Abstract
Background: Dipterans exhibit a remarkable diversity of chromosome end structures in contrast to the conserved system defined by telomerase and short repeats. Within dipteran families, structure of chromosome termini is usually conserved within genera. With the aim to assess whether or not the evolutionary distance between genera implies chromosome end diversification, this report exploits two representatives of Sciaridae, Rhynchosciara americana, and Trichomegalosphys pubescens. Methods: Probes and plasmid microlibraries obtained by chromosome end microdissection, in situ hybridization, cloning, and sequencing are among the methodological approaches employed in this work. Results: The data argue for the existence of either specific terminal DNA sequences for each chromosome tip in T. pubescens, or sequences common to all chromosome ends but their extension does not allow detection by in situ hybridization. Both sciarid species share terminal sequences that are significantly underrepresented in chromosome ends of T. pubescens. Conclusions: The data suggest an unusual terminal structure in T. pubescens chromosomes compared to other dipterans investigated. A putative, evolutionary process of repetitive DNA expansion that acted differentially to shape chromosome ends of the two flies is also discussed.
Keywords:
diptera; repetitive DNA; Rhynchosciara; Sciaridae; subtelomeres; telomeres; Trichomegalosphys 1. Introduction
Telomeric structure composed of short tandem repeats synthesized by telomerase is broadly conserved across taxa. However, telomerase is not present in Diptera [1], an order which displays unexpected diversity in relation to chromosome ends. Telomeric sequences vary even between species of the same genus and provide an example of telomere diversification. Dipteran telomeric structures diverge depending on DNA repeat types and how they are organized at the very end of the chromosomes. In Drosophila melanogaster, chromosome termini are composed of specific retrotransposons [2,3,4]. In D. virilis, complex terminal satellites, in addition to retrotransposons, were identified at chromosome ends [5,6]. Among lower dipterans (suborder Nematocera), tandem arrays of complex repeats (>120 bp) have been characterized at telomeres of Chironomus species [7,8,9,10,11,12], and in Anopheles gambiae [13,14].
In the nematoceran Rhynchosciara americana, apparently long homopolymer dA/dT tracts [15,16,17,18,19] and reverse transcriptase-related proteins are regularly associated with chromosome ends [18]. These components are uncommon in Diptera and led us to initiate the characterization of terminal sequences in R. americana chromosomes. Complex (414 bp) tandem repeats were first characterized in this sciarid fly that were found to be sub-telomeric rather than truly terminal repeats [20]. A second repeat type named M-22 is unusually short for a repeat tandemly arrayed at the chromosome ends of dipterans. Additional data showed that M-22 tandem repeats lie distal to sub-telomeric repeat arrays [21].
Continued investigation of chromosome ends in R. americana identified other short tandem repeat types. One composed of 16 nucleotides and named M-16 is frequently intermingled within M-22 arrays. The occurrence of segmental duplications, as deduced by sequence analyses involving islands of M-22 and M-16 repeats that appear to reach the very end of chromosomes, might have implications for the process of chromosome end maintenance in this species [22]. T-14 repeats, only 14 bp long, were cloned eventually in R. americana. This is the shortest tandem repeat composing chromosome termini that have been characterized in dipterans to date [23]. M-22, M-16, and T-14 short tandem repeats represent a divergent chromosome end structure compared to long (complex) terminal repeats that have been characterized in chironomids and in Anopheles. D. melanogaster constitutes a third divergent structure composed of specific mobile elements in place of tandem DNA repeats.
By analyzing distinct Rhynchosciara species, the results argue for differential occupation of repetitive sequences at chromosome ends during the evolution of this genus [16,20,21,22,23]. Nevertheless, data restricted to a single genus have not given information on how chromosome ends are structured in other representatives of Sciaridae. Such a limitation also applies to Chironomus and Drosophila, dipterans that have long been studied with a focus on telomeres. Therefore, insights into the chromosome end evolution within dipteran families through the analysis of distinct genera have been lacking.
Taken the above gap into consideration, Trichomegalosphys pubescens was exploited in this work together with Rhynchosciara americana, the only sciarid fly that have been studied with a focus on the telomere to date. Relationships within the family Sciaridae have long been controversial. In an early report, Trichomegalosphys and Rhynchosciara appear in distinct tribes, Megalosphyni and Sciarini respectively [24]. Still considering early classifications, another study placed the two genera in different groups, I and II [25]. More recently, molecular phylogeny as well as cladograms on sciarid relationships have been produced [26] but analyses including T. pubescens and R. americana have not been available yet. Despite the lack of molecular data, genera placed in distinct tribes or even groups strongly suggest that they are significantly divergent.
Results obtained with homologous as well as heterologous probes indicated that R. americana and T. pubescens, while having different chromosome end structures, share DNA sequences that appear to be significantly underrepresented in the latter species. The data also suggest two possible structures for chromosome termini in T. pubescens as well as a putative process that led to the evolutionary divergence between chromosome ends of T. pubescens and R. americana.
2. Materials and Methods
2.1. Animals
Larvae of R. americana were collected in the region of Mongaguá, state of São Paulo, Brazil. Larvae were kept in the laboratory at 18–22 °C. T. pubescens (Diptera: Sciaridae) larvae were collected either on the campus of the University of São Paulo or in the region of Mogi das Cruzes, state of São Paulo Brazil. This species was formerly named Trichosia pubescens [27]. Laboratory cultures were kept at 18–22 °C to a 12/12 h light/dark cycle.
2.2. Preparation of Chromosome Spreads
Salivary glands were dissected in Ringer buffer or 1×PBS and briefly fixed in ethanol-acetic acid (3:1). After squashing the salivary glands in 50% acetic acid, the slides were frozen on dry ice for 10 min. The coverslips were pried off with a razor blade and the slides were then kept in absolute ethanol at −20 °C until microdissection or hybridization procedures. Chromosome spreads of Chironomus riparius (Diptera: Chironomidae) were prepared as a control as described above and were kindly sent by Dr. J.L. Martinez-Guitarte. This species was formerly named Chironomus thummi thummi [28].
2.3. Microdissection
Chromosome ends were microdissected with the aid of microneedles made from solid glass tubes with the aid of a microforge (De Fonbrune). Microdissection was carried out on a Zeiss Axiovert S100 inverted microscope coupled with a manual micro-manipulator (Narishige). The microdissected fragment was recovered from air-dried slides with the tip of a micro-needle that was subsequently broken into a polymerase chain reaction (PCR) tube.
2.4. DOP-PCR
The DNA in the chromosome fragment was amplified by polymerase chain reaction (PCR) using degenerate oligonucleotides (DOP-PCR) [29] in a thermocycler (Mastercycler, Eppendorf), omitting topoisomerase and proteinase K treatments. The following reaction mixture was added to the PCR tube: 0.2 mM of each dNTP; 1× Taq buffer; 2.5 U Taq DNA polymerase (Life Tech, Carlsbad, CA, USA); 1 mM MgCl2; 75 pM DOP primer, H2O to a final volume of 50 µL. Prior to the amplification rounds, the mixture was briefly centrifuged. Cycle conditions for the first amplification were 93 °C for 4 min followed by 8 cycles of 94 °C for 1 min, 30 °C for 90 s, and 72 °C for 3 min. The first amplification was completed with additional 28 cycles of 94 °C for 1 min, 56 °C for 1 min, 72 °C for 3 min. For the second amplification round, 5 µL from the volume of the first amplification reaction were added to 50 µL of a new reaction mixture exactly as described above. Cycle conditions were 93 °C for 4 min followed by 26 cycles of 94 °C for 1 min, 56 °C for 1 min, 72 °C for 3 min, and a final extension at 72 °C for 5 min.
The amplification products were checked in agarose gels before cloning or labelling procedures.
2.5. Cloning and Sequencing of PCR Products
The purified PCR products were ligated in pGEM-T Easy vector according to standard protocols (Promega, Madison, WI, USA). Procedures for transformation and plating of E. coli DH5 strain were described elsewhere [30]. Individual colonies (134) were randomly chosen to grow in liquid medium for plasmid DNA extraction using Concert Rapid Plasmid Miniprep System (Life Tech) to be subsequently analyzed by Southern-blot hybridization using salivary gland DNA of T. pubescens as a probe. Sequencing reactions of both strands of plasmid DNAs were performed with Big Dye Terminator Cycle Sequencing Ready Reaction as recommended by the manufacturer (Applied Biosystems, Foster City, CA, USA) using M13 Forward or Reverse primers (Life Tech) and subsequently run in the ABI PRISM 310 Genetic Analyzer (Applied Biosystems). GenBank searches were done with BLAST [31].
2.6. Southern-Blot Hybridization
Plasmid or genomic DNA were then cut with EcoRI, the digests run in agarose gels prepared according to current protocols [30] and transferred to Hybond N+ or Zeta Probe membranes according to standard procedures (Bio-Rad). Salivary gland DNA (200 ng) and pTp-36 insert (50 ng) were labelled by random priming with 32P-dATP following current protocols (Life Tech). Hybridization was carried out overnight at 60 °C in 0.5 M Na2HPO4, 2% SDS. The membranes were washed twice at 60 °C for 30 min in 40 mM Na2HPO4, 2% SDS. Restriction enzymes and DNA size markers were purchased from New England Biolabs.
2.7. Non-Radioactive Labelling of the Probes
Probe labelling was done with 0.1 mM biotinylated-11 dUTP (Sigma, Burlington, MA, USA) replacing dTTP in 50 µL of PCR reaction mixture as described previously for the second amplification round of DOP-PCR using 2 µL of total DOP-PCR products. PTp-4 plasmid insert was labelled replacing DOPs by M-13 Forward and Reverse primers (Life Tech). A second probe derived from the pTp-4 plasmid was made as described above with one modification. Forward and reverse primers were replaced by two primers (Tp-4a and Tp-4b) flanking the tandem array contained in the insert. Tp-4a: 5′GTTGGAAATAATCTTTAACC3′. Tp4-4b: 5′AATAGTGTTCACTTTCCTTTTC3′. Plasmids (pTp-1, pTp-2, pTp-3) and phage DNA were labelled by nick translation using DIG Nick Translation Mix (Roche, Basel, Switzerland) according to the instructions of the manufacturer.
2.8. In Situ Hybridization
The probe mixture consisted of labelling reaction product (50 µL), 40% formamide, 2X SSC and 0.1% SDS to a final volume of 110 µL. The probe mixture (5–10 µL) was applied on each air-dried slide and covered with a plastic coverslip. The slides were steam heated at 75 °C for 5–10 min and immediately kept in a closed box. Hybridization was carried out overnight at either 32 °C or 37 °C. The slides were then washed in 0.5× SSC, 0.2% SDS at either 32 °C or 37 °C for 30 min, followed by incubation at room temperature in 1× TBS, 0.1% Triton X-100 (TBST) for 10–20 min. For fluorescent detection, goat IgG anti-biotin (Sigma) diluted 1:50 in TBST was applied on the slides for 1 h at room temperature followed by washes in TBST and incubation for 45 min with rhodamine-labelled rabbit IgG anti-goat (Sigma) diluted 1:100 in TBST. Chromosomal DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). For detection with alkalyne phosphatase, slides were incubated with anti-digoxigenin conjugated with alkalyne phosphatase (Roche) diluted 1:100 in TBST for 1 h at room temperature. The slides were then washed in TBST for 10 min and finally in 1× TBS for 5 min. For fluorescence inspection, the slides were mounted in antifading medium (Vectashield, Vector Labs, Burlingame, CA, USA) and inspected under a Nikon Eclipse 80i microscope equipped with epifluorescence optics. Images were captured using the NIS-Elements software package (Nikon, Tokyo, Japan).
2.9. Genomic Library
The T. pubescens genomic library, kindly provided by Dr. E.M.B. Dessen, was prepared with DNA from T. pubescens salivary glands using Lambda EMBL4 as a vector. Procedures for screening, phage DNA extraction, and sub-cloning of restriction fragments were carried out according to usual protocols [30].
3. Results
3.1. Polytene Chromosomes of R. americana and T. pubescens
Chromosome description of the species studied in this work is limited in the literature, particularly for T. pubescens [32]. For this reason, the reintroduction of the polytene chromosomes of the two sciarid species enables comparisons of how evolutionary change shaped the chromosomes of R. americana and T. pubescens. Three of the four polytene chromosomes of R. americana (B, C, and X) display proximal heterochromatin blocks within sections B-15, C-11, and X-12 that hybridize to rDNA probes [33]. Polytene chromosomes of this species have thus three centromeric or pericentric heterochromatic ends and five non-centromeric ends, corresponding to the sections A-1, A-18, B-1, C-1, and X-1 (Figure 1).
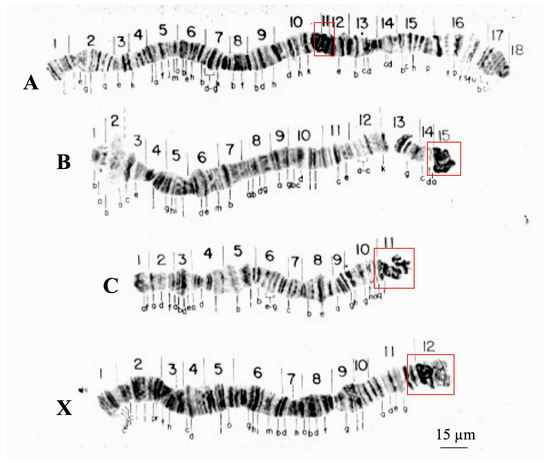
Figure 1.
Polytene chromosomes of R. americana modified from [16]. Boxes (red) are signaling chromosome sections comprising centromeric and pericentric regions. Chromosome nomenclature (A–D) as well as section numbering was described previously [16].
Although T. pubescens also has four polytene chromosomes, there are significant structural differences in relation to Rhynchosciara. T. pubescens chromosomes lack centromeric or pericentric ends as well as conspicuous heterochromatic blocks associated with these regions as seen in Rhynchosciara species. Polytene pericentric regions of T. pubescens are usually identified by chromosome breakpoints as a result of local DNA under-replication (Figure 2).
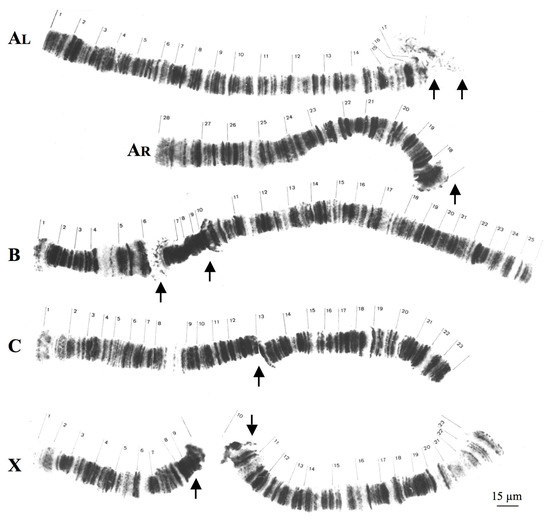
Figure 2.
Polytene chromosomes of T. pubescens modified from [32]. AL and AR correspond respectively to long and short arms of chromosome A. The arrows indicate the most frequent breakpoint regions in each chromosome. Chromosome nomenclature (A–D) as well as section numbering was described previously [32].
In T. pubescens, ribosomal DNA (rDNA) probes hybridized at the mid portion of the chromosome X, very close to the breakpoint of the polytene X chromosome [33]. Hence, centromeres and pericentric regions occupy different positions in chromosomes of Rhynchosciara and T. pubecens in addition to being morphologically distinct.
3.2. Localization of DNA Sequences from the B-1 Tip of R. americana in R. americana Chromosomes
The results of in situ hybridization were obtained with a probe synthesized with DNA from a single B-1 polytene chromosome tip of this species and are presented for the first time. Significant hybridization signals were seen in all telomeres but displaying variable signal intensity at certain chromosome ends. A-18, B-1, and C-1 tips always appeared more intensely labelled than A-1 and X-1 tips. Hybridization was also detected at centromeric ends, pericentric regions of all chromosomes and at several interstitial sites (Figure 3).
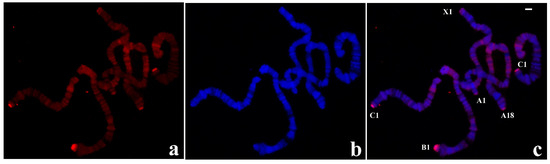
Figure 3.
(a) Localization of the probe (red signal) synthesized with DNA from a microdissected B-1 chromosome end of R. americana in polytene chromosomes of R. americana. (b) The corresponding image stained with DAPI (blue signal) and (c) the merged signals. Non centromeric chromosome ends labelled by the probe were identified. Bar = 20 µm.
Use of individual microdissected polytene chromosome ends other than B-1 has resulted in probes that are able to reproduce essentially the same results as observed in Figure 3, namely the probe from any single chromosome end hybridized to the five non centromeric ends of this species.
3.3. Localization of DNA Sequences from Chromosome Tips of T. pubescens in T. pubescens Chromosomes
The experiments performed in this species were initiated with a probe synthesized by PCR from a single X-1 tip of T. pubescens that was labelled and used for in situ hybridization. In contrast to the results obtained in R. americana chromosomes with the B-1 probe described above, no hybridization signal was seen at chromosome ends other than X-1. Labelling of lower intensity was also observed in chromosome breakpoints and at interstitial sites (Figure 4).
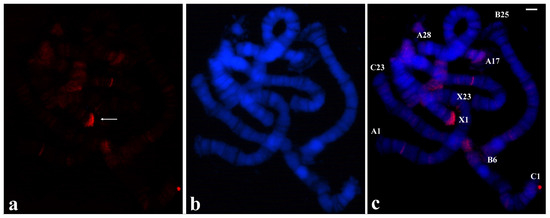
Figure 4.
(a) Polytene chromosomes of T. pubescens after in situ hybridization (red signal) using the probe made of DNA from the microdissected X-1 chromosome end of T. pubescens. The arrow points to the main hybridization signal at the chromosome end X-1. (b) The same chromosomes stained with DAPI and (c) the merged signals. Bar =15 µm.
Given the results in disagreement with what is usually expected, microdissection procedures were repeated in order to discard technical artifacts. The results obtained with probes from distinct chromosome ends reproduced the same hybridization pattern as shown in Figure 4, namely there is a main signal corresponding to the microdissected end but no significant labelling, if any, was seen at other chromosome termini. The possible DNA requirement from more than one microdissected chromosome end was considered as all the T. pubescens probes, as done in R. americana, were made with a single chromosome tip. Use of a probe made of two distinct tips of T. pubescens chromosomes resulted in labelling restricted to two chromosome ends corresponding to microdissected tips used for the probe synthesis (Figure 5).
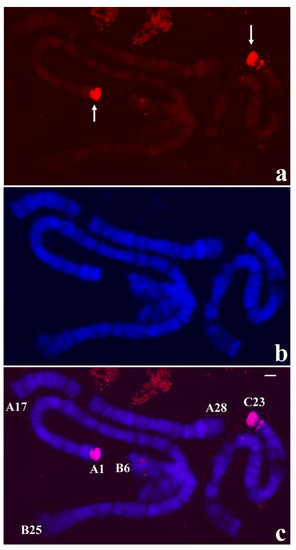
Figure 5.
(a) Localization of the probe (red signal) synthesized with DNA from two microdissected chromosome ends A-1 and C-23 of T. pubescens in polytene chromosomes of T. pubescens. The arrows point to the main hybridization signals. (b) Chromosomes stained with DAPI and (c) the corresponding merged signals. Image superimposition shows chromosome end labelling at A-1 and C-23. Bar = 15 µm.
3.4. Localization of Heterologous Probes in T. pubescens and R. americana Chromosomes
Although T. pubescens and R. americana are distantly related sciarid flies, and in addition probes from microdissection produced disparate hybridization results, the possibility that probes from chromosome tips of T. pubescens could hybridize to R. americana chromosomes was considered. The probe made with A-1 tip DNA of T. pubescens was clearly localized at all non-centromeric ends of R. americana. Significant labelling was also observed in the pericentric heterochromatin and interstitial bands in all the experiments performed (Figure 6).
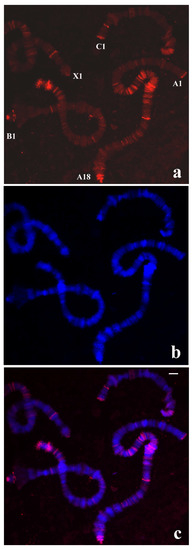
Figure 6.
(a) Localization of the probe (red signal) synthesized with DNA from a microdissected chromosome end A-1 of T. pubescens in polytene chromosomes of R. americana. (b) Chromosomes stained with DAPI and (c) the corresponding merged signals. Chromosome ends labelled by the probe are identified. Bar = 20 µm.
Results obtained with the probe from the X-1 end of T. pubescens on chromosomes of R. americana were essentially those obtained with the A-1 tip probe of T. pubescens although producing less intense signals and also distinct localizations with regard to interstitial chromosome sites (Supplementary Figure S1).
The results described above showed that T. pubescens and R. americana share sequences at chromosome ends as well as in other genomic regions despite divergent labelling patterns in chromosomes of the two species. For this reason, the possibility that the R. americana B-1 probe could hybridize to chromosomes of T. pubescens was tested. In the first attempts, weak labelling was observed in polytene chromosomes of T. pubescens when hybridization and washes were performed at 37 °C. However, when the hybridization temperature and washings was lowered to 32 °C, high intensity signals were detected at B-25 ends. Less intense staining was also observed at interstitial regions of the chromosomes of this species (Figure 7).
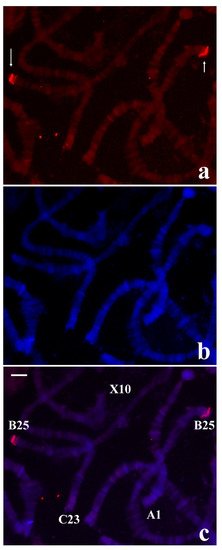
Figure 7.
(a) Localization of the probe (red signal) synthesized with DNA from the microdissected B-1 tip of R. americana in chromosomes of T. pubescens. (b) The same chromosomes stained with DAPI and (c) the corresponding merged signals. The arrows point to the main labelling restricted to chromosome ends B-25. Bar = 15 µm.
The above results raised the possibility that repeats from R. americana chromosome ends characterized previously might be detected in T. pubescens chromosomes under low stringent hybridization and washings. Nevertheless, use of the R. americana sub-telomeric DNA probe [20] and terminal repeats localized distally [21,22,23] did not produce results in T. pubescens chromosomes even by lowering the temperature of hybridization and washings. Controls using R. americana polytene chromosomes showed that the probes used for hybridization were labelled and reproduced in this species the expected results as already published.
3.5. Controls for Microdissection Procedures
Microdissection procedures involving chromosomes of R. americana and T. pubescens were separated by a five-year interval in order to discard the possibility of contamination and always performed with newly sterilized material. When the results led to suspect that either contamination or another methodological problem had occurred, additional microdissections were carried out using a new reagent set. The results of in situ hybridization presented in this work were reproduced in a large number of slides.
Another control was introduced in this work by performing in situ hybridization using a probe obtained by DOP-PCR in chromosomes of Chironomus riparius (Diptera: Chironomidae), a nematoceran that has extensively been studied with a focus on the telomere. Telomeric puffing induced by heat shock is a very rare phenomenon that has been observed in polytene chromosomes of C. riparius [34]. The efficacy of the methods employed in this work was assessed by microdissecting a single terminal puff (TBRIII) of this species for use in DOP-PCR and subsequent probe preparation. The ability of the above procedure, employed for the first time in this species, to amplify telomeric sequences was confirmed since the probe synthesized from a single TBRIII DNA hybridized to the expected seven non-centromeric ends of C. riparius. Interstitial bands and certain pericentromeric regions appeared labelled less intensely than telomeres, probably indicating that non-telomeric DNA sequences were microdissected together with TBRIII DNA (Figure 8).
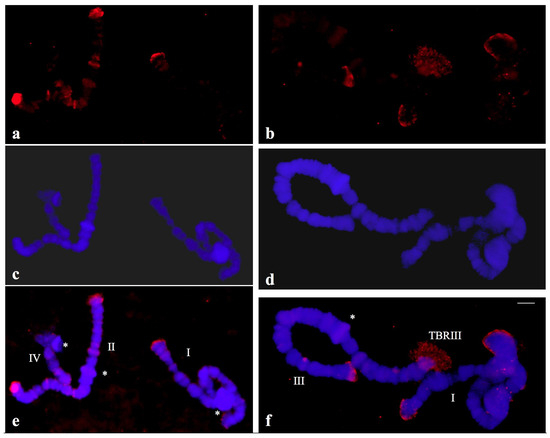
Figure 8.
(a,b) Localization of the probe (red signal) synthesized with DNA from a microdissected telomeric puff (TBRIII) of Chironomus riparius in polytene chromosomes of C. riparius. (c,d) the same chromosomes stained with DAPI and (e,f) the merged hybridization and DAPI signals. The asterisks indicate the centromeric regions of the chromosomes identified individually (I, II, III, IV).
3.6. Screening of a Plasmid Library Constructed with X-1 Tip DNA of T. pubescens
DOP-PCR performed with DNA from the X-1 tip of T. pubescens was used for the construction of a plasmid library. Plasmid DNA from 134 colonies was digested with EcoRI and analyzed by Southern-blot hybridization using salivary gland DNA of T. pubescens as the probe. Controls included ribosomal DNA (rDNA) inserts as references for moderately repetitive sequences. Such a procedure was employed in previous reports [7,8,20,21,22,23,35] and its rationale exploits the genomic representation of repetitive DNA. Total genomic DNA labelled for hybridization is enriched with highly repetitive DNA. Therefore, it would be able to produce, by comparison, to single copy or even middle repetitive DNA, significant hybridization signals in plasmid inserts representing highly repetitive sequences.
No hybridization signal indicating the existence of cloned inserts with highly repetitive sequences was detected in T. pubescens, only two inserts showed signal intensity comparable to middle repetitive DNA. In addition, in situ hybridization results using these short inserts showed no significant signal in polytene chromosomes of either T. pubescens or in R. americana, reinforcing that they do not correspond to repetitive DNA in the genomes of the two species. Inserts that displayed hybridization signals apparently representing middle repetitive DNA together with others that were randomly chosen were sequenced. Most results showed no significant similarity with sequences deposited in data banks except for the two inserts containing features described above (Supplementary Figure S2). The pTp-29 insert is 67% similar to a stretch identified in the genomic chimera named “RaTART” analyzed in detail previously [35], which in turn displays high identity with sequences encoding reverse transcriptase of insect mobile elements.
The second insert, pTp-36, is 97% similar to that found in the pRa-43 insert from the chromosome end of R. americana [21]. The two sequences contain short microssatellite arrays and also a 47 bp repeat unit named M-47 that is not tandemly organized in the R. americana genome. It was characterized as sub-telomeric in a previous work and appeared enriched in chromosome ends A-18 and X-1 of R. americana [21]. The size and sequences of pTp-36 and pRa-43 inserts are not exactly identical, and the two libraries were constructed with a time separation of five years. The micro-libraries of the two species have also never been manipulated simultaneously and the pTp-36 insert hybridized to genomic DNA of T. pubescens during the screening of the microlibrary. An additional control was made by performing Southern-blot hybridization under high stringent conditions using salivary gland DNA of T. pubescens and the pTp-36 as a probe (data not shown). These results confirmed that sequences within the pTp-36 insert are present in the T. pubescens genome.
3.7. Screening of a Genomic Library of T. pubescens with pTp-36 Insert
As the pTp-36 insert was the only cloned DNA that represented a DNA stretch shared by chromosome ends of T. pubescens and R. americana, it was used as a probe to isolate genomic clones from a phage library in an attempt to provide information on the structure of telomeres/sub-telomeres of T. pubescens. Four genomic clones were isolated and produced multiple hybridization signals in chromosomes of T. pubescens (data not shown). Attention was given to the phage 2 insert that was the only genomic clone that also produced hybridization signals in chromosomes of R. americana (Supplementary Figure S3). Phage 2 DNA was cut with restriction enzymes and four fragments were sub-cloned for further analyses.
After sequencing two sub-clones, pTp2.2 and pTp2.3, no significant similarity with sequences deposited in databases was observed. In addition, both sub-clones produced very weak hybridization signals dispersed in all chromosomes of T. pubescens, showing no preferential localization at chromosome ends that were devoid of hybridization signals. Both probes did not produce hybridization signals in chromosomes of R. americana. Therefore, as the two sub-clones did not contribute with information to the aim of this investigation, they were not included as figures.
Hybridization results using the pTp-2.1 were also included in this report since part of its sequence could be identified in databases and, in addition, very clear hybridization results were obtained in polytene chromosomes of T. pubescens, displaying localization in multiple chromosome sites, although no enrichment at chromosome ends was observed (Supplementary Figure S4). Conceptual translation of part of its sequence using BLASTX showed identity with structural proteins from densoviruses (Supplementary Figure S5).
At last, after sequencing the whole pTp-2.4 insert, no similarity with sequences deposited in data banks was found. However, a visual inspection showed a stretch that seemed to contain repetitive DNA (Figure 9a). A manual alignment of sequences composing repeat candidates was made and enabled the identification of a 31 bp repeat unit organized as a tandem array (Figure 9b), named M-31.

Figure 9.
pTp2.4 insert sequence (a); stretches comprising tandem repeats are underlined. (b) The manual alignment was done with repeats taken individually; a single divergence in relation to the most frequent base within a column was identified in red. Gaps (-) were introduced to optimize the alignment.
The pTp-2.4 plasmid was labelled and then used for in situ hybridization in polytene chromosomes of T. pubescens. The most intense hybridization signals were detected in several interstitial chromosome regions namely, no apparent preference for chromosome ends was observed (Figure 10).
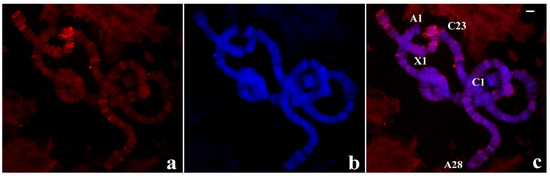
Figure 10.
(a) Localization of the pTp-2.4 probe (red signal) in chromosomes of T. pubescens. (b) The same chromosomes stained with DAPI and (c) the merged hybridization and DAPI signals. Bar = 15 µm.
As the M-31 represents a novel satellite-like sciarid tandem repeat, the probe was also used in chromosomes of R. americana to check its possible occurrence in the genome of the latter species. Interestingly, the probe hybridized to the R. americana chromosomes showing the most intense signals restricted to chromosome ends A-18 and X-1 of this species and also weak labelling dispersed by interstitial sites (Figure 11).
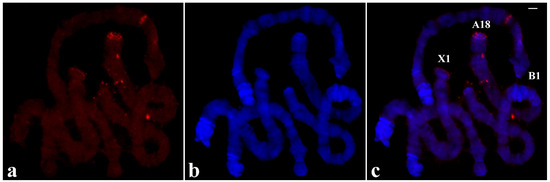
Figure 11.
(a) Localization of the pTp-2.1 probe (red signal) in chromosomes of R.americana. (b) The same chromosomes stained with DAPI and (c) the merged hybridization and DAPI signals. Chromosome tips identified in the figure show the probe labelling restricted to A-18 and X-1. Bar = 15 µm.
In order to discard that the probe signals came from sequences that are not contained in the tandem array of the pTp-2.4 insert, a second probe was made by PCR using the same plasmid, but instead two primers flanking the tandem repeat so that other sequences were excluded from the probe. The hybridization results obtained in polytene chromosomes of the two species were the same as those showed in Figure 10 and Figure 11, confirming that chromosomal location of the probe corresponds specifically to the tandem repeat in the pTp-2.4.
4. Discussion
Given the diversity of sequences composing chromosome termini in Diptera, massive sequencing methods cannot help to provide information on chromosome ends of species in this order. Microdissection followed by cloning procedures have been proven to be useful to this aim and have long been employed in telomere biology of nematocerans [8]. This report represents the first attempt to compare chromosome ends of two genera that belong to the same dipteran family using the procedures described above.
Probes obtained by microdissection of a single chromosome tip of R. americana indicated that sequences are shared by non-centromeric ends of this species. Localization results that were seen in the past, but showed for the first time in this work, allowed the characterization of repeats composing telomeres and sub-telomeres [20,21,22,23] from micro-libraries constructed with microdissected DNA. Tandem DNA repeats were found to be shared by all non-centromeric ends of R. americana. In this sense, R. americana is not an exception since terminal repeats of chironomids were also found to be specific to non-centromeric ends [7,8,9,10,11,12].
Nevertheless, when the same methodological approach was applied to T. pubescens, the results unexpectedly showed that sequences within a given chromosome tip do not seem to be present at other chromosome ends. The data allow to suggest two possibilities for the structure of chromosome termini of this species. The first possibility could be seen as quite unusual and implies specific sequences for each chromosome tip. The second, more conservative possibility argues for the existence of DNA sequences common to all chromosome ends in T. pubescens. However, the short extension of these sequences impedes detection in chromosome ends by in situ hybridization. In any case, the data imply an unusual chromosome end structure T. pubescens in relation to dipteran data available in the literature.
Positive results of heterologous probe hybridization, although showing very different localization patterns, argue for sequence conservation to variable extents within chromosome ends of R. americana and T. pubescens. Since consistent labelling by the T. pubescens probe occurred in non-centromeric ends, pericentric regions and interstitial sites of R. americana, the data strongly suggest in addition that T. pubescens sequences from a single chromosome tip are over-represented in the genome of R. americana. On the other hand, the R. americana B-1 tip probe produced hybridization results that were preferentially detected at B-25 ends of T. pubescens by lowering the hybridization stringency. These data argue that probe and genomic sequences are dissimilar and, in addition, that B-1 and B-25 ends are syntenic. Whether the putative short extension of telomeric DNA in T. pubescens does not allow detection by in situ hybridization as hypothesized above, the synteny suggested would be representative of sub-telomeric DNA that also seems specific for each chromosome end of T. pubescens and conserved to some extent at B-1 tip of R. americana. As homologous and heterologous probes used separately are predominantly located respectively in chromosomes X and B of T. pubescens, these features represent further support for the peculiar structure of chromosome ends in this species.
A comparison with data from the screening of the R. americana library made in the past with a single B-1 tip DNA may give clues about differences in the chromosome end structure of the two sciarid species. Three inserts displaying a high degree of reiteration were found within 54 inserts from the B-1 micro-library of R. americana that were chosen at random [20]. Another insert from the same micro-library (pRa-43) representing moderately repetitive DNA was characterized eventually [21]. On the other hand, no highly repetitive DNA insert was found after screening more than 100 inserts from the T. pubescens microlibrary that were also chosen by chance, only two representing middle repetitive DNA were identified. The data from the two screenings suggest that the representation of repetitive DNA at chromosome ends of T. pubescens is significantly lower than that of terminal regions of R. americana chromosomes. Such an assumption agrees with in situ hybridization results using T. pubescens probes obtained by microdissection and reinforces the divergent telomere/sub-telomere structure in T. pubescens compared to those that have been characterized in flies up to now.
Among the results obtained after screening the genomic library of T. pubescens, the identification of a plasmid insert (pTp-2.1) containing sequences related to densovirus proteins is possibly far from helping in the characterization of sequences composing chromosome ends of T. pubescens. Densoviruses have been described in a number of organisms but insertion of viral DNA in genomes has been seldom reported [36]. In situ hybridization data in T. pubescens using pTp-2.1 as probe were included in this report because they are likely to be the first chromosomal visualization of densovirus DNA insertion in a dipteran genome. The second plasmid of interest from the screening (pTp-2.4) contains a tandem repeat that is a novel minisatelite-like sequence in sciarid genomes. Named M-31, it is the first repeat characterized in T. pubescens and, in addition, the first tandem repeat detected by in situ hybridization in two sciarid genera. M-31 arrays appeared in many chromosome regions of T. pubescens without displaying preferential localization in the genome of this species. In contrast, M-31 repeats are clearly enriched in chromosome ends A-18 and X-1 of R. americana that also correspond to the main location of M-47, sub-telomeric repeats described previously in this species [21]. Interestingly, the M-47 sequence is part of the pTp-36 insert used to screen the genomic library of T. pubescens and made possible the characterization of M-31 repeats.
M-31 arrays were detected in certain chromosome ends of T. pubescens. However, if the hypothesis on short tandem repeats in genomes that are below the detection limits is considered, the occurrence of M-31 arrays at chromosome ends devoid of labelling remains conjectural. Despite this apparent lack of M-31 DNA in some chromosome ends of T. pubescens, the disparate chromosomal localization of M-31 in the two species studied in this work together with the data that have been obtained in this work suggest that T. pubescens and R. americana genomes underwent an evolutionary process that led to divergent chromosome end structures in the two species as hypothesized in the following paragraphs.
Sequences from a single chromosome end of T. pubescens that were found to be over-represented in R. americana indicate a repetitive DNA expansion in the latter species. This process seems to have acted more particularly at chromosome ends and in proximal heterochromatin of R. americana according to the in situ hybridization data. In T. pubescens, localization results using homologous and heterologous chromosome tip probes suggest that repeat expansion has been inactive or may have acted with extremely low intensity during the evolution of this species.
Chromosome end repeats of R. americana, characterized prior to this work, have not been detected by in situ hybridization in T. pubescens chromosomes. The results argue for the absence of these sequences in the T. pubescens genome or, alternatively, for their genomic under-representation, thus impeding visualization by hybridization in chromosomes of this species. Sequencing results from the screening of R. americana and T. pubescens micro-libraries constructed with chromosome end DNA also point to a higher representation of repetitive DNA at chromosome ends of R. americana. These data reinforce preferential repeat expansion at chromosome ends of R. americana.
The pTp-36 insert, from the chromosome end X-1 of T. pubescens, could not be detected by in situ hybridization in its expected genomic location. In contrast, the pRa-43 insert, displaying highly similar sequences and characterized previously in R. americana, was detected at chromosome ends A-18 and X-1 of this species [21]. It is worth mentioning that the pRa-43 insert, whose origin is the microdissected chromosome tip B-1 of R. americana, did not produce hybridization signals at chromosome end B-1 of this species. The data argue for repeat expansion as a requirement for detection by in situ hybridization in the two species studied in this work and moreover converge to the repeat expansion hypothesis in chromosome ends of R. americana.
M-31 repeats characterized in this work display preferential localization in interstitial sites of T. pubescens chromosomes as inferred by in situ hybridization results, not being detected at all chromosome ends of this species. In contrast, M-31 arrays appeared clearly concentrated at chromosome ends A-18 and X-1 of R. americana. The data represent further support for an evolutionary process of repeat expansion, toward accumulating tandem and dispersed DNA repeats at chromosome ends of R. americana.
5. Conclusions
In summary, there are sequences shared by chromosome ends of two distantly related sciarid genera. Part of these sequences may be dissimilar and are differentially represented in both genomes, particularly at chromosome ends of the two species by the action of a putative process expanding locally repetitive DNA in the genome of R. americana. Compared to other organisms displaying non-canonical chromosome ends, the results introduce an unusual terminal structure in T. pubescens chromosomes as synthesized by two hypotheses. One argues for terminal DNA sequences that are specific for each chromosome tip, being absent in other chromosome termini. According to the second hypothesis, there are DNA sequences common to all chromosome ends in T. pubescens but DNA extension limits prevent detection by in situ hybridization. In any case, how the two putative structures work to counterbalance the terminal DNA loss in the absence of telomerase, remains to be elucidated.
By exploiting a single sciarid species, T. pubescens, this investigation indicates that chromosome end diversification may occur in distinct genera within a dipteran family. Flies that have been studied with a focus on chromosome ends are far from representing the organism diversity within this order. To broaden this scenario, basic studies are necessary in order to keep other dipterans in the laboratory, a task that is not always possible. An additional issue that persists is that cytogenetic methods necessary for telomere research cannot always be performed in several species. However, there are genera still unexploited within Drosophilidae, Chironomidae, Culicidae, Psychodidae, and Sciaridae that can be raised in the laboratory and also allow chromosome research. The introduction of new dipteran genera in chromosome end studies may show structures other than those that have already been characterized to date.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/11/2425/s1, Figure S1: In situ hybridization using the X-1 probe in chromosomes of R. americana; Figure S2: Sequences from the X-1 library of T. pubescens; Figure S3: Localization of phage 2 probe in chromosomes of R. americana; Figure S4: Localization of pTp-1 probe in T. pubescens chromosomes; Figure S5. Partial sequence of pTp-2.1 plasmid.
Funding
This research was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), grant 2017/02588-6.
Acknowledgments
I thank J.L. Martinez for providing chromosome spreads of C. riparius, A.J. Stocker for T. pubescens chromosome identification, A.Y. Silvestre and M.E. Cesário for technical assistance.
Conflicts of Interest
The author declares no conflict of interest.
References
- Mason, J.M.; Randall, T.A.; Frydrychová, R. Čapková Telomerase lost? Chromosoma 2015, 125, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Frydrychova, R.C.; Biessmann, H. Drosophila telomeres: An exception providing new insights. BioEssays 2008, 30, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Villasante, A.; De Pablos, B.; Mendez-Lago, M.; Abad, J.P. Telomere maintenance in Drosophila: Rapid transposon evolution at chromosome ends. Cell Cycle 2008, 7, 2134–2138. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, V.; Grozeva, S.; Gokhman, V. Telomere structure in insects: A review. J. Zool. Syst. Evol. Res. 2019, 58, 127–158. [Google Scholar] [CrossRef]
- Biessmann, H.; Zurovcova, M.; Yao, J.G.; Lozovskaya, E.; Walter, M.F. A telomeric satellite in Drosophila virilis and its sibling species. Chromosoma 2000, 109, 372–380. [Google Scholar] [CrossRef]
- Casacuberta, E.; Pardue, M.-L. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc. Natl. Acad. Sci. USA 2003, 100, 3363–3368. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.J.; Morcillo, G.; Galler, R.; Martínez-Salas, E.; De La Campa, A.G.; Diez, J.L.; Edström, J.E. Cloning and molecular characterization of a telomeric sequence from a temperature-induced Balbiani ring. Chromosoma 1985, 92, 108–115. [Google Scholar] [CrossRef]
- Saiga, H.; Edström, J. Long tandem arrays of complex repeat units in Chironomus telomeres. EMBO J. 1985, 4, 799–804. [Google Scholar] [CrossRef]
- Nielsen, L.; Schmidt, E.R.; Edström, J.-E. Subrepeats result from regional DNA sequence conservation in tandem repeats in Chironomus telomeres. J. Mol. Biol. 1990, 216, 577–584. [Google Scholar] [CrossRef]
- Lopez, C.C.; Nielsen, L.; E Edström, J. Terminal long tandem repeats in chromosomes form Chironomus pallidivittatus. Mol. Cell. Biol. 1996, 16, 3285–3290. [Google Scholar] [CrossRef]
- Rosen, M.; Edstrom, J.-E. DNA structures common for chironomid telomeres terminating with complex repeats. Insect Mol. Biol. 2000, 9, 341–347. [Google Scholar] [CrossRef]
- Martinez, J.L.; Edström, J.-E.; Morcillo, G.; Díez, J.L. Telomeres in Chironomus thummi are characterized by different families of complex DNA repeats. Chromosoma 2001, 110, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Biessmann, H.; Donath, J.; Walter, M.F. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol. Biol. 1996, 5, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Biessmann, H.; Kobeski, F.; Walter, M.F.; Kasravi, A.; Roth, C.W. DNA organization and length polymorphism at the 2L telomeric region of Anopheles gambiae. Insect Mol. Biol. 1998, 7, 83–93. [Google Scholar] [CrossRef]
- Jones, K.W.; Bishop, J.O.; Brito-Da-Cunha, A. Complex formation between Poly-r (U) and various chromosomal loci in Rhynchosciara. Chromosoma 1973, 43, 375–390. [Google Scholar] [CrossRef]
- Stocker, A.J.; Gorab, E.; Amabis, J.M.; Lara, F.J.S. A molecular cytogenetic comparison between Rhynchosciara americana and Rhynchosciara hollaenderi (Diptera: Sciaridae). Genome 1993, 36, 831–843. [Google Scholar] [CrossRef]
- Stocker, A.J.; Gorab, E. Local enrichment with homopolymeric (dA/dT) DNA in genomes of some lower dipterans and Drosophila melanogaster. Chromosoma 2003, 111, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Gorab, E. Reverse transcriptase-related proteins in telomeres and in certain chromosomal loci of Rhynchosciara (Diptera: Sciaridae). Chromosoma 2003, 111, 445–454. [Google Scholar] [CrossRef]
- Gorab, E.; Amabis, J.M.; Stocker, A.J.; Drummond, L.; Stollar, B.D. Potential sites of triple-helical nucleic acid formation in chromosomes of Rhynchosciara (Diptera: Sciaridae) and Drosophila melanogaster. Chromosom. Res. 2009, 17, 821–832. [Google Scholar] [CrossRef]
- Madalena, C.R.G.; Gorab, E. A chromosome end satellite of Rhynchosciara americana (Diptera: Sciaridae) resembling nematoceran telomeric repeats. Insect Mol. Biol. 2005, 14, 255–262. [Google Scholar] [CrossRef]
- Rossato, R.M.; Madalena, C.R.G.; Gorab, E. Unusually short tandem repeats in the chromosome end structure of Rhynchosciara (Diptera: Sciaridae). Genetica 2007, 131, 109–116. [Google Scholar] [CrossRef]
- Madalena, C.R.G.; Amabis, J.M.; Gorab, E. Unusually short tandem repeats appear to reach chromosome ends of Rhynchosciara americana (Diptera: Sciaridae). Chromosoma 2010, 119, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Madalena, C.R.G.; Gorab, E. Cloning and characterisation of a novel chromosome end repeat enriched with homopolymeric (dA)/(dT) DNA in Rhynchosciara americana (Diptera: Sciaridae). Chromosom. Res. 2012, 20, 435–445. [Google Scholar] [CrossRef]
- Enderlein, G. Die phyletischen Beziehungen der Lycoriiden (Sciariden) zu den Fungivoriden (Mycetophiliden) und Itonididen (Cecidomyiiden) und ihre systematische Gliederung. Archiv. Naturgesch. 1911, 77, 116–201. [Google Scholar]
- Frey, R. Entwurf einer neuen Klassifikation der Mückenfamilie Sciaridae (Lycoriidae). Notul. Ent. 1942, 22, 5–44. [Google Scholar]
- Shin, S.; Jung, S.; Menzel, F.; Heller, K.; Lee, H.; Lee, S. Molecular phylogeny of black fungus gnats (Diptera: Sciaroidea: Sciaridae) and the evolution of larval habitats. Mol. Phylogenetics Evol. 2013, 66, 833–846. [Google Scholar] [CrossRef]
- Amorim, D.S. A catalogue of the family Sciaridae (Diptera) of the Americas south of the United States. Revta. Bras. Ent. 1992, 36, 55–77. [Google Scholar]
- Michailova, P. The polytene chromosomes and their significance to the systematics of the family Chironomidae, Diptera. Acta Zool. Fennica 1989, 186, 1–107. [Google Scholar]
- Telenius, H.; Carter, N.P.; Bebb, C.E.; Nordenskjöld, M.; Ponder, B.A.; Tunnacliffe, A. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerate primer. Genomics 1992, 13, 718–725. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Altschul, S.F.; Gisg, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Amabis, J.M. The polytene chromosomes of the salivary gland of Trichosia pubescens (Diptera: Sciaridae) Brazil. J. Genet. 1983, 6, 415–424. [Google Scholar]
- Madalena, C.R.G.; Amabis, J.M.; Stocker, A.J.; Gorab, E. The localization of ribosomal DNA in Sciaridae (Diptera: Nematocera) reassessed. Chromosom. Res. 2007, 15, 409–416. [Google Scholar] [CrossRef]
- Díez, J.L.; Martínez-Guitarte, J.L.; Gorab, E.; Morcillo, G. Transcriptional activity at the telomeres of Chironomus (Diptera). Its possible role in the lengthening of the telomeres. In Telomeres; Function, Shortening and Lengthening; Mancini, L., Ed.; Nova Publishers: New York, NY, USA, 2009; pp. 315–331. [Google Scholar]
- Madalena, C.R.G.; Fernandes, T.; Villasante, A.; Gorab, E. Curiously composite structures of a retrotransposon and a complex repeat associated with chromosome ends of Rhynchosciara americana (Diptera: Sciaridae). Chromosom. Res. 2010, 18, 587–598. [Google Scholar] [CrossRef]
- Carlson, J.; Suchman, E.; Buchatsky, L. Densoviruses for Control and Genetic Manipulation of Mosquitoes. Int. Rev. Cytol. 2006, 68, 361–392. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).