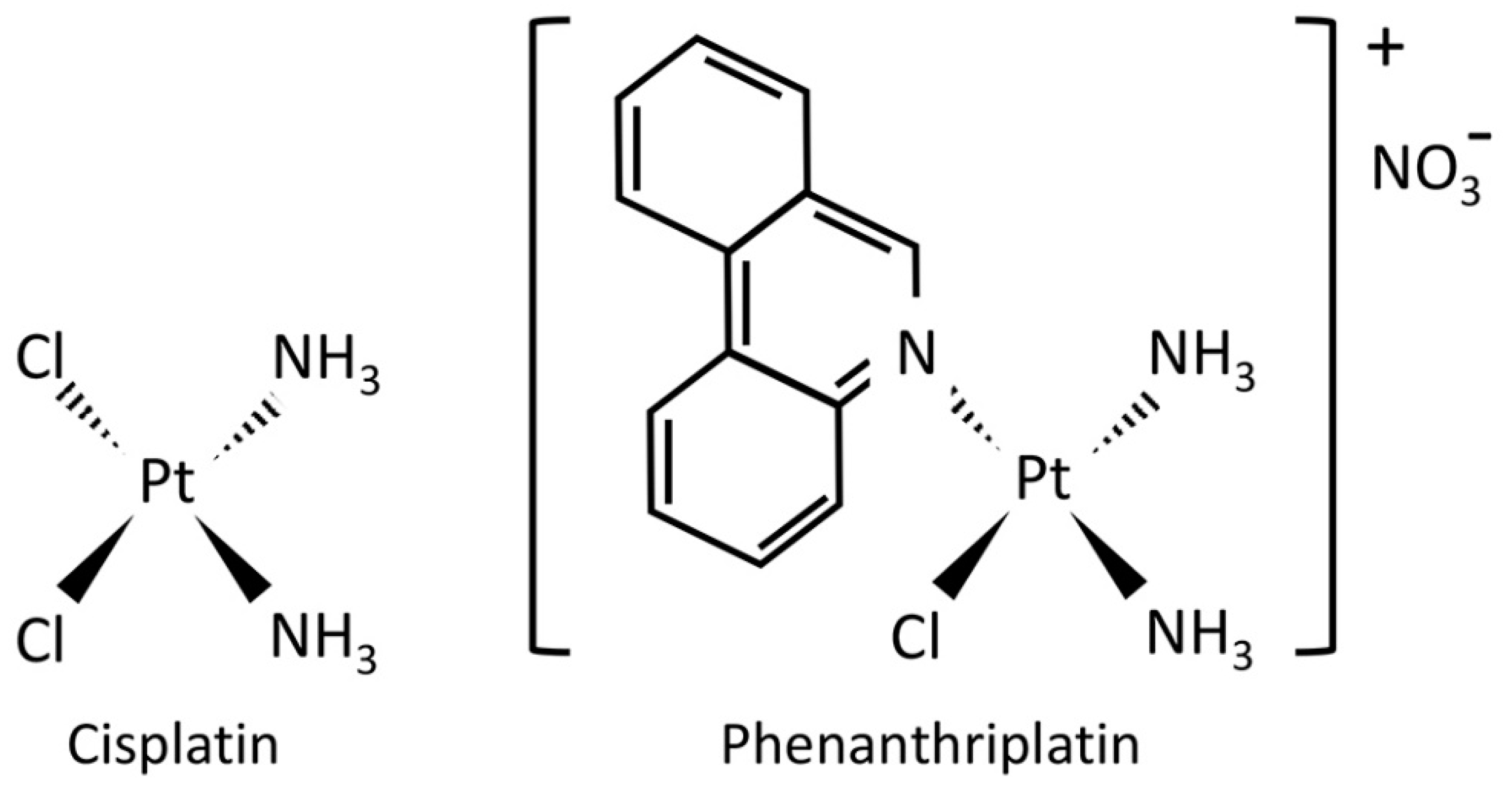
RNA-Seq Analysis of Cisplatin and the Monofunctional Platinum(II) Complex, Phenanthriplatin, in A549 Non-Small Cell Lung Cancer and IMR90 Lung Fibroblast Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. RNA Isolation and Library Preparation
2.3. Next Generation Sequencing
2.4. Droplet Digital Polymerase Chain Reaction Analysis
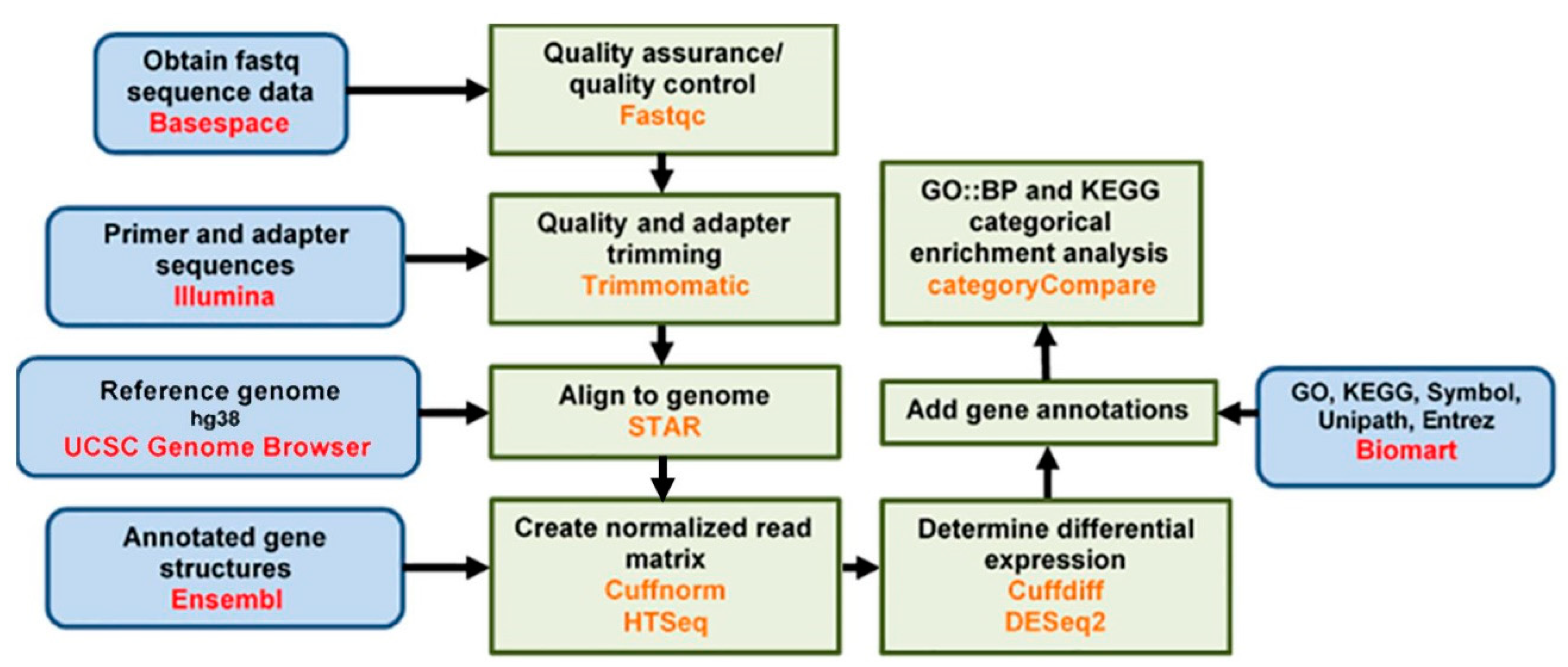
2.5. Data Analysis
2.6. Statistical Analysis
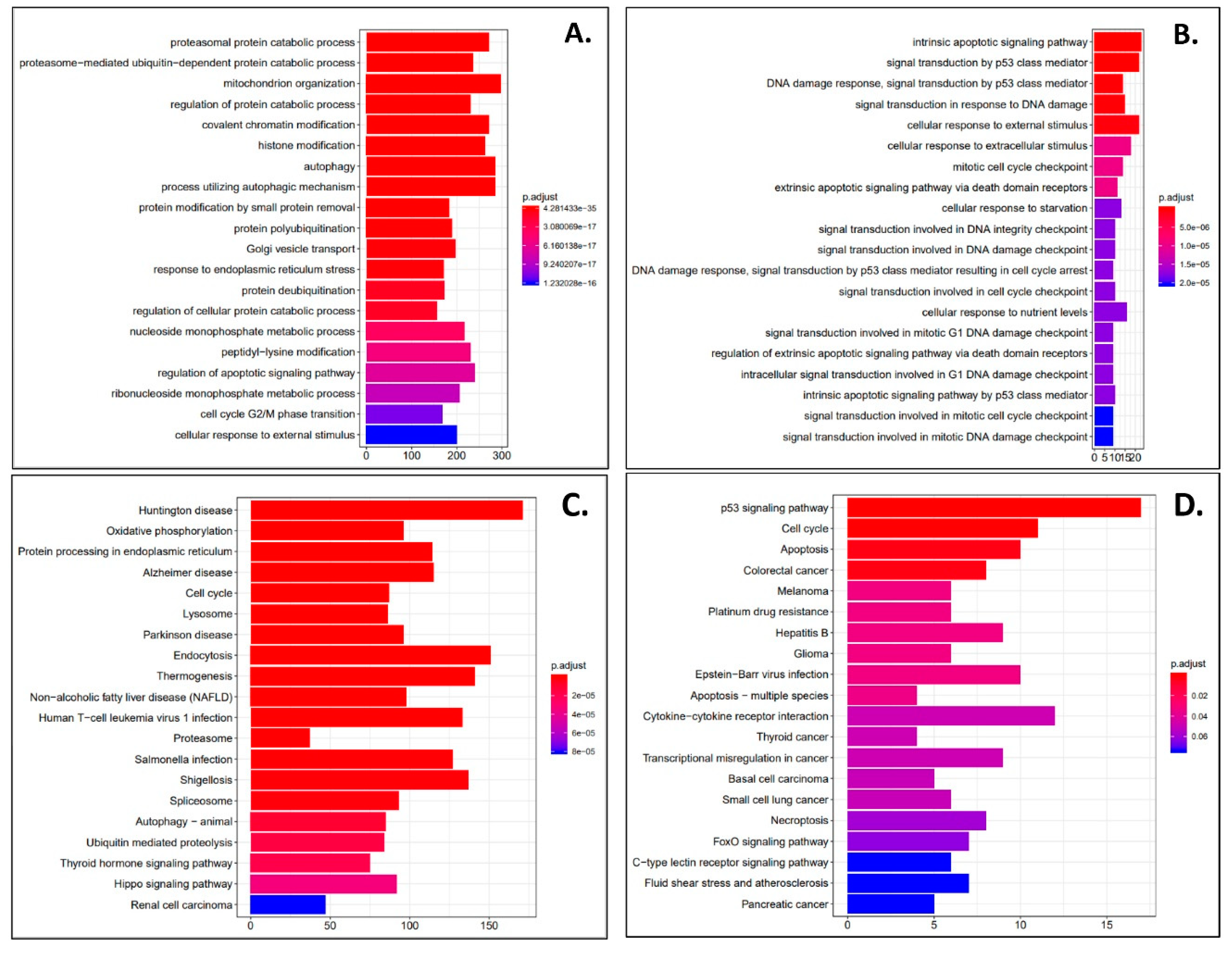
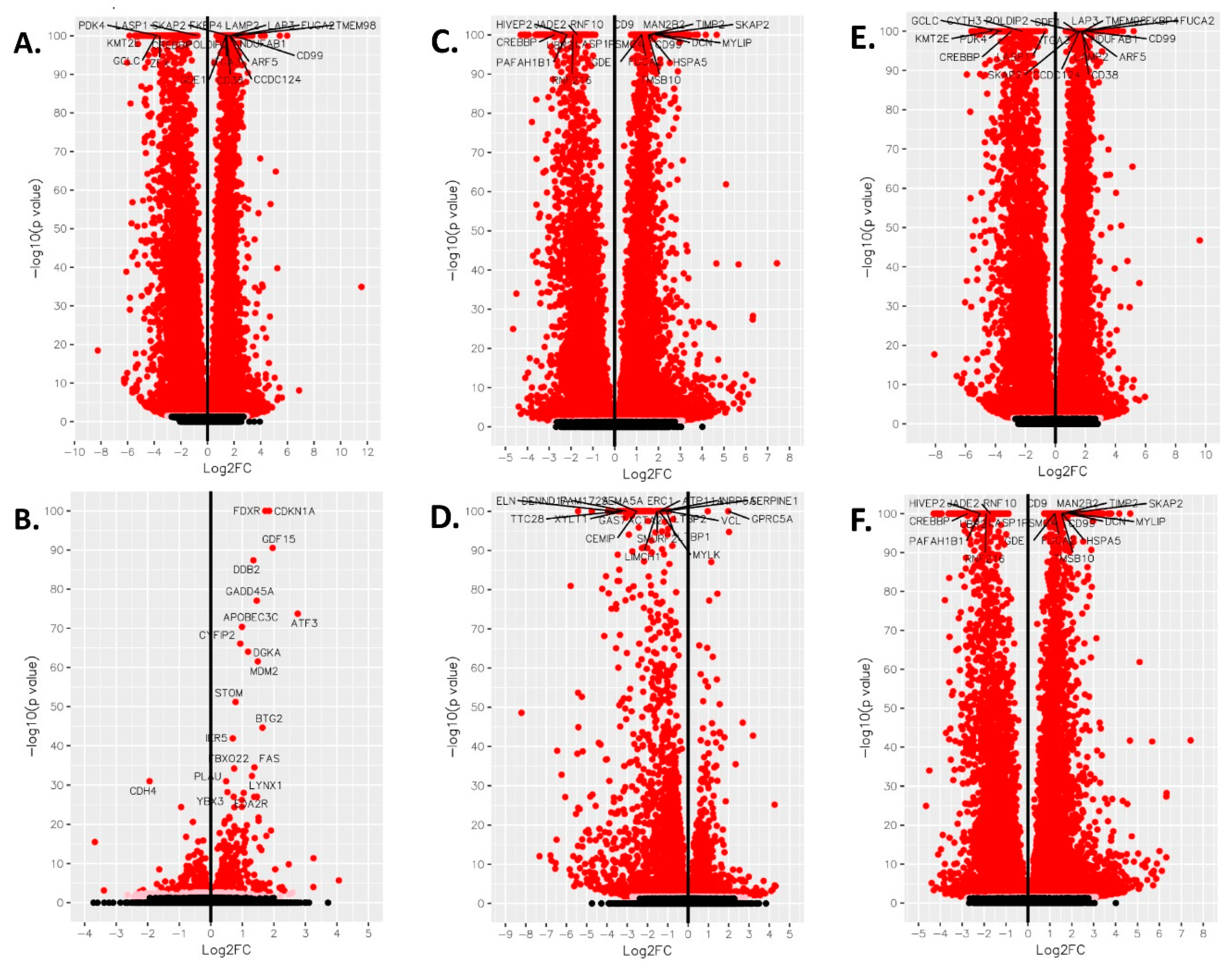
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cepeda, V.; Fuertes, M.A.; Castilla, J.; Alonso, C.; Quevedo, C.; Pérez, J.M. Biochemical Mechanisms of Cisplatin Cytotoxicity. Anticanc. Agents Med. Chem. 2007, 7, 3–18. [Google Scholar] [CrossRef]
- Todd, R.C.; Lippard, S.J. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009, 1, 280–291. [Google Scholar] [CrossRef]
- Carminati, P.O.; Mello, S.S.; Fachin, A.L.; Junta, C.M.; Sandrin-Garcia, P.; Carlotti, C.G.; Donadi, E.A.; Passos, G.A.; Sakamoto-Hojo, E.T. Alterations in Gene Expression Profiles Correlated With Cisplatin Cytotoxicity in the Glioma U343 Cell Line. Genet. Mol. Biol. 2010, 33, 159–168. [Google Scholar] [CrossRef]
- Yamano, Y.; Uzawa, K.; Saito, K.; Nakashima, D.; Kasamatsu, A.; Koike, H.; Kouzu, Y.; Shinozuka, K.; Nakatani, K.; Negoro, K.; et al. Identification of Cisplatin-Resistance Related Genes in Head and Neck Squamous Cell Carcinoma. BMC Cancer 2006, 6, 224. [Google Scholar] [CrossRef]
- Cheng, L.; Lu, W.; Kulkarni, B.; Pejovic, T.; Yan, X.; Chiang, J.H.; Hood, L.; Odunsi, K.; Lin, B. Analysis of chemotherapy response programs in ovarian cancers by the next-generation sequencing technologies. Gynecol. Oncol. 2010, 117, 159–169. [Google Scholar] [CrossRef]
- Gabriel, M.; Delforge, Y.; Deward, A.; Habraken, Y.; Hennuy, B.; Piette, J.; Klinck, R.; Chabot, B.; Colige, A.; Lambert, C. Role of the splicing factor SRSF4 in cisplatin-induced modifications of pre-mRNA splicing and apoptosis. BMC Cancer 2015, 15, 227. [Google Scholar] [CrossRef] [PubMed]
- Motamedian, E.; Ghavami, G.; Sardari, S. Investigation on Metabolism of Cisplatin Resistant Ovarian Cancer Using a Genome Scale Metabolic Model and Microarray Data. Iran J. Basic Med. Sci. 2015, 18, 267–276. [Google Scholar]
- Wang, S.; Xie, J.; Li, J.; Liu, F.; Wu, X.; Wang, Z. Cisplatin Suppresses the Growth and Proliferation of Breast and Cervical Cancer Cell Lines by Inhibiting Integrin β5-mediated Glycolysis. Am. J. Cancer Res. 2016, 6, 1108–1117. [Google Scholar]
- Zhang, Y.; Du, H.; Li, Y.; Yuan, Y.; Chen, B.; Sun, S. Elevated TRIM23 expression predicts cisplatin resistance in lung adenocarcinoma. Cancer Sci. 2020, 111, 637–646. [Google Scholar] [CrossRef]
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin Nephrotoxicity. Toxins (Basel) 2010, 2, 2490–2518. [Google Scholar] [CrossRef]
- Karasawa, T.; Steyger, P.S. An integrated view of cisplatin-induced nephrotoxicity and ototoxicity. Toxicol. Lett. 2015, 237, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Park, G.Y.; Wilson, J.J.; Song, Y.; Lippard, S.J. Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc. Natl. Acad. Sci. USA 2012, 109, 11987–11992. [Google Scholar] [CrossRef] [PubMed]
- Monroe, J.D.; Hruska, H.L.; Ruggles, H.K.; Williams, K.M.; Smith, M.E. Anti-cancer characteristics and ototoxicity of platinum(II) amine complexes with only one leaving ligand. PLoS ONE 2018, 13, e0192505. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Park, G.Y.; Lippard, S.J. Understanding and improving platinum anticancer drugs--phenanthriplatin. Anticancer Res. 2014, 34, 471–476. [Google Scholar]
- Kellinger, M.W.; Park, G.Y.; Chong, J.; Lippard, S.J.; Wang, D. Effect of a monofunctional phenanthriplatin-DNA adduct on RNA polymerase II transcriptional fidelity and translesion synthesis. J. Am. Chem. Soc. 2013, 135, 13054–13061. [Google Scholar] [CrossRef]
- Riddell, I.A.; Johnstone, T.C.; Park, G.Y.; Lippard, S.J. Nucleotide Binding Preference of the Monofunctional Platinum Anticancer-Agent Phenanthriplatin. Chemistry 2016, 22, 7574–7581. [Google Scholar] [CrossRef]
- Guinea Viniegra, J.; Hernández Losa, J.; Sánchez-Arévalo, V.J.; Parada Cobo, C.; Fernández Soria, V.M.; Ramón y Cajal, S.; Sánchez-Prieto, R. Modulation of PI3K/Akt pathway by E1a mediates sensitivity to cisplatin. Oncogene 2002, 21, 7131–7136. [Google Scholar] [CrossRef][Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol 2010, 11, R106. [Google Scholar] [CrossRef]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Hucke, A.; Park, G.Y.; Bauer, O.B.; Beyer, G.; Köppen, C.; Zeeh, D.; Wehe, C.A.; Sperling, M.; Schröter, R.; Kantauskaitè, M.; et al. Interaction of the New Monofunctional Anticancer Agent Phenanthriplatin With Transporters for Organic Cations. Front. Chem. 2018, 6, 180. [Google Scholar] [CrossRef]
- Küng, A.; Strickmann, D.B.; Galanski, M.; Keppler, B.K. Comparison of the binding behavior of oxaliplatin, cisplatin and analogues to 5′-GMP in the presence of sulfur-containing molecules by means of capillary electrophoresis and electrospray mass spectrometry. J. Inorg. Biochem. 2001, 86, 691–698. [Google Scholar] [CrossRef]
- Li, J.; Wood, W.H., 3rd; Becker, K.G.; Weeraratna, A.T.; Morin, P.J. Gene expression response to cisplatin treatment in drug-sensitive and drug-resistant ovarian cancer cells. Oncogene 2007, 26, 2860–2872. [Google Scholar] [CrossRef]
- Du, A.; Jiang, Y.; Fan, C. NDRG1 Downregulates ATF3 and Inhibits Cisplatin-Induced Cytotoxicity in Lung Cancer A549 Cells. Int. J. Med. Sci. 2018, 15, 1502–1507. [Google Scholar] [CrossRef]
- Jiao, P.; Hou, J.; Yao, M.; Wu, J.; Ren, G. SNHG14 silencing suppresses the progression and promotes cisplatin sensitivity in non-small cell lung cancer. Biomed. Pharmacother. 2019, 117, 109164. [Google Scholar] [CrossRef]
- Wu, J.; Hu, C.P.; Gu, Q.H.; Li, Y.P.; Song, M. Trichostatin A sensitizes cisplatin-resistant A549 cells to apoptosis by up-regulating death-associated protein kinase. Acta Pharmacol. Sin. 2010, 31, 93–101. [Google Scholar] [CrossRef]
- Li, F.L.; Liu, J.P.; Bao, R.X.; Yan, G.; Feng, X.; Xu, Y.P.; Sun, Y.P.; Yan, W.; Ling, Z.Q.; Xiong, Y.; et al. Acetylation accumulates PFKFB3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat. Commun. 2018, 9, 508. [Google Scholar] [CrossRef]
- Liu, X.; Yue, P.; Khuri, F.R.; Sun, S.Y. Decoy receptor 2 (DcR2) is a p53 target gene and regulates chemosensitivity. Cancer Res. 2005, 65, 9169–9175. [Google Scholar] [CrossRef]
- Gonzalez-Rajal, A.; Hastings, J.F.; Watkins, D.N.; Croucher, D.R.; Burgess, A. Breathing New Life into the Mechanisms of Platinum Resistance in Lung Adenocarcinoma. Front. Cell Dev. Biol. 2020, 8, 305. [Google Scholar] [CrossRef]
- Kontro, H.; Cannino, G.; Rustin, P.; Dufour, E.; Kainulainen, H. DAPIT Over-Expression Modulates Glucose Metabolism and Cell Behaviour in HEK293T Cells. PLoS ONE 2015, 10, e0131990. [Google Scholar] [CrossRef]
- Dai, D.; Shi, R.; Han, S.; Jin, H.; Wang, X. Weighted gene coexpression network analysis identifies hub genes related to KRAS mutant lung adenocarcinoma. Medicine (Baltimore) 2020, 99, e21478. [Google Scholar] [CrossRef]
- Zhao, D.; Qiao, J.; He, H.; Song, J.; Zhao, S.; Yu, J. TFPI2 suppresses breast cancer progression through inhibiting TWIST-integrin α5 pathway. Mol. Med. 2020, 26, 27. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, K.; Nakamura, Y.; Kitagawa, Y.; Suzuki, Y.; Shibagaki, Y.; Hattori, S.; Ohta, Y. AGAP1 Regulates Subcellular Localization of FilGAP and Control Cancer Cell Invasion. Biochem. Biophys. Res. Commun. 2020, 522, 676–683. [Google Scholar] [CrossRef]
- Grueb, S.S.; Muhs, S.; Popp, Y.; Schmitt, S.; Geyer, M.; Lin, Y.N.; Windhorst, S. The Formin Drosophila Homologue of Diaphanous2 (Diaph2) Controls Microtubule Dynamics in Colorectal Cancer Cells Independent of Its FH2-domain. Sci. Rep. 2019, 9, 5352. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chen, T.; Li, Y.; Gao, L.; Zhang, S.; Wang, T.; Chen, M. Downregulation of miR-25 modulates non-small cell lung cancer cells by targeting CDC42. Tumour. Biol. 2015, 36, 1903–1911. [Google Scholar] [CrossRef]
- Zhao, C.; Li, Y.; Qiu, W.; He, F.; Zhang, W.; Zhao, D.; Zhang, Z.; Zhang, E.; Ma, P.; Liu, Y.; et al. C5a Induces A549 Cell Proliferation of Non-Small Cell Lung Cancer via GDF15 Gene Activation Mediated by GCN5-dependent KLF5 Acetylation. Oncogene 2018, 37, 4821–4837. [Google Scholar] [CrossRef]
- Tarfiei, G.A.; Shadboorestan, A.; Montazeri, H.; Rahmanian, N.; Tavosi, G.; Ghahremani, M.H. GDF15 induced apoptosis and cytotoxicity in A549 cells depends on TGFBR2 expression. Cell Biochem. Funct. 2019, 37, 320–330. [Google Scholar] [CrossRef]
- Feng, G.; Zhang, Y.; Yuan, H.; Bai, R.; Zheng, J.; Zhang, J.; Song, M. DNA methylation of trefoil factor 1 (TFF1) is associated with the tumorigenesis of gastric carcinoma. Mol. Med. Rep. 2014, 9, 109–117. [Google Scholar] [CrossRef]
- Soutto, M.; Chen, Z.; Saleh, M.A.; Katsha, A.; Zhu, S.; Zaika, A.; Belkhiri, A.; El-Rifai, W. TFF1 activates p53 through down-regulation of miR-504 in gastric cancer. Oncotarget 2014, 5, 5663–5673. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Sun, B.; Liao, Y.; Xu, D.; Guo, W.; Wang, T.; Jing, B.; Hu, M.; Li, K.; Yao, F.; et al. GPRC5A Deficiency Leads to Dysregulated MDM2 via Activated EGFR Signaling for Lung Tumor Development. Int. J. Cancer 2019, 144, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Sarin, N.; Engel, F.; Kalayda, G.V.; Mannewitz, M.; Cinatl, J.; Rothweiler, F.; Michaelis, M.; Saafan, H.; Ritter, C.A.; Jaehde, U.; et al. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS ONE 2017, 12, e0181081. [Google Scholar] [CrossRef]
- Khamas, A.; Ishikawa, T.; Mogushi, K.; Iida, S.; Ishiguro, M.; Tanaka, H.; Uetake, H.; Sugihara, K. Genome-wide Screening for Methylation-Silenced Genes in Colorectal Cancer. Int. J. Oncol. 2012, 41, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.M.; Chan, P.L.; Yau, W.L.; Chan, H.K.; Chan, K.C.; Yu, Z.Y.; Kwong, F.M.; Miller, L.D.; Liu, E.T.; Yang, L.C.; et al. Monochromosome Transfer and Microarray Analysis Identify a Critical Tumor-Suppressive Region Mapping to Chromosome 13q14 and THSD1 in Esophageal Carcinoma. Mol. Cancer Res. 2008, 6, 592–603. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rui, Y.N.; Xu, Z.; Fang, X.; Menezes, M.R.; Balzeau, J.; Niu, A.; Hagan, J.P.; Kim, D.H. The Intracranial Aneurysm Gene THSD1 Connects Endosome Dynamics to Nascent Focal Adhesion Assembly. Cell Physiol. Biochem. 2017, 43, 2200–2211. [Google Scholar] [CrossRef] [PubMed]

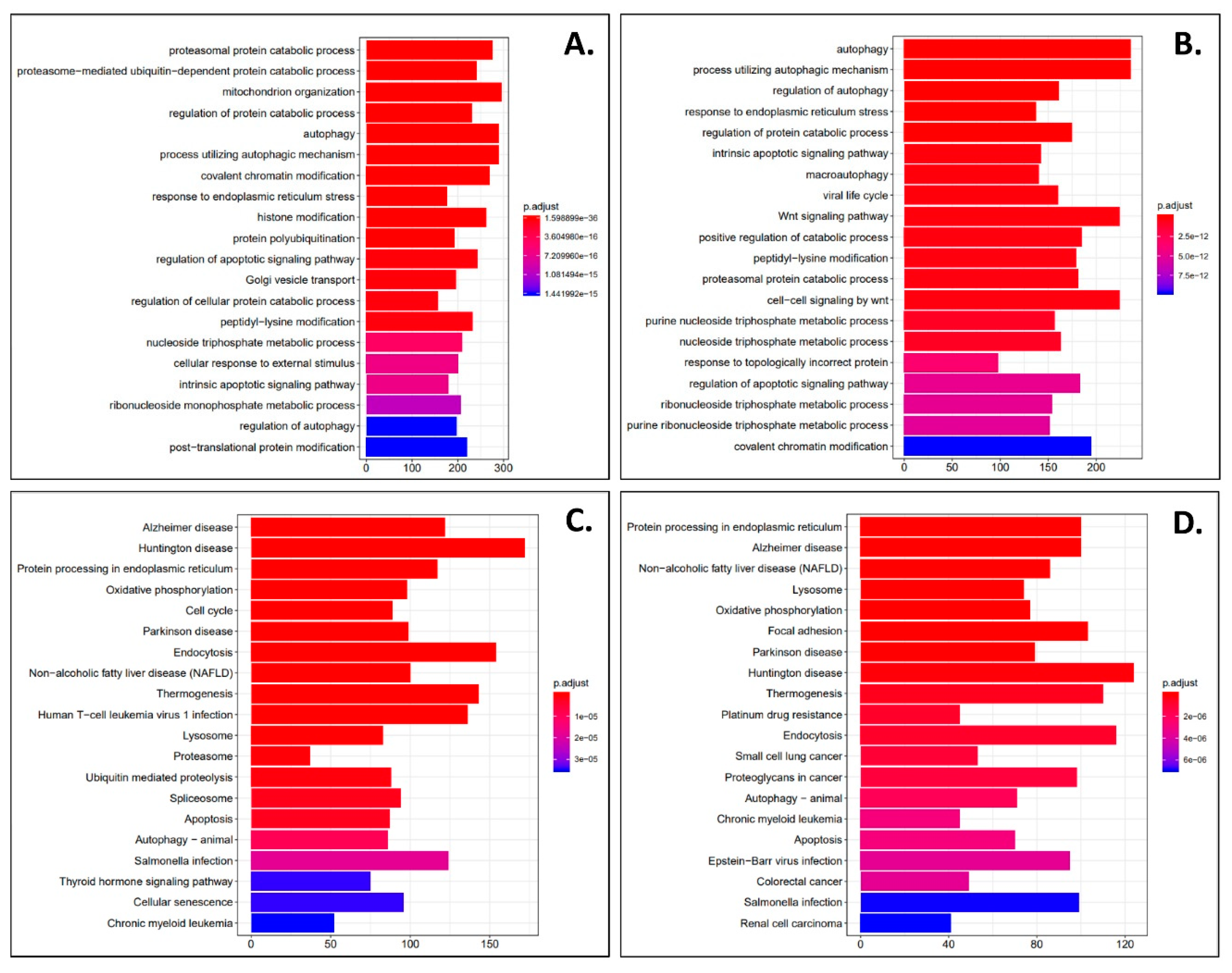
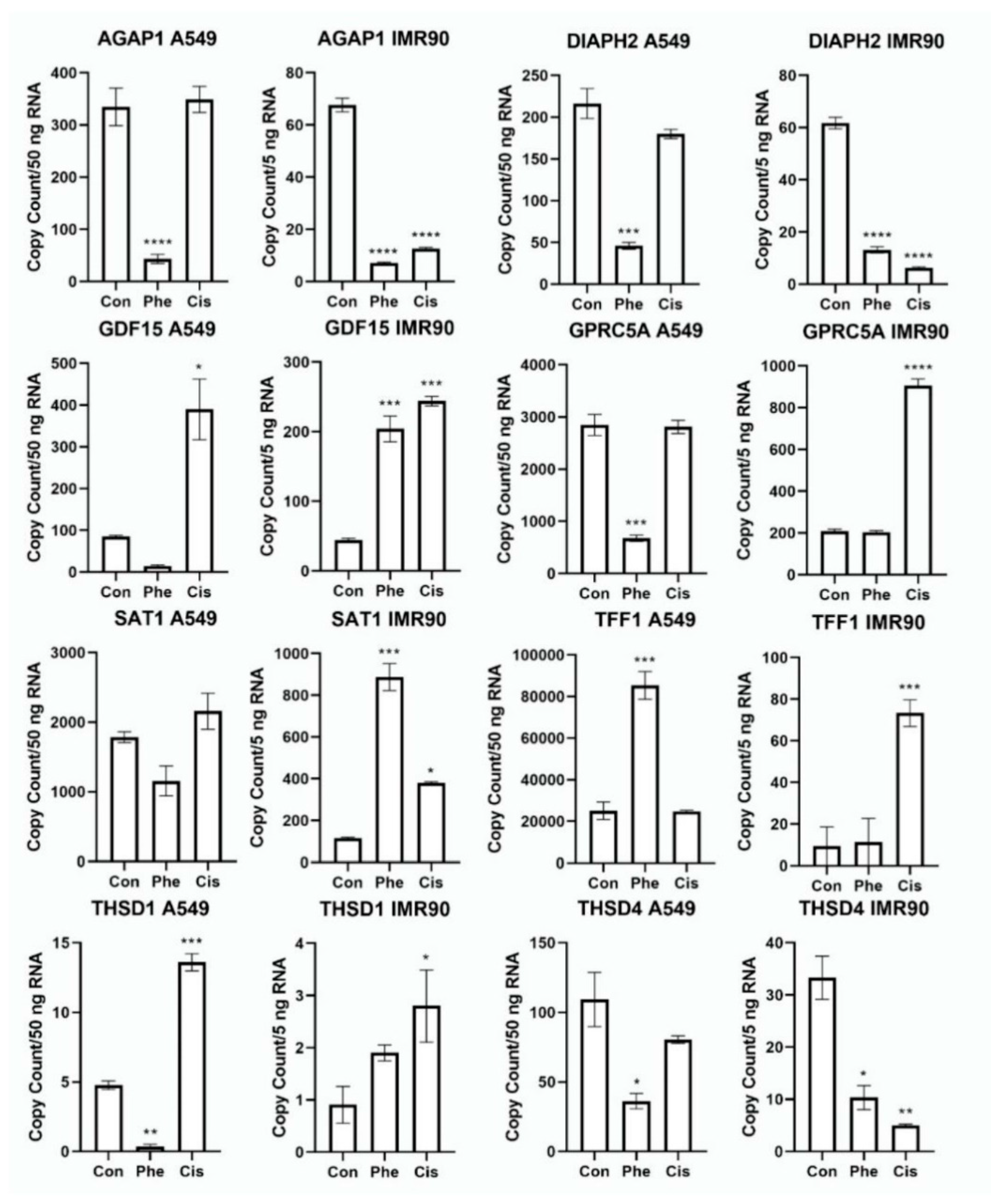
| Gene (↑) | Log2FC | p-Value | q-Value | Gene (↓) | Log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| ATF3 | 5.986 | ≤0.0001 | ≤0.0001 | PFKFB3 | −4.236 | ≤0.0001 | ≤0.0001 |
| TFF1 | 4.067 | ≤0.0001 | ≤0.0001 | LPCAT1 | −4.240 | ≤0.0001 | ≤0.0001 |
| RBP4 | 3.324 | ≤0.0001 | ≤0.0001 | DAPK1 | −4.305 | ≤0.0001 | ≤0.0001 |
| S100A6 | 2.806 | ≤0.0001 | ≤0.0001 | RAI1 | −4.330 | ≤0.0001 | ≤0.0001 |
| TNFRSF10D | 2.760 | ≤0.0001 | ≤0.0001 | ZNF609 | −4.484 | ≤0.0001 | ≤0.0001 |
| C12orf75 | 2.695 | ≤0.0001 | ≤0.0001 | PDE4D | −4.590 | ≤0.0001 | ≤0.0001 |
| TFPI2 | 2.691 | ≤0.0001 | ≤0.0001 | B4GALT5 | −4.665 | ≤0.0001 | ≤0.0001 |
| ATP5MD | 2.678 | ≤0.0001 | ≤0.0001 | SIN3A | −4.809 | ≤0.0001 | ≤0.0001 |
| SNHG14 | 2.667 | ≤0.0001 | ≤0.0001 | HMOX1 | −4.968 | ≤0.0001 | ≤0.0001 |
| S100A4 | 2.659 | ≤0.0001 | ≤0.0001 | TNFRSF1A | −5.852 | ≤0.0001 | ≤0.0001 |
| Gene (↑) | Log2FC | p-Value | q-Value | Gene (↓) | Log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| ATF3 | 2.757 | 1.880 × 10−74 | 4.441 × 10−71 | MCM5 | −0.482 | 7.133 × 10−11 | 9.723 × 10−9 |
| GDF15 | 1.962 | 2.950 × 10−91 | 1.393 × 10−87 | SLIT3 | −0.530 | 6.219 × 10−13 | 1.101 × 10−10 |
| CDKN1A | 1.852 | 2.829 × 10−142 | 4.010 × 10−138 | TRAPPC9 | −0.565 | 2.361 × 10−21 | 1.014 × 10−18 |
| FDXR | 1.728 | 2.276 × 10−111 | 1.613 × 10−107 | SAPCD2 | −0.686 | 1.617 × 10−12 | 2.730 × 10−10 |
| BTG2 | 1.639 | 2.434 × 10−45 | 2.875 × 10−42 | KCNMA1 | −0.710 | 2.770 × 10−12 | 4.620 × 10−10 |
| MDM2 | 1.490 | 2.982 × 10−62 | 4.227 × 10−59 | MSRA | −0.769 | 4.236 × 10−10 | 5.177 × 10−8 |
| SESN2 | 1.467 | 8.864 × 10−28 | 5.815 × 10−25 | MCM6 | −0.782 | 1.516 × 10−17 | 4.478 × 10−15 |
| GADD45A | 1.458 | 8.776 × 10−78 | 2.488 × 10−74 | GPC6 | −0.940 | 3.773 × 10−25 | 2.057 × 10−22 |
| FAS | 1.384 | 3.439 × 10−35 | 3.482 × 10−32 | CDH4 | −1.940 | 1.156 × 10−31 | 9.432 × 10−29 |
| DDB2 | 1.350 | 4.346 × 10−88 | 1.540 × 10−84 | LSAMP | −3.677 | 3.272 × 10−16 | 8.434 × 10−14 |
| Gene (↑) | Log2FC | p-Value | q-Value | Gene (↓) | Log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| ATF3 | 5.364 | ≤0.0001 | ≤0.0001 | HMGA2 | −3.439 | ≤0.0001 | ≤0.0001 |
| SAT1 | 4.094 | ≤0.0001 | ≤0.0001 | ASAP1 | −3.543 | ≤0.0001 | ≤0.0001 |
| GDF15 | 3.657 | ≤0.0001 | ≤0.0001 | GSK3B | −3.549 | ≤0.0001 | ≤0.0001 |
| GADD45A | 3.433 | ≤0.0001 | ≤0.0001 | LBH | −3.571 | ≤0.0001 | ≤0.0001 |
| NPTX1 | 2.941 | 7.459 × 10−307 | 2.830 × 10−304 | NRG1 | −3.680 | ≤0.0001 | ≤0.0001 |
| PCNA | 2.344 | ≤0.0001 | ≤0.0001 | IGF2BP2 | −4.246 | ≤0.0001 | ≤0.0001 |
| PEG10 | 2.312 | ≤0.0001 | ≤0.0001 | COL1A1 | −4.330 | ≤0.0001 | ≤0.0001 |
| TM4SF1 | 2.103 | ≤0.0001 | ≤0.0001 | SLC38A2 | −4.409 | ≤0.0001 | ≤0.0001 |
| TFPI2 | 2.007 | ≤0.0001 | ≤0.0001 | ZMIZ1 | −4.422 | ≤0.0001 | ≤0.0001 |
| TNFRSF10D | 2.006 | ≤0.0001 | ≤0.0001 | SMURF2 | −4.866 | ≤0.0001 | ≤0.0001 |
| Gene (↑) | Log2FC | p-Value | q-Value | Gene (↓) | Log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| THSD1 | 2.690 | 9.073 × 10−47 | 5.963 × 10−45 | PLXDC2 | −2.875 | 7.181 × 10−249 | 9.512 × 10−246 |
| GDF15 | 2.016 | 1.914 × 10−95 | 3.330 × 10−93 | TRAPPC9 | −2.988 | 3.254 × 10−240 | 3.503 × 10−237 |
| GPRC5A | 1.975 | 5.924 × 10−103 | 1.214 × 10−100 | PRKCA | −3.083 | 3.001 × 10−224 | 2.720 × 10−221 |
| CYP1B1 | 1.556 | 2.229 × 10−44 | 1.396 × 10−42 | EXOC4 | −3.317 | 9.567 × 10−293 | 2.353 × 10−289 |
| PLCXD1 | 1.553 | 1.755 × 10−51 | 1.244 × 10−49 | PTPRG | −3.364 | ≤0.0001 | ≤0.0001 |
| SAT1 | 1.444 | 1.116 × 10−79 | 1.467 × 10−77 | LRBA | −3.455 | 4.344 × 10−275 | 8.312 × 10−272 |
| FDXR | 1.417 | 8.633 × 10−58 | 7.182 × 10−56 | NAALADL2 | −3.479 | 3.697 × 10−212 | 3.184 × 10−209 |
| MDM2 | 1.246 | 4.780 × 10−44 | 2.919 × 10−42 | THSD4 | −3.693 | ≤0.0001 | ≤0.0001 |
| CDKN1A | 1.212 | 1.385 × 10−63 | 1.332 × 10−61 | AGAP1 | −3.956 | ≤0.0001 | ≤0.0001 |
| PLK2 | 1.176 | 1.313 × 10−46 | 8.603 × 10−45 | DIAPH2 | −4.744 | 3.869 × 10−244 | 4.760 × 10−241 |
| Gene (↑) | Log2FC | p-Value | q-Value | Gene (↓) | Log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| TFF1 | 4.503 | ≤0.0001 | ≤0.0001 | PFKFB3 | −4.165 | ≤0.0001 | ≤0.0001 |
| RBP4 | 3.360 | ≤0.0001 | ≤0.0001 | DAPK1 | −4.210 | ≤0.0001 | ≤0.0001 |
| CST1 | 2.984 | ≤0.0001 | ≤0.0001 | NR3C1 | −4.246 | ≤0.0001 | ≤0.0001 |
| SNHG14 | 2.741 | ≤0.0001 | ≤0.0001 | RAI1 | −4.316 | ≤0.0001 | ≤0.0001 |
| TFPI2 | 2.740 | ≤0.0001 | ≤0.0001 | PDE4D | −4.425 | ≤0.0001 | ≤0.0001 |
| SERF2 | 2.733 | ≤0.0001 | ≤0.0001 | ZNF609 | −4.451 | ≤0.0001 | ≤0.0001 |
| S100A6 | 2.722 | ≤0.0001 | ≤0.0001 | MDM2 | −4.538 | ≤0.0001 | ≤0.0001 |
| S100A4 | 2.675 | ≤0.0001 | ≤0.0001 | SIN3A | −4.629 | ≤0.0001 | ≤0.0001 |
| FSTL3 | 2.661 | ≤0.0001 | ≤0.0001 | B4GALT5 | −4.724 | ≤0.0001 | ≤0.0001 |
| COX8A | 2.654 | ≤0.0001 | ≤0.0001 | TNFRSF1A | −5.596 | ≤0.0001 | ≤0.0001 |
| Gene (↑) | Log2FC | p-Value | q-Value | Gene (↓) | Log2FC | p-Value | q-Value |
|---|---|---|---|---|---|---|---|
| THSD4 | 3.658 | ≤0.0001 | ≤0.0001 | PUM1 | −2.792 | ≤0.0001 | ≤0.0001 |
| NPTX1 | 3.163 | ≤0.0001 | ≤0.0001 | TRAM2 | −2.797 | ≤0.0001 | ≤0.0001 |
| P4HA3 | 2.809 | ≤0.0001 | ≤0.0001 | CDC42EP3 | −2.833 | ≤0.0001 | ≤0.0001 |
| CEMIP | 2.668 | ≤0.0001 | ≤0.0001 | PSME4 | −2.860 | ≤0.0001 | ≤0.0001 |
| GADD45A | 2.616 | ≤0.0001 | ≤0.0001 | FOXP1 | −3.229 | 2.013 × 10−283 | 8.117 × 10−281 |
| SERPINE2 | 2.045 | ≤0.0001 | ≤0.0001 | GSK3B | −3.275 | 4.903 × 10−290 | 2.314 × 10−287 |
| DCN | 1.906 | ≤0.0001 | ≤0.0001 | SMURF2 | −3.359 | ≤0.0001 | ≤0.0001 |
| COL4A2 | 1.875 | ≤0.0001 | ≤0.0001 | NRG1 | −3.561 | ≤0.0001 | ≤0.0001 |
| TFPI2 | 1.692 | ≤0.0001 | ≤0.0001 | IGF2BP2 | −3.569 | 4.865 × 10−272 | 1.711 × 10−269 |
| PEG10 | 1.689 | ≤0.0001 | ≤0.0001 | SLC38A2 | −4.173 | ≤0.0001 | ≤0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monroe, J.D.; Moolani, S.A.; Irihamye, E.N.; Speed, J.S.; Gibert, Y.; Smith, M.E. RNA-Seq Analysis of Cisplatin and the Monofunctional Platinum(II) Complex, Phenanthriplatin, in A549 Non-Small Cell Lung Cancer and IMR90 Lung Fibroblast Cell Lines. Cells 2020, 9, 2637. https://doi.org/10.3390/cells9122637
Monroe JD, Moolani SA, Irihamye EN, Speed JS, Gibert Y, Smith ME. RNA-Seq Analysis of Cisplatin and the Monofunctional Platinum(II) Complex, Phenanthriplatin, in A549 Non-Small Cell Lung Cancer and IMR90 Lung Fibroblast Cell Lines. Cells. 2020; 9(12):2637. https://doi.org/10.3390/cells9122637
Chicago/Turabian StyleMonroe, Jerry D., Satya A. Moolani, Elvin N. Irihamye, Joshua S. Speed, Yann Gibert, and Michael E. Smith. 2020. "RNA-Seq Analysis of Cisplatin and the Monofunctional Platinum(II) Complex, Phenanthriplatin, in A549 Non-Small Cell Lung Cancer and IMR90 Lung Fibroblast Cell Lines" Cells 9, no. 12: 2637. https://doi.org/10.3390/cells9122637
APA StyleMonroe, J. D., Moolani, S. A., Irihamye, E. N., Speed, J. S., Gibert, Y., & Smith, M. E. (2020). RNA-Seq Analysis of Cisplatin and the Monofunctional Platinum(II) Complex, Phenanthriplatin, in A549 Non-Small Cell Lung Cancer and IMR90 Lung Fibroblast Cell Lines. Cells, 9(12), 2637. https://doi.org/10.3390/cells9122637







