA and PALB2-related cancers in the precision medicine era. Genet. Mol. Boil. 2019, 42, 215–231. [Google Scholar] [CrossRef]
- Soria-Bretones, I.; Sáez, C.; Ruíz-Borrego, M.; Japón, M.; Huertas, P. Prognostic value of CtIP/RBBP8 expression in breast cancer. Cancer Med. 2013, 2, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Luengo, A.; Gui, D.Y.; Heiden, M.G.V. Targeting Metabolism for Cancer Therapy. Cell Chem. Boil. 2017, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.L.; Hsieh, C.-M.; Chan, N.-L.; Hiasa, H. Topoisomerases as anticancer targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef]
- Lord, C.J.; Tutt, A.N.; Ashworth, A. Synthetic Lethality and Cancer Therapy: Lessons Learned from the Development of PARP Inhibitors. Annu. Rev. Med. 2015, 66, 455–470. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, Y.; Lu, Z. Metabolic features of cancer cells. Cancer Commun. 2018, 38, 65. [Google Scholar] [CrossRef]
- Jaiswal, A.S.; Banerjee, S.; Aneja, R.; Sarkar, F.H.; Ostrov, D.A.; Narayan, S. DNA Polymerase β as a Novel Target for Chemotherapeutic Intervention of Colorectal Cancer. PLoS ONE 2011, 6, e16691. [Google Scholar] [CrossRef]
- Barakat., K.; Gajewski, M.M.; Tuszynski, J.A. DNA polymerase beta (pol β) inhibitors: A comprehensive overview. Drug Discov. Today 2012, 17, 913–920. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; Jones, D.; Lee, S.-H.; Williamson, E.A.; Hromas, R. Drugging the Cancers Addicted to DNA Repair. J. Natl. Cancer Inst. 2017, 109, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Feng, W.; Lim, P.X.; Kass, E.M.; Jasin, M. Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu. Rev. Cancer Boil. 2017, 2, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Palmbos, P.L.; Hussain, M.H. Targeting PARP in Prostate Cancer: Novelty, Pitfalls, and Promise. Oncol. (Williston Park. N.Y.) 2016, 30, 377–385. [Google Scholar]
- Mirza, M.; Pignata, S.; Ledermann, J. Latest clinical evidence and further development of PARP inhibitors in ovarian cancer. Ann. Oncol. 2018, 29, 1366–1376. [Google Scholar] [CrossRef]
- Rottenberg, S.; Jaspers, J.E.; Kersbergen, A.; Van Der Burg, E.; Nygren, A.O.H.; Zander, S.A.L.; Derksen, P.W.B.; De Bruin, M.; Zevenhoven, J.; Lau, A.; et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. USA 2008, 105, 17079–17084. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Waggoner, S.; Vidal, G.A.; Mita, M.; Moroney, J.W.; Holloway, R.; Van Le, L.; Sachdev, J.C.; Chapman-Davis, E.; Colon-Otero, G.; et al. Single-Arm Phases 1 and 2 Trial of Niraparib in Combination With Pembrolizumab in Patients With Recurrent Platinum-Resistant Ovarian Carcinoma. JAMA Oncol. 2019, 5, 1141. [Google Scholar] [CrossRef]
- Gornstein, E.L.; Sandefur, S.; Chung, J.H.; Gay, L.M.; Holmes, O.; Erlich, R.L.; Soman, S.; Martin, L.K.; Rose, A.V.; Stephens, P.J.; et al. BRCA2 Reversion Mutation Associated With Acquired Resistance to Olaparib in Estrogen Receptor-positive Breast Cancer Detected by Genomic Profiling of Tissue and Liquid Biopsy. Clin. Breast Cancer 2018, 18, 184–188. [Google Scholar] [CrossRef]
- Naldini, L.; Trono, D.; Verma, I.M. Lentiviral vectors, two decades later. Science 2016, 353, 1101–1102. [Google Scholar] [CrossRef]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef]
- Carroll, D. Genome Engineering with Zinc-Finger Nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef]
- Mussolino, C.; Cathomen, T. TALE nucleases: Tailored genome engineering made easy. Curr. Opin. Biotechnol. 2012, 23, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Mussolino, C.; Cathomen, T. RNA guides genome engineering. Nat. Biotechnol. 2013, 31, 208–209. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef]
- Lederman, M.M.; Penn-Nicholson, A.; Cho, M.; Mosier, N. Biology of CCR5 and Its Role in HIV Infection and Treatment. JAMA 2006, 296, 815. [Google Scholar] [CrossRef] [PubMed]
- Cornu, T.I.; Mussolino, C.; Bloom, K.; Cathomen, T. Editing CCR5: A Novel Approach to HIV Gene Therapy. Adv. Exp. Med. Biol. 2015, 848, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Patsali, P.; Turchiano, G.; Papasavva, P.; Romito, M.; Loucari, C.C.; Stephanou, C.; Christou, S.; Sitarou, M.; Mussolino, C.; Cornu, T.I.; et al. Correction of IVS I-110(G>A) β-thalassemia by CRISPR/Cas-and TALEN-mediated disruption of aberrant regulatory elements in human hematopoietic stem and progenitor cells. Haematologica 2019, 104, e497–e501. [Google Scholar] [CrossRef]
- Patsali, P.; Mussolino, C.; Ladas, P.; Floga, A.; Kolnagou, A.; Christou, S.; Sitarou, M.; Antoniou, M.N.; Cathomen, T.; Lederer, C.W.; et al. The Scope for Thalassemia Gene Therapy by Disruption of Aberrant Regulatory Elements. J. Clin. Med. 2019, 8, 1959. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, V.G.; Orkin, S.H. The Switch from Fetal to Adult Hemoglobin. Cold Spring Harb. Perspect. Med. 2012, 3, a011643. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, V.G.; Menne, T.F.; Xu, J.; Akie, T.E.; Lettre, G.; Van Handel, B.; Mikkola, H.K.A.; Hirschhorn, J.N.; Cantor, A.B.; Orkin, S.H. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science 2008, 322, 1839–1842. [Google Scholar] [CrossRef]
- Psatha, N.; Reik, A.; Phelps, S.; Zhou, Y.; Dalas, D.; Yannaki, E.; Levasseur, D.N.; Urnov, F.D.; Holmes, M.C.; Papayannopoulou, T. Disruption of the BCL11A Erythroid Enhancer Reactivates Fetal Hemoglobin in Erythroid Cells of Patients with β-Thalassemia Major. Mol. Ther. Methods Clin. Dev. 2018, 10, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Salmaninejad, A.; Valilou, S.F.; Bayat, H.; Ebadi, N.; Daraei, A.; Yousefi, M.; Nesaei, A.; Mojarrad, M. Duchenne muscular dystrophy: An updated review of common available therapies. Int. J. Neurosci. 2018, 128, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Ousterout, D.G.; Perez-Pinera, P.; Thakore, P.I.; Kabadi, A.M.; Brown, M.T.; Qin, X.; Fedrigo, O.; Mouly, V.; Tremblay, J.P.; Gersbach, A.C. Reading Frame Correction by Targeted Genome Editing Restores Dystrophin Expression in Cells From Duchenne Muscular Dystrophy Patients. Mol. Ther. 2013, 21, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.; Suresh, S.; Guo, D.; Daman, K.; Chen, J.C.J.; Liu, P.; Zieger, M.; Luk, K.; Roscoe, B.P.; Mueller, C.; et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 2019, 568, 561–565. [Google Scholar] [CrossRef]
- Grajcarek, J.; Monlong, J.; Nishinaka-Arai, Y.; Nakamura, M.; Nagai, M.; Matsuo, S.; Lougheed, D.; Sakurai, H.; Saito, M.; Bourque, G.; et al. Genome-wide microhomologies enable precise template-free editing of biologically relevant deletion mutations. Nat. Commun. 2019, 10, 4856. [Google Scholar] [CrossRef] [PubMed]
- Vengalil, S.; Preethish-Kumar, V.; Polavarapu, K.; Mahadevappa, M.; Sekar, D.; Purushottam, M.; Thomas, P.T.; Nashi, S.; Nalini, A. Duchenne Muscular Dystrophy and Becker Muscular Dystrophy Confirmed by Multiplex Ligation-Dependent Probe Amplification: Genotype-Phenotype Correlation in a Large Cohort. J. Clin. Neurol. 2017, 13, 91–97. [Google Scholar] [CrossRef]
- Mendell, J.R.; Campbell, K.; Rodino-Klapac, L.; Sahenk, Z.; Shilling, C.; Lewis, S.; Bowles, D.; Gray, S.; Li, C.; Galloway, G.; et al. Dystrophin immunity in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2010, 363, 1429–1437. [Google Scholar] [CrossRef]
- Gundry, M.; Brunetti, L.; Lin, A.; Mayle, A.; Kitano, A.; Wagner, D.L.; Hsu, J.I.; Hoegenauer, K.A.; Rooney, C.M.; Goodell, M.A.; et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep. 2016, 17, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Rouet, P.; Smih, F.; Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 1994, 91, 6064–6068. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014, 3, 04766. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Shi, Z.; Guo, X.; Jiang, B.; Wang, G.; Luo, D.; Chen, Y.; Zhu, Y.-S. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR–Cas9 and ssODN in human cancer cells. Cell Biosci. 2018, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Khedher, A.H.Y.; Menoret, S.; Brion, A.; Lamribet, K.; Dardillac, E.; Boix, C.; Perrouault, L.; Tesson, L.; Geny, S.; et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018, 9, 1133. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, B.S.; Mandal, P.K.; Frock, R.L.; Boyraz, B.; Yadav, R.; Upadhyayula, S.; Gutierrez-Martinez, P.; Ebina, W.; Fasth, A.; Kirchhausen, T.; et al. Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat. Biomed. Eng. 2017, 1, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Canver, M.; Orkin, S.H. Customizing the genome as therapy for the β-hemoglobinopathies. Blood 2016, 127, 2536–2545. [Google Scholar] [CrossRef] [PubMed]
- Porteus, M.H. Genome Editing for the β-Hemoglobinopathies BT-Gene and Cell Therapies for Beta-Globinopathies; Malik, P., Tisdale, J., Eds.; Springer: New York, NY, USA, 2017; pp. 203–217. ISBN 978-1-4939-7299-9. [Google Scholar]
- Dever, D.P.; Porteus, M. The changing landscape of gene editing in hematopoietic stem cells. Curr. Opin. Hematol. 2017, 24, 481–488. [Google Scholar] [CrossRef]
- Agudelo, D.; Duringer, A.; Bozoyan, L.; Huard, C.C.; Carter, S.; Loehr, J.; Synodinou, D.; Drouin, M.; Salsman, J.; Dellaire, G.; et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods 2017, 14, 615–620. [Google Scholar] [CrossRef] [PubMed]
- De Ravin, S.S.; Li, L.; Wu, X.; Choi, U.; Allen, C.; Koontz, S.; Lee, J.; Theobald-Whiting, N.; Chu, J.; Garofalo, M.; et al. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci. Transl. Med. 2017, 9, eaah3480. [Google Scholar] [CrossRef] [PubMed]
- De Ravin, S.S.; Reik, A.; Liu, P.-Q.; Li, L.; Wu, X.; Su, L.; Raley, C.; Theobald, N.; Choi, U.; Song, A.H.; et al. Targeted gene addition in human CD34+ hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 2016, 34, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.-K. Off-target effects of engineered nucleases. FEBS J. 2016, 283, 3239–3248. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, M.P.; Krishnakumar, R.; Timlin, J.A.; Carney, J.P.; Butler, K.S. Gene editing and CRISPR in the clinic: Current and future perspectives. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carusillo, A.; Mussolino, C. DNA Damage: From Threat to Treatment. Cells 2020, 9, 1665. https://doi.org/10.3390/cells9071665
Carusillo A, Mussolino C. DNA Damage: From Threat to Treatment. Cells. 2020; 9(7):1665. https://doi.org/10.3390/cells9071665
Chicago/Turabian StyleCarusillo, Antonio, and Claudio Mussolino. 2020. "DNA Damage: From Threat to Treatment" Cells 9, no. 7: 1665. https://doi.org/10.3390/cells9071665
APA StyleCarusillo, A., & Mussolino, C. (2020). DNA Damage: From Threat to Treatment. Cells, 9(7), 1665. https://doi.org/10.3390/cells9071665



