Abstract
Heteromorphic sex chromosomes, particularly the ZZ/ZW sex chromosome system of birds and some reptiles, undergo evolutionary dynamics distinct from those of autosomes. The W sex chromosome is a unique karyological member of this heteromorphic pair, which has been extensively studied in snakes to explore the origin, evolution, and genetic diversity of amniote sex chromosomes. The snake W sex chromosome offers a fascinating model system to elucidate ancestral trajectories that have resulted in genetic divergence of amniote sex chromosomes. Although the principal mechanism driving evolution of the amniote sex chromosome remains obscure, an emerging hypothesis, supported by studies of W sex chromosomes of squamate reptiles and snakes, suggests that sex chromosomes share varied genomic blocks across several amniote lineages. This implies the possible split of an ancestral super-sex chromosome via chromosomal rearrangements. We review the major findings pertaining to sex chromosomal profiles in amniotes and discuss the evolution of an ancestral super-sex chromosome by collating recent evidence sourced mainly from the snake W sex chromosome analysis. We highlight the role of repeat-mediated sex chromosome conformation and present a genomic landscape of snake Z and W chromosomes, which reveals the relative abundance of major repeats, and identifies the expansion of certain transposable elements. The latest revolution in chromosomics, i.e., complete telomere-to-telomere assembly, offers mechanistic insights into the evolutionary origin of sex chromosomes.
1. Introduction
A fundamental aspect of the life history of sexually reproducing organisms is the fusion of haploid gametes produced by meiosis through the mechanisms of independent assortment and genetic recombination of chromosomes from two parental genomes to form a zygote. This contributes to the phenotypic diversity at population and species levels arising from natural selection during the course of evolution. Sex determination is the process by which organisms develop as either male or female, and it exhibits remarkable mechanistic diversity and turnover among species. The mechanisms range from environmental sex determination (ESD), where sexes do not differ in genotype, to genotypic sex determination (GSD) resulting from homomorphic to highly differentiated heteromorphic sex chromosomes as either male heterogametic (XX/XY) or female heterogametic (ZZ/ZW) [1,2,3].
Amniotes diverged into two major lineages comprising Synapsida, including all living mammals, and Sauropsida, including all extant non-avian reptilian and avian species, with substantial variation in sex determination mode [4,5]. Sex chromosomes classically evolve from a pair of autosomes (proto sex chromosomes) after one autosome acquires a sex-determining locus [6]. This locus is located on the Y or W sex chromosome and is restricted to a single sex, which affects subsequent processes in the adjacent region as sexually antagonistic genes. To produce a novel allele or gene, a genetic variant must gain control over the sex determination cascade, which is subject to a master sex-determining gene [6,7]. This might have occurred through a point mutation causing gene knockout and loss-of-function, or creating a novel function and regulatory change [8,9,10,11,12,13]. The master sex-determining gene can act in a dominant fashion on the Y or W sex chromosomes, where one copy is needed to determine maleness (on a Y sex chromosome) or femaleness (on a W sex chromosome), such as SRY in mammals or in a dose-dependent manner on the X or Z sex chromosomes, where two functional copies are needed for femaleness (on the X sex chromosome) or maleness (on the Z sex chromosome) [6,7]. Regions around this master sex-determining locus progressively stop recombination with their respective homologous regions on the X/Z counterparts [6,7]. This suppression of recombination might be involved with the selective advantage and preservation of linkage disequilibrium between sex-determining and sexually antagonistic genes, leading to multiple formations of evolutionary strata in sex chromosome evolution and differentiation [6,14,15,16,17,18,19].
Cessation of recombination triggers structural changes, predominantly on the Y or W sex chromosomes, including accumulation of deleterious mutations, degradation of gene content, accumulation of repeats, heterochromatinization, and changes in gene expression [20,21,22,23,24,25,26,27,28,29,30,31,32]. Deleterious mutations might accumulate in the nonrecombining region through Muller’s ratchet or genetic drift, causing Y or W genes to lose their function or disappear altogether [33]. Simultaneously, strong selection acting on the sex-determining region can induce background selection, genetic hitchhiking, and selective sweeps that reduce genetic variability in the adjacent regions [34]. Chromosomal inversions surrounding the master sex-determining region probably occurred on the Y or W chromosomes, thereby preventing chromosome pairing and crossing over with the homologous X- or Z-linked inverted regions, as observed in chicken and Japanese quail [35,36,37,38]. The degree of divergence between X and Y or Z and W sex chromosomes is independently observed across amniote lineages with remarkable variation [39]. Synonymous substitution rates of XY (or ZW) gametologous genes, which are homologous genes located in the nonrecombining region of differentiated sex chromosomes, can be used to trace the evolutionary history of sex chromosomes [14,16,32,40,41,42,43,44,45,46,47,48].
Despite considerable research efforts and recent advances in omic technologies, prediction of the ancestral and transition states between particular sex determination modes and sex chromosomes in amniotes remains uncertain [18,49,50,51]. The ancestral state might be ESD or polygenic sex determination, changing to GSD later on [18,52,53,54,55]. By contrast, it might include the presence of multiple transitions from GSD to ESD. Sex chromosomes evolved independently multiple times within amniotes and remained notably stable after their emergence in mammals, birds, and many lineages of reptiles [16,17,18,56,57]. This scenario is supported by evidence of the same linkage homology blocks that perform the role of sex chromosomes in several amniote lineages, or by unrelated sex chromosomes sharing partial linkage homologies across distantly related groups [18,25,27,28,30,31,32,58]. This was probably caused by multiple random selections from a limited number of linkage homologies, or a stronger tendency for a linkage homology to be co-opted owing to its gene content, particularly as a result of enrichment of the genes involved in gonad differentiation, and the possibility of homologous sex-determining systems [7]. The molecular machinery of the sex determination pathway is observed across ESD amniote lineages, which concurs with independent co-option of the same epigenetic process [59]. Interestingly, recent comparative genomic analyses indicate that the majority of the squamate reptile chromosome 2 (SR2) and the snake W sex chromosomes share partial sex chromosomal linkage homologies with sex-related elements of other amniotes, despite their apparent diversity of sex-determining mechanisms [18,25,27,28,30,31,32,58]. Hypothetically, the SR2 and snake W sex chromosomes may have been part of a larger ancestral amniote super-sex chromosome with a GSD system that subsequently split into many sex chromosomes across several amniote lineages by multiple chromosomal rearrangements such as fission [27,28,30]. This hypothesis suggests an incredible diversity of sex-determining systems, raising questions for many models, including: (i) whether several lineages co-opted the same chromosome pair, or at least parts of them, to function as sex chromosomes?; (ii) whether these co-options resulted from the lack of alternatives, with only a limited number of chromosomes in the ancestral karyotype to form sex chromosomes, or certain unique characteristics of these chromosomes found in SR2 and snake W sex chromosomes, and particularly if the content of genes involved in gonad differentiation predisposed certain chromosomes to become sex chromosomes?; and (iii) what drives some sex chromosomes to be maintained over millions of years and differentiate fully, while others are replaced by new sex-determining chromosomes before differentiation has occurred?
Here, we review evidence pertaining to different sex chromosomal profiles in amniotes obtained from chromosomics and show that correlation with snake W sex chromosomes is a relict of an ancestral super-sex chromosome. Using data sourced from a recent near-complete chromosome-level assembly of the Indian cobra (Naja naja) genome [60], we also report the comparative repeatomic landscape of Z and W chromosomes and highlight the genomic abundance of major repeated elements on sex chromosomes. Evolutionary dynamics of repeat-mediated sex chromosome formation are also discussed.
2. Turnover of Sex Chromosomes in Amniotes
Sex chromosomes carry important sex-determining genes and/or genes that specifically influence male or female fitness, and may have facilitated their recruitment for sex determination [6,7]. Sex chromosome turnover occurs when the existing master sex-determining gene physically moves onto an autosome and retains its control over sex determination [61]. Sex determination systems and/or sex chromosomes have evolved independently numerous times, with frequent turnover from one system to another, exhibiting both inter- and intra-specific variation across many species of amniotes, whereas the X and Y (or Z and W) chromosomes of mammals and birds are conserved [2,40,51,56,58,62,63,64,65,66,67,68]. There are two possible explanations for the emergence of new sex-determining genes and sex chromosomes across amniote species [2,69]. First, when a new sex-determining locus arises on an autosome, it converts the autosome into a ‘proto-sex-chromosome’, and the ancestral sex chromosome reverts to an autosome (Figure 1) [70,71]. Turnover can occur when a new master sex-determining gene arises de novo on an autosome (termed ‘non-homologous turnover’) [61,72,73]. The emergence of a new master sex-determining locus can have very different consequences depending on how it interacts with the previous sex determination system [61,73]. If the new sex-determining locus was associated with a gain in fitness, turnover is more likely to result in different sex chromosomal linkages between species. By contrast, when a new sex-determining gene arises on the existing sex chromosome (termed ‘homologous turnover’) [61,73], turnover between XY and ZW determination systems on the same chromosome arises in the course of evolution. Caenophidian snakes share the same ancestral ZW chromosomes, with varying degrees of W degeneration; however, pythons have an XY system, leading to the emergence of a new sex-determining locus, although only a few specimens have been examined [41,47]. The question of how and why these turnovers arise remains unclear but is assumed to result from sexual conflict, genetic drift, and mutation accumulation [73,74,75,76,77,78,79,80]. Second, autosomes can translocate (by simple translocation, centric fusion, or insertion) to sex chromosomes and create ‘neo-sex-chromosomes’ as observed in stickleback and black muntjac [6,81,82]. In the fusion process between sex chromosomes and autosomes, chromosomes harboring such genes may often be involved in the formation and turnover of sex chromosomes, forming neo-sex-chromosomes [6,83]. It is likely that sex-specific selection pressures (including sexual antagonism) are the primary evolutionary contributors to sex determination pathways, evolutionary turnover in sex chromosomes, and the fixation of neo-sex-chromosomes [18,75,76,84,85,86,87,88,89].
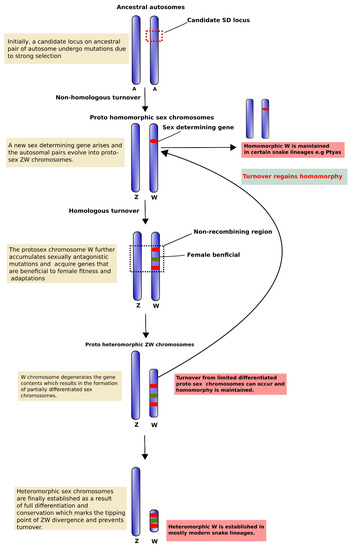
Figure 1.
Schematic diagram of different phases in ZW sex chromosome evolution. We propose a hypothetical evolutionary model to illustrate the origin and evolution of ZW sex chromosomes. First, owing to strong selection of an evolutionary hotspot region, an ancestral autosomal pair undergoes mutation to become a sex determination region, and transformation into homomorphic proto-sex chromosomes. This is followed by heteromorphic differentiation resulting in formation of a proto-W chromosome with cessation of recombination and gain of female beneficial sequences for fitness and adaptation. The proto-W chromosome subsequently undergoes structural changes, such as rearrangements, gene degradation, repeat accumulations, and heterochromatinization, to form a neo-ZW chromosome system with limited differentiation. In some cases, during this stage turnover cycles might convert the partially differentiated heteromorphic sex chromosomes into homomorphic sex chromosomes, as in certain snake species, such as Ptyas species. To achieve full heteromorphy the neo-ZW chromosomes escape this evolutionary trap, and the young W chromosome undergoes severe degeneration with lineage-specific sequence variation and evolves into a mature and stable sex chromosome.
Key questions include ‘why do some lineages maintain and conserve sex chromosome/sex determination?’ and ‘why do other lineages show frequent recurrent turnover?’ The answers might be informed by the mechanism of the ‘evolutionary trap’ hypothesis [54]. Sex chromosomes may undergo cycles of turnover by default unless a tipping point of differentiation is crossed. Sex chromosomes then are stably maintained and fully differentiate, which prevents frequent transition from GSD to TSD or turnover to different GSD systems [40,54]. The transition between GSD and TSD requires traversing a group of fitness-related genes, where individuals are produced carrying suboptimal or lethal WW or YY genotypes. Sex chromosome turnover involves the fixation of a new sex-determining locus in the population, varying the effective population size of the species [73,79,90,91]. By contrast, reptiles possess homomorphic sex chromosomes that appear to be evolutionarily young, owing to frequent turnover [3,92]. The transition from GSD to ESD, as well as turnover of sex chromosomes within GSD, requires an intermediate step of sex reversal, producing individuals with a mismatch between phenotypic and ancestral genotypic sex [51]. Sex-reversed individuals should lack a specialized sex-specific combination of sex chromosomes in lineages with differentiated sex chromosomes and thus show lower fitness. Such sex-reversed amniotes with differentiated sex chromosomes are infertile or possess atypical sex-specific phenotypes [93,94,95]. Homomorphic sex chromosomes are maintained by occasional XY or ZW recombination in sex-reversed XY females or ZW males, known as the ‘fountain of youth’ hypothesis. This is possible if recombination suppression is independent of phenotypic sex assignment [96,97,98]. This might enable escape from the trap and independent evolution in the lineage. Pleurodonts and the sister group corytophanids, a family of iguanian lizards, harbor different partial sex chromosomal linkage groups within each lineage [99,100,101]. However, the tendency for recurrence of sex chromosomal groups might result in homoplasy. New data emerging from non-model sex chromosome systems may provide interesting exceptions to the hypothesis on how sex chromosomes originate and evolve, and suggest diversity in the process not previously acknowledged. Systematic differences between amniote lineages and their frequency of acquisition of stable sex chromosome/sex determination require further investigation to obtain more conclusive evidence.
3. Sex Chromosomal Linkage Homology in Relation to SR2 and Snake W Sex Chromosome
Comparison of genome assemblies and chromosome maps among amniotes have revealed a high degree of linkage homology and elucidated the process of chromosomal rearrangement over millions of years [20,21,25,27,28,30,102,103,104,105,106,107,108,109,110,111,112,113,114]. In a few cases, sex chromosomes share homology among some amniote lineages; however, genome sequence analyses and cross-species chromosome mapping have revealed that unrelated sex chromosomes share linkage homologies across distantly related taxa, and might involve genomic regions orthologous to SR2 and the snake W sex chromosome [18,25,27,28,30,58,60]. These overlaps of partial sex chromosomal linkage homology may be part of a hypothetical ancestral super-sex chromosome (Box 1). Portions of an ancestral super-sex chromosome probably exist in amniotes, with multiple chromosomal rearrangements, such as fissions or insertions, as evolutionary sources of various sex chromosomal linkages [18,25,27,28,30,31,32,58,115]. Under this concept of a super-sex chromosome, sex-specific nonrecombining regions of Y or W sex chromosomes are likely to be ‘super-segments’, enabling adaptation to sex-specific functions or sex-biased expression [33,116]. Co-localization of these genes/regions might reflect the co-option of particularly favorable genes/segments. The same sex-determining genes, as orthologous or paralogous states, have been used repeatedly in distantly related amniotes, such as DMRT1, SOX3, or AMH in a sex-determining function [16,117]. Particular linkage homology is often associated with sexual development in distantly related amniote lineages because of genetic hitchhiking [118].
Alternatively, the occurrence of a super-sex chromosome might reflect group sex-determining gene interactions. The majority of sex-determining genes in one species are part of the conserved sex-determining network in all amniote lineages. Several genes have been independently recruited to the first step of the sex-determining pathway in different species, where each is probably necessary and sufficient for sex determination [2,119]. Their physical proximity may facilitate biochemical interaction of the products of these genes to bring about sex determination. In some cases, such as Drosophila, sex is determined according to the ratio of X chromosomes to autosome sets. Key genes involved in the sex-determining pathway include Sxl, tra, and dsx, and expression of these genes is regulated by several transcription factors encoded on the X chromosome [120,121,122]. Another plausible hypothesis concerns chromosome territory, which could make their physical translocation more likely, as in the case of translocations between chromosomes bearing nucleolus organizing regions (NORs) [123]. Chromosomes occupy highly conserved territories in somatic cells of mammals, birds [124,125,126], and mammalian germ cells [127]. The positions of these territories are associated with the gene content of chromosomes; sex chromosomes with a low gene density are more frequently located at the periphery [128,129]. In addition to several models for the origin of a super-sex chromosome, an underlying principle of sex determination in amniote lineages is the sharing of linkage homology. Sequences such as repeats were once linked in a super-sex chromosome that was broken up by different means. Many changes to genomic content occur once an autosomal pair becomes a sex chromosome pair. As well as deletion, selection of sex-specific traits on the sex-specific chromosome (Y or W) and changes in the genomic content of the partner sex chromosome (X or Z) reflect the hemizygous state in one sex [6,130,131]. Particular sequences, such as 18S–28S ribosomal RNA genes, may play roles in sex chromosome regulation or create novel sex chromosomes [18,58,132]. Opossum and kangaroo sex chromosome pairs have independently fused with a segment carrying the NOR, whereas platypus sex chromosomes are frequently arrayed around the nucleolus during meiosis, which brings them into close proximity to the NOR-bearing chromosome 6 that shows homology with the human X-conserved region [133,134]. Proximity to the site of RNA synthesis might facilitate epigenetic processes involving long noncoding RNAs [135]. Minimally differentiated XY chromosomes are observed in three cryptodiran turtles (Staurotypus crassicollis, S. triporcatus, and S. salvinii), in which the Y chromosomes are smaller than the X chromosomes owing to a difference in copy number of 18S–28S rRNA genes [136,137]. SR2 is highly conserved among squamate reptiles [58,102,103,132,138], and NORs are generally located on a pair of microchromosomes or chromosome 2 in iguanas and some snakes [139,140]. The NORs are located on the ZW microchromosomes in the bearded dragon (Pogona vitticeps), which share a common ancestry with SR2 [3,58,109,115,132,141].
From a different perspective, recent studies of many amniotes have revealed a striking difference of the gene and repeat content of their Y/W sex chromosomes, with substantial disparity even between closely related species [16,17,47,51,142,143,144,145,146,147,148,149,150]. This is despite the prolonged stability of sex determination systems in these lineages and the extensive between-species homology of their X/Z-specific gene contents [17,42,151]. However, genomic regions of snake W sex chromosomes show substantial homology with sex chromosomal linkage homologies and repeat content in amniotes [25,27,28]. Singchat et al. [25,27,28] asserted that 16 bacterial artificial chromosomes (BACs) showing partial homology with sex chromosomes of several amniotes were mapped on the heterochromatic W sex chromosomes of different species, including Siamese cobra (Naja kaouthia), Russell’s viper (Daboia russelii), and the common tiger snake (Telescopus semiannulatus), based on hybridization signals such as repeats. In comparison, two chicken BACs located on Gallus gallus chromosome Z (GGAZ) that showed a high abundance of the long interspersed nuclear element (LINE) and long terminal repeat (LTR) transposable elements (TEs) were mapped on SR2 and the snake W sex chromosome [25,27,28]. This suggests that repeats on the snake W sex chromosome also share sex chromosomal linkage homology to SR2 and GGAZ, and the snake W sex chromosome might include a genomic region involving sex chromosome conformation in amniotes. The Y or W sex chromosomes showing accumulation of satellites and amplification of telomeric or microsatellite repeats [(GATA)n, (AAGG)n, (AATC)n, and (ACAG)n] are commonly observed in snake W chromosomes and in other amniotes [20,22,23,24,25,29,115,152,153,154,155]. One microsatellite amplified on the W sex chromosome in several caenophidian snakes is the banded krait minor satellite (Bkm), which consists of a microsatellite repeat motif (AGAT)n or (GACA)n sequence, and is associated with the degree of ZW differentiation [156]. This might result from rapid and independent amplification of repeats on W sex chromosomes, and suggests that frequent amplification of the repeats has a structural role in heterochromatinization and promotes further sex chromosome differentiation [25,27,28,29,30]. Amplification of repeats has occurred independently in each lineage and might represent convergent sex chromosomal differentiation among amniotes [18,25,30]. Interestingly, bird and snake W sex chromosomes share blocks of three repeats (Bkm repeats, 18S–28S rRNA-related repeats, and DMRT-related repeats) [23]. This suggests that repeats are shared partially between the sex chromosomes of chicken and snakes, and supports the hypothesis that SR2 and the snake W sex chromosome were associated with a larger ancestral amniote super-sex chromosome [18,25,27,28,30,58]. Many studies have identified convergent genomic patterns in independently formed sex chromosomes [34,157], and causes of the repeated origins of these unique regions of the genome have been suggested [6]. Amplified repeats were possibly retained in the sex chromosomes of an amniote common ancestor, and subsequent reshuffling led to the appearance of sex chromosomes in each lineage. Convergent evolution of sex chromosomes across distantly related taxa might lead to genomic elements, such as repeats, which are particularly adept in a sex-determining role. However, the majority of repeats or genomic regions are more likely to be associated with snake W sex chromosomes [25,27,28]. Most of these explored orthologous regions have been cytogenetically mapped to better understand candidate sequences such as BACs or other repeats; however, further chromosomic level studies will elucidate the possible occurrence of linked genes in shared chromosomal regions [30,31,32,49].
4. Repeats: A Driver for Sex Chromosome Conformation after the Split of an Ancestral Amniote Super-Sex Chromosome
During the process of sex chromosome differentiation, heterochromatin is enriched at repeats (TEs and satellites), and its loss can result in de-repression and mobilization of silenced TEs. The number of repeats can differ substantially between sexes owing to the presence of a highly repeated (and normally poorly assembled) Y or W sex chromosome in the heterogametic sex individual [29,92,155,158]. Transposable elements are located at the boundaries of recombining and nonrecombining regions, which suggests their causal role [159,160]. Insertion of TEs near the sex-determining locus can act to suppress recombination by creating a divergence between sex chromosomes, and TEs are often assumed to accumulate following suppression of recombination. This would invoke host mechanisms to silence TEs, resulting in suppressed recombination at hotspots adjacent to TE insertions [161]. Weaker selection against the insertion of additional TEs leads to their accumulation under a lack of recombination. Moreover, TEs can promote ectopic recombination, facilitating genomic rearrangement to further suppress recombination [162]. The heterochromatic regions in amniotes are also predominantly accumulated by satellite DNA, in a class of repeats characterized by a tandem arrangement with highly repeated monomeric units longer than 100 bp, or simple repeats, such as mini- (>10 and <100 bp) and microsatellites (usually <10 bp) [163,164]. These satellites are often abundant on sex chromosomes in amniotes [25,27,28,29,30,115,165,166,167,168]. Lacertid lizards have highly differentiated ZZ/ZW sex chromosomes, and the W sex chromosome is indicated to be enriched in satellite motifs in Acanthodactylus lineomaculatus, Eremias velox, and several species from the genera Lacerta and Timon [169,170,171,172,173,174]. The primary function of the satellites is unknown; however, they may contribute to the suppression of recombination, heterochromatinization, and changes in gene expression. Different types of these repeats are randomly accumulated on sex chromosomes and largely reflect historical contingency [175,176,177,178]. The important functional role of such sequences implies that the pattern of the distribution of their accumulation should be relatively well conserved across species of the same lineage. In snake, PBI-DdeI (196 bp) satellite DNA is located in the centromeric region of the Burmese python [168]. Interestingly, PBI-DdeI satellites are frequently localized to the W sex chromosome of Siamese cobra. Localization of high copy numbers in female rather than male individuals suggests that PBI-DdeI might act as an evolutionary driver with several repeats, facilitating W sex chromosome differentiation and heterochromatinization [29]. Transposable elements and satellites may play a critical role in the early stages of recombination suppression, with the ability to shuffle genes and alter expression patterns. Repeats may simultaneously promote the turnover of sex chromosomes and sex-determining genes, initiating suppression of recombination, chromosomal rearrangements, and eventual recruitment of sex chromosomes [38].
A well-known example is the genome of the Indian cobra, which is closely related to the Siamese cobra [179]. This genome encompasses a total size of 1.79 Gb with W and Z chromosomes spanning 52.1 Mb and 154.6 Mb, respectively [60]. This shows that the Z sex chromosome is almost three times larger than the W sex chromosome in genomic content. Ideally, the genes occurring on the W sex chromosome might play a different role in determining female-associated phenotypes, while changing female-biased selective forces might strongly affect the evolution of the W sex chromosome [180]. Although several predictions have been proposed, the W sex chromosome’s functional role is unknown, except for a few W-linked genes that have been studied mostly in birds [180]. Here, we present a functional view of the W sex chromosome of Indian cobra to corroborate the hypothesis that the W sex chromosome might be involved in multiple functions of cellular processes in addition to sex determination. The W sex chromosome harbors a diverse set of genes and, based on a gene ontology enrichment analysis (Supplementary Note 1), the W sex chromosome is enriched with genes coding for brain development, microtubule organization, histone deacetylation, DNA repair, signaling, and transport (Figure 2 and Supplementary Dataset 1). In addition, the repeat contents of Indian cobra sex chromosomes are presented (see Supplementary Note 1). The comparative repeatomic landscape of the highly repeated Indian cobra Z and W assembled chromosomes showed remarkable differences between TE abundance and the overall higher enrichment of repeats; with total repeats of 22.57% on Z chromosomes compared with 15.39% on W chromosomes. A similar pattern might be observed in other snakes with a smaller W sex chromosome [181]. The majority of TEs such as retroelements (LINEs and short interspersed nuclear elements) and DNA transposons are highly abundant on the Z sex chromosome. The most abundant elements are L2/CR1/Rex, which constitute 9.99% and 7.44% of Z and W sex chromosomes, respectively, with certain elements (CRE/SLACS, PiggyBac, Mirage, and P-elements) completely absent on the W sex chromosome (Figure 3 and Table 1). Did the Z sex chromosome become larger as the result of insertions of specific repeat elements, or did the W sex chromosome experience depletion of these elements? Chromosome mapping has indicated a high accumulation of repeats as telomeric repeats, microsatellites, satellites, and TEs on the snake W sex chromosome, with none or fewer on the Z sex chromosome [25,27,28,29]. Chicken BAC sequences mapped on the snake W sex chromosome show nonhomology to the Indian cobra W sex chromosome-level genome assembly [25,27,28,60]. Comparative repeatomic analysis supports our hypothesis that the Z sex chromosome of Indian cobra might have experienced a recent explosion of TEs that could have contributed to further gain in genetic contents. The Kimura substitution landscape TE model also indicated that the W sex chromosome contained many ancient/degenerated copies of LINEs, whereas the Z sex chromosome accumulated many recent or less divergent copies of these elements with two peaks/rounds of TE insertions (Figure 3). Collectively, these results suggest that repeats on the snake W sex chromosome might be inherited from an ancestral amniote sex chromosome with high differentiation via nonhomologous recombination, which has resulted in the evolution of heteromorphic Z and W sex chromosomes in advanced snakes [30,32]. This finding agrees with the results of BAC fluorescent in situ hybridization (FISH) mapping on the Siamese cobra, Russell’s viper, and common tiger snake [25,27,28]. These TEs may have undergone molecular degeneration, rending their identities senescent in the Indian cobra genome. A time estimation model and molecular evolutionary analyses of TE insertions could further advance our understanding and solve the complex issue of whether a recent new explosion of TEs occurred in the Indian cobra genome. Comparison of Siamese cobra and Indian cobra genomes would provide further insights into the possible occurrence of repeated elements in closely related species, and allow in-depth comparisons of repeat element density and distribution of autosomes versus sex chromosomes.
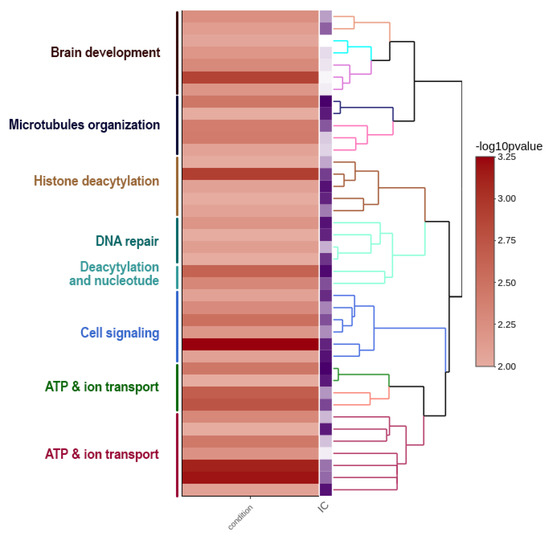
Figure 2.
Gene ontology (GO) enrichment of annotated genes on the W chromosome of Indian cobra. Clustering heatmap plot with log10 (p-value) from functional enrichment tests and information content (IC). A higher log10 (p-value) represents a more enriched function. The results show that W chromosomes carry an enriched set of genes associated with development, histone deacetylation, signaling, and transport.
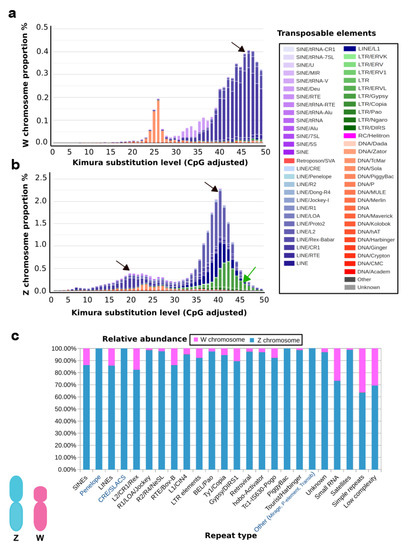
Figure 3.
Comparative genomic characterization of repeated DNA contents between the W and Z sex chromosomes. Repeat landscape of (a) W and (b) Z sex chromosomes. Histogram plots show the degree of sequence divergence of each transposable element (TE) derived from its consensus (X-axis) in relation to the percentage of its copies in the total genetic contents of the chromosome (Y-axis). Peaks represent waves of insertion (black arrows) of elements into the sex chromosome. Older insertions of TEs are shown as a peak wave on the right side (K-value > 25) of the plot, whereas younger elements are depicted on the left side (K-value < 25). Different colors show distinct element types, as described on the right. A higher abundance of LTRs (green) of the Z sex chromosome landscape as indicated by a green arrow, is evident. The Y-axis percentage difference and a recent wave of expansion on the Z chromosome are evident. (c) Comparative analysis of Z and W localized repeat contents. Each column represents the copy number percentage stacked for the repeated element. Different proportions for Z and W sex chromosomes are indicated in blue and red, respectively. Elements with higher proportions (in blue) show expansion; those TEs present exclusively on Z chromosomes, and absent on W chromosomes are highlighted in blue text on the X-axis.

Table 1.
Comparative list of repeat contents localized on the Indian cobra Z and W sex chromosomes and the relative abundance of each identified element. Note: For certain TEs (PiggyBac and Mirage, P-element), the percentage was considered negligible and rounded to 0 (less than 0.01%).
5. Diversity and Stability of Snake Sex Chromosomes
Snakes represent about one-third of all reptilian species, with almost 3800 extant species classified into three major lineages: the Caenophidia, the likely paraphyletic Scolecophidia, and Henophidia [182,183,184,185]. Caenophidia is the most species-rich and diverse group, including more than 3100 species [186]. Scolecophidia contains approximately 400 species of blind snakes with a worm-like body shape and fossorial lifestyle, and Henophidian snakes comprise about 200 species [186]. The largest family is Colubridae, commonly termed colubrids [187,188], and is the most extensively studied for cytogenetic investigation. We observe that colubrids exhibit higher variability in chromosome number and genome size compared with those of other snake families, such as Boidae and Viperidae. (Figure 4). This high degree of variation might have contributed to the remarkable diversity and speciation of colubrids. The diploid chromosome number across all snakes is 2n = 24–56 [152,189]. Variation involving macro- and microchromosome numbers have been reported in different families across snake lineages [152,189]. However, phylogenetic reconstruction reveals that the ancestral snake karyotype consisted of 2n = 36 chromosomes with 16 macro- and 20 microchromosomes. This is the karyotype commonly observed in the majority of snake species [189]. Female heterogamety (ZZ/ZW system) occurs in caenophidian snakes [20,23,24,25,27,28,41,44,45,46], whereas for noncaenophidian snakes (i) facultative parthenogenesis in pythons and boas leads to exclusively female progeny [190,191,192,193], (ii) inheritance of a color mutation in the ball python (Python regius) indicates a XX/XY sex determination [194], and (iii) a recent study suggests that a transition from ZW to XY may have occurred for Boa imperator and Python bivittatus based on male-specific genetic markers as well as transcriptomic and genomic data [41]. A report of heteromorphic ZZ/ZW sex chromosomes in the Madagascar boa based on conventional cytogenetics was recently confirmed by molecular cytogenetic methods in Acrantophis sp. cf. dumerili [47,195]; however, the sex chromosomes of many snake species remain undifferentiated, with no large morphological changes (such as Boidae and Phytonidae), and a low degree of differentiation between Z and W or X and Y sex chromosomes [41,47,196,197]. In comparison to the long-term stability of the Z chromosome across all snakes, the sex determination systems in noncaenophidian snakes are likely far less stable and more dynamic [47,196,197]. A recent study showed that the scolecophidian long-nosed worm snake (Myriopholis macrorhyncha) may have heteromorphic ZZ/ZW sex chromosomes, which are likely nonhomologous to sex chromosomes of caenophidian snakes [198]. The Z sex chromosome is indicated to share the same gene content across caenophidian snakes [20,153,199], without large morphological modifications [200]. By contrast, several repeats are known to be the primary source of differentiation of W sex chromosomes in caenophidian snakes [24,25,27,28,29], with highly degenerated and heterochromatic accumulations of repeats, and variable topology and degree of accumulation among species [181,201]. Notwithstanding conventional and molecular cytogenetic approaches, the snake W sex chromosome can remain undetectable for genomic content based on recent omic technology, except for the Indian cobra for which partial information on sex chromosomes is detectable [60]. The study of the Indian cobra genome provides an overall view of evolution by focusing on comparative genomics, thereby unlocking the diversity of toxin genes that lack in-depth sex chromosome investigation.
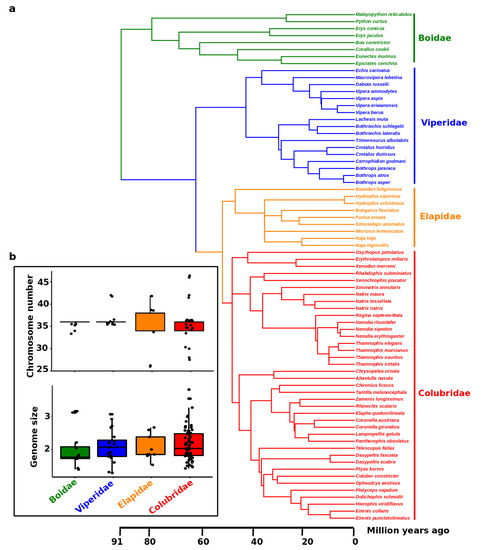
Figure 4.
(a) Phylogeny of 108 snake species, with available data for karyotypes and genome size, belonging to the families Boidae, Viperidae, Elapidae, and Colubridae. (b) Boxplots show the distribution of chromosome number and genome size (C-value) for the four families. Each dot represents the species as given in the phylogeny. The phylogenetic tree was sourced from TimeTree databases (http://www.timetree.org) [202] and shows each species with information on chromosome number (2n) and genome size variation. Data were sourced from the Animal Genome Size Database (http://www.genomesize.com) [203].
6. Chromosomics of Snake W Sex Chromosomes: Bridging the Gap between Genomes and Chromosomes
Snakes have unique genomic features that make them particularly interesting to study. The sequencing of snake genomes is increasing our understanding of their molecular evolution and genetic diversity. Evolutionary studies of venomous organisms provide sources of medical information to catalog venom proteins for drug and antivenom development. A decade ago, it was a major feat to sequence the first snake genome [204]. Subsequent advances in sequencing technology have made the sequencing of many more snake genomes attainable [60]. Technologies based on genome sequencing have the potential to resolve profiles of genetic differences between sexes at the nucleotide level, making it possible to reveal sex-specific loci or sex-specific genes in species where these had not previously been identified [40,44,45,46,205,206]. Recent advances in genome sequencing technology have assisted in the assembly of heterochromatic and/or low-complexity genomic regions, such as centromeres and differentiated W sex chromosomes. Current high-throughput sequencing methodologies and bioinformatic tools have replaced conventional molecular biological investigation techniques [29,60]. We currently have limited knowledge of whether the between-species variability of the snake W genomic content and repeat content is exceptional [25,27,28,29,30], as research to date among amniotes have been restricted predominantly to a small number of studied lineages, or whether it is common during sex chromosome differentiation.
Physical anchoring of chromosome sequences is required to validate a chromosome-level assembly. Once chromosome-level assemblies have been achieved for a greater number of snake species, investigation of changes in the packaging and interactions between chromosomes will contribute to an understanding of the role genome architecture has played during snake and amniote sex chromosome evolution. Technological advances in genomic sequencing, particularly long-read (PacBio; [207,208] and ultra-long-read (Oxford NanoPore Technologies; [209]) sequencing platforms, have provided exceptional improvement in scaffold sizes of genome assemblies. A combination of short- and long-read sequencing can provide chromosome-scale descriptions of repeat landscapes of sex chromosomes using all available genome sequence data from snakes [60,210], but currently available data for snakes is taxonomically limited and elucidation of the basic molecular machinery across snake lineages is required. Despite the improvement in long-read sequencing over short reads in genome assembly, new genome sequences often fail to produce ‘chromosome-level’ assemblies, where contigs represent a complete chromosome. In this regard, advances in cytogenetics and innovations in sequencing technology are useful for providing higher-resolution genome assemblies, and will be important for implementation in snake research moving forward. Without chromosome-level assemblies, the ability critically to examine evolutionary questions, including basic questions surrounding genome evolution and function, as well as adaptation and speciation, is limited [30,49,211,212]. Cytogenetic approaches, such as FISH, will help to generate physical maps to confirm experimentally the correct orientation of scaffolded genome sequences scaled into chromosomes. Such techniques will also enable the analysis of breakpoints and gene order, positioning of centromeric and telomeric sequences, and structural variation. A novel approach was developed in birds, where a set of chicken BAC clones was bioinformatically identified and empirically validated as a set of universal avian probes to anchor sequence scaffolds rapidly to chromosomes of sequenced species [110,213]. In this manner, it would be possible to anchor the increasing number of snake or other amniote genomes being sequenced. Appropriate samples can be obtained for the preparation of chromosomes [25,27,28], and these probe sets will provide high-resolution sequence arrangements on chromosomes. Some of the technical difficulties of FISH mapping include low-throughput data, limited microscopic resolution, and probe specificity for each analysis; in addition, examination of multiple loci simultaneously is technically challenging. Development of high-throughput, next-generation chromosome conformation capture technologies, such as the Hi-C approach [214], will provide more information on interactions and chromosomal conformations in three-dimensional genomic structures. Integration with cytogenetics mapping will allow orientation of the assembled contigs into chromosomes [215]. Bionano optical-mapping technology can also be used to acquire long-range data throughout the genome, which are highly suited to filling gaps and improving fragmented genomes in ways that are not possible using classical cytogenetics [216]. Despite substantial advances in the aforementioned technologies, no single genome has been completed with end-to-end chromosome assembly. Assembling the X chromosome telomere-to-telomere will resolve many gaps and long arrays of complex repetitive regions in the human genome using high-coverage ultra-long (Oxford nanopore) reads complemented with optical mapping [217]. This achievement will revolutionize the field of chromosomics, and high-resolution data produced from combinations of these approaches will elucidate further novelties regarding evolution of sex chromosomes [218,219].
7. Concluding Remarks
Sex chromosomes were discovered by Nettie Stevens in 1905. She observed that in mealworms, male cells carried chromosomes that were smaller than the rest, whereas female cells carried equally sized chromosomes [220]. The availability of genomic data for many nonmodel species, and the development of methods to detect sex-linked sequences in species with both differentiated and undifferentiated sex chromosomes, have provided a global overview of the diversity of sex-determining systems in amniotes. Recent progress on the evolution of sex chromosomes in several amniotes has supported long-standing hypotheses and, for many other amniotes, has revealed that there is no single narrative for how these regions form and evolve. Sex chromosomes show convergent genomic signatures, suggesting broader trends in their formation. The hypothesized scenario of a super-sex chromosome in the ancestral state of amniotes is followed by multiple fission to form products in the evolutionary lineages. Unpredictably, the snake W sex chromosome shows the remnants of sex chromosomal linkage homology shared among amniotes, as well as large abundances of satellites and TEs. The snake W sex chromosome may retain the most ancestral state from an ancestral super-sex chromosome in amniotes. Homologous sex chromosome turnover might occur in small clades under pressure of selection. However, the diversity of sex chromosomes reveals a remarkable number of exceptions and, therefore, a parallel diversity of underlying mechanisms. The evolutionary trap is another potential hypothesis, followed by turnover of homologous sex chromosomes in species with homomorphic sex chromosomes. Nonhomologous turnover could alternatively maintain ancestral heterogamety (that is, XY to XY transitions, or ZW to ZW), or induce a shift in heterogamety (XY to ZW, or vice versa). Although the incredible diversity of sex chromosomes and sex-determining systems has been revealed, much less progress has been achieved in understanding the evolutionary forces that have shaped this diversity. Studying ongoing or extremely recent turnovers and the possibility of a super-sex chromosome is therefore required to elucidate the causal turnover mechanisms further. A deeper characterization of sex determination in clades, where both homomorphic and heteromorphic sex chromosomes are present, will help to determine the differentiation and conservation of sex chromosomes. Studies of snake sex chromosomes of recent origin may also provide data on the formative processes, although such studies are extremely difficult given that divergence between sex chromosomes is slight. Snakes are an excellent model with which to examine hypotheses of sex chromosome evolution, which can occur rapidly; thus, population-based approaches are useful for understanding the mechanisms and patterns involved. Cytogenetic studies have presented the first glimpses of ancestral amniote super-sex evolution; however, integration of multiple NGS platforms is required to attain an in-depth understanding. The novel procedure of telomere-to-telomere assembly will further enable the mechanisms involved in reshaping sex chromosome evolution to be deciphered.
Box 1. What is the super-sex chromosome hypothesis? What can we learn from snake sex chromosomes?
The term “super-sex chromosome” was first proposed by Ezaz et al. [18] to express an ancestral donor source of sequences contributing to the evolutionary diversification of amniote sex chromosomes. The hypothesis of the existence of an ancient super-sex chromosome emerged from the results of extensive cytogenetic studies carried out in several amniote species, as a means of understanding the organization of candidate BACs and repeated element sequences mapped on sex chromosomes [25,28,30,58]. This hypothesis has also been postulated in genome-wide SNP studies to identify sex-specific regions [31,32] to suggest that the sex chromosomes of diverse amniote lineages exhibit sequence homology, and that a homologous super-sex portion might exist on an ancient super-sex chromosome that experienced several rearrangements including multiple fissions and repeat element insertions [28,30]. Snake sex chromosomes offer an excellent model, exhibiting a ZZ/ZW sex chromosome system, with different phases of evolutionary degeneration or amplification of the W chromosome [20,21,22,23,24,25,28,29,30,32]. To test this hypothesis, we mapped different BAC sequences on the snake W sex chromosome found to be partially homologous to other amniotes [25,28]. Recently, comparative cytogenetic analysis has identified homologies of sex chromosomes across ancestral (Henophidia) and more recent (Caenophidia) snakes [202]. The principal concept of a super-sex chromosome hypothesis is based on cytogenetics mapping; however, whether the evidence of partial homology may be exclusively linked to a super-sex segment remains unclear, with the possibility of random repeated element distribution throughout the genome. The mapped sequences, which form the basis of the original hypothesis, also represent a set of candidate loci that span a very small portion, as a few bp to several kb of the genome, compared to the total genome content of sex chromosomes that spans several megabases in amniotes. Therefore, considering this limitation, we propose to further explore the super-sex chromosome hypothesis using modern tools including genome-wide mapping of whole-sex chromosomes among diverse amniote lineages.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/11/2386/s1, Supplementary note 1: Gene ontology enrichment analysis and comparative repeatomic landscaping of Indian cobra Z and W chromosomes [60,221,222,223], Supplementary dataset 1: Complete list of gene ontology terms together with function descriptions for significantly enriched genes on the W chromosome detected in the enrichment analysis.
Author Contributions
Conceptualization, W.S., S.F.A. and K.S.; methodology and data analysis, W.S., S.F.A. and K.S.; visualization, W.S., S.F.A, T.P. and K.S.; Writing—original draft preparation, W.S., S.F.A. and K.S.; Writing—review and editing, W.S., S.F.A., N.L., A.S., T.P., D.K.G. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by grants from the Center for Advanced Studies in Tropical Natural Resources, National Research University-Kasetsart University (CASTNAR, NRU-KU, Thailand) (nos. 6/2558 and 9/2559) awarded to K.S.; Thailand Research Fund (TRF) (nos. RSA6180075, PHD60I0014, and PHD60I0082) awarded to K.S., W.S. and A.S.; the e-ASIA Joint Research Program (no. P1851131) awarded to K.S. and W.S.; Thailand Research Fund-Newton Fund Ph.D. Placement (no. GA/PhD/scholar/Year5/007) awarded to W.S.; Thailand Research Fund-Newton Fund Placement, Travel Grant for Ph.D. Supervisors (GA/PhD/Sup/Year4/003) awarded to K.S.; Postdoctoral Researcher at Kasetsart University awarded to S.F.A. and K.S., Science Achievement Scholarship of Thailand (no. 5917400296) through a grant from the Office of the Higher Education Commission, Thailand awarded to N.L.; and a grant from the Graduate Scholarship Program of the Graduate School, Kasetsart University, Thailand awarded to T.P. and K.S. This work was also funded in part by the Biotechnology and Biological Sciences Research Council (BB/K008161/1) of the University of Kent (to D.K.G.).
Acknowledgments
The Center for Agricultural Biotechnology (CAB) at Kasetsart University, Kamphaeng Saen Campus, provided support with server services for repeatomic data analysis. The Faculty of Science and Faculty of Forestry at Kasetsart University supported the research facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hake, L.; O’Connor, C. Genetic mechanisms of sex determination. Nature 2008, 1, 25. [Google Scholar]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Tree of Sex Consortium Sex determination: Why so many ways of doing it. PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, T.; Quinn, A.E.; Sarre, S.D.; O’Meally, D.; Georges, A.; Marshall Graves, J.A. Molecular marker suggests rapid changes of sex-determining mechanisms in Australian dragon lizards. Chromosome Res. 2009, 17, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Vallender, E.J.; Lahn, B.T. Multiple independent origins of sex chromosomes in amniotes. Proc. Natl. Acad. Sci. USA 2006, 103, 18031–18032. [Google Scholar] [CrossRef] [PubMed]
- Donoghue, P.C.J.; Benton, M.J. Rocks and clocks: Calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 2007, 22, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.E.; Dean, R.; Zimmer, F.; Mank, J.E. How to make a sex chromosome. Nat. Commun. 2016, 7, 12087. [Google Scholar] [CrossRef]
- Johnson, N.A.; Lachance, J. The genetics of sex chromosomes: Evolution and implications for hybrid incompatibility. Ann. N. Y. Acad. Sci. 2012, 1256, 1–22. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Okada, E.; Umemoto, H.; Tamura, K.; Uno, Y.; Nishikida-Umehara, C.; Matsuda, Y.; Takamatsu, N.; Shiba, T.; Ito, M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 2008, 105, 2469–2474. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Roeszler, K.N.; Ohnesorg, T.; Cummins, D.M.; Farlie, P.G.; Doran, T.J.; Sinclair, A.H. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 2009, 461, 267–271. [Google Scholar] [CrossRef]
- Herpin, A.; Braasch, I.; Kraeussling, M.; Schmidt, C.; Thoma, E.C.; Nakamura, S.; Tanaka, M.; Schartl, M. Transcriptional rewiring of the sex determining dmrt1 gene duplicate by transposable elements. PLoS Genet. 2010, 6, e1000844. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Kai, W.; Tasumi, S.; Oka, A.; Matsunaga, T.; Mizuno, N.; Fujita1, M.; Suetake, H.; Suzuki, S.; Hosoya1, S.; et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 2012, 8, e1002798. [Google Scholar] [CrossRef]
- Myosho, T.; Otake, H.; Masuyama, H.; Matsuda, M.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Hamaguchi, S.; Sakaizumi, M. Tracing the emergence of a novel sex-determining gene in Medaka, Oryzias luzonensis. Genetics 2012, 191, 163–170. [Google Scholar] [CrossRef]
- Harkess, A.; Zhou, J.; Xu, C.; Bowers, J.E.; Van der Hulst, R.; Ayyampalayam, S.; Tang, H. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Lahn, B.T.; Page, D.C. Four evolutionary strata on the human X chromosome. Science 1999, 286, 964–967. [Google Scholar] [CrossRef]
- Bergero, R.; Charlesworth, D. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 2009, 24, 94–102. [Google Scholar] [CrossRef]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nature 2014, 508, 488–493. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, J.; Bachtrog, D.; An, N.; Huang, Q.; Jarvis, E.D.; Gilbert, M.T.; Zhang, G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 2014, 346, 6215. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, T.; Srikulnath, K.; Graves, J.A.M. Origin of amniote sex chromosomes: An ancestral super-sex chromosome, or common requirements. J. Hered. 2017, 108, 94–105. [Google Scholar] [CrossRef]
- Xu, S.; Stapley, J.; Gablenz, S.; Boyer, J.; Appenroth, K.J.; Sree, K.S.; Gershenzon, J.; Widmer, A.; Huber, M. Low genetic variation is associated with low mutation rate in the giant duckweed. Nat. Commun. 2019, 10, 1–6. [Google Scholar] [CrossRef]
- Matsubara, K.; Tarui, H.; Toriba, M.; Yamada, K.; Nishida-Umehara, C.; Agata, K.; Matsuda, Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 18190–18195. [Google Scholar] [CrossRef]
- Matsubara, K.; Kuraku, S.; Tarui, H.; Nishimura, O.; Nishida, C.; Agata, K.; Kumazawa, Y.; Matsuda, Y. Intra-genomic GC heterogeneity in sauropsids: Evolutionary in sights from cDNA mapping and GC3 profiling in snake. BMC Genom. 2012, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; O’Meally, D.; Azad, B.; Georges, A.; Sarre, S.D.; Graves, J.A.; Matsuda, Y.; Ezaz, T. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 2015, 125, 111–123. [Google Scholar] [CrossRef]
- O’Meally, D.; Patel, H.R.; Stiglec, R.; Sarre, S.D.; Georges, A.; Graves, J.A.M.; Ezaz, T. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 2010, 18, 787–800. [Google Scholar] [CrossRef]
- Augstenová, B.; Mazzoleni, S.; Kratochvíl, L.; Rovatsos, M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Singchat, W.; O’Connor, R.E.; Tawichasri, P.; Suntronpong, A.; Sillapaprayoon, S.; Suntrarachun, S.; Muangmai, N.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; et al. Chromosome map of the Siamese cobra: Did partial synteny of sex chromosomes in the amniote represent “a hypothetical ancestral super-sex chromosome” or random distribution? BMC Genom. 2018, 19, 939. [Google Scholar] [CrossRef]
- Singchat, W.; Kraichak, E.; Tawichasri, P.; Tawan, T.; Suntronpong, A.; Sillapaprayoon, S.; Phatcharakullawarawat, R.; Muangmai, N.; Suntrarachun, S.; Baicharoen, S.; et al. Dynamics of telomere length in captive Siamese cobra (Naja kaouthia) related to age and sex. Ecol. Evol. 2019, 9, 6366–6377. [Google Scholar] [CrossRef] [PubMed]
- Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Indananda, C.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Do sex chromosomes of snakes, monitor lizards, and iguanian lizards result from multiple fission of an “ancestral amniote super-sex chromosome”? Chromosome Res. 2020, 28, 209–228. [Google Scholar] [CrossRef]
- Singchat, W.; Ahmad, S.F.; Sillapaprayoon, S.; Muangmai, N.; Duengkae, P.; Peyachoknagul, S.; O’Connor, R.E.; Griffin, D.K.; Srikulnath, K. Partial amniote sex chromosomal linkage homologies shared on snake W sex chromosomes support the possibility of ancestral super-sex chromosome evolution in amniotes. Front. Genet. 2020, 11, 948. [Google Scholar] [CrossRef]
- Thongchum, R.; Singchat, W.; Laopichienpong, N.; Tawichasri, P.; Kraichak, E.; Prakhongcheep, O.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Suntrarachun, S.; et al. Diversity of PBI-DdeI satellite DNA in snakes correlates with rapid independent evolution and different functional roles. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of paradigm shift with repeat landscapes in reptiles: Powerful facilitators of chromosomal rearrangements for diversity and evolution. Genes 2020, 11, 827. [Google Scholar] [CrossRef]
- Koomgun, T.; Laopichienpong, N.; Singchat, W.; Panthum, T.; Phatcharakullawarawat, R.; Kraichak, E.; Sillapaprayoon, S.; Ahmad, S.F.; Muangmai, N.; Peyachoknagul, S.; et al. Genome complexity reduction high-throughput genome sequencing of green iguana (Iguana iguana) reveal a paradigm shift in understanding sex-chromosomal linkages on homomorphic X and Y sex chromosomes. Front. Genet. 2020, 11, 1272. [Google Scholar] [CrossRef]
- Laopichienpong, N.; Kraichak, E.; Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Suntrarachun, S.; Baicharoen, S.; Peyachoknagul, S.; Chanhome, L.; Srikulnath, K.; et al. Genome-wide SNP analysis of Siamese cobra (Naja kaouthia) reveals the molecular basis of transitions between Z and W sex chromosomes and supports the presence of an ancestral super-sex chromosome in amniotes. Genomics. In Press. [CrossRef]
- Mank, J.E. The W, X, Y and Z of sex chromosome dosage compensation. Trends Genet. 2009, 25, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Bachtrog, D.; Kirkpatrick, M.; Mank, J.E.; McDaniel, S.F.; Pires, J.C.; Rice, W.; Valenzuela, N. Are all sex chromosomes created equal? Trends Genet. 2011, 27, 350–357. [Google Scholar] [CrossRef]
- Haldane, J.B.S. More precise expressions for the cost of natural selection. J. Genet. 1960, 57, 351–360. [Google Scholar] [CrossRef]
- Shibusawa, M.; Minai, S.; Nishida-Umehara, C.; Suzuki, T.; Mano, T.; Yamada, K.; Namikawa, T.; Matsuda, Y. A comparative cytogenetic study of chromosome homology between chicken and Japanese quail. Cytogenet. Cell Genet. 2001, 95, 103–109. [Google Scholar] [CrossRef]
- Hooper, D.M.; Price, T.D. Chromosomal inversion differences correlate with range overlap in passerine birds. Nat. Ecol. Evol. 2017, 1, 1526–1534. [Google Scholar] [CrossRef]
- Ponnikas, S.; Sigeman, H.; Abbott, J.K.; Hansson, B. Why do sex chromosomes stop recombining? Trends Genet. 2018, 34, 492–503. [Google Scholar] [CrossRef]
- Deakin, J.E.; Ezaz, T. Tracing the evolution of amniote chromosomes. Chromosoma 2014, 123, 201–216. [Google Scholar] [CrossRef]
- Gamble, T.; Coryell, J.; Ezaz, T.; Lynch, J.; Scantlebury, D.P.; Zarkower, D. Restriction Site-Associated DNA Sequencing (RAD-seq) Reveals an Extraordinary Number of Transitions among Gecko Sex-Determining Systems. Mol. Biol. Evol. 2015, 32, 1296–1309. [Google Scholar] [CrossRef]
- Gamble, T.; Castoe, T.A.; Nielsen, S.V.; Banks, J.L.; Card, D.C.; Schield, D.R.; Schuett, G.W.; Booth, W. The Discovery of XY Sex Chromosomes in a Boa and Python. Curr. Biol. 2017, 27, 2148–2153. [Google Scholar] [CrossRef]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Pokorná, M.J. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef]
- Sabath, N.; Itescu, Y.; Feldman, A.; Meiri, S.; Mayrose, I.; Valenzuela, N. Sex determination, longevity, and the birth and death of reptilian species. Ecol. Evol. 2016, 6, 5207–5220. [Google Scholar] [CrossRef] [PubMed]
- Laopichienpong, N.; Muangmai, N.; Chanhome, L.; Suntrarachun, S.; Twilprawat, P.; Peyachoknagul, S.; Srikulnath, K. Evolutionary dynamics of the gametologous CTNNB1 gene on the Z and W chromosomes of snakes. J. Hered. 2017, 108, 142–151. [Google Scholar] [CrossRef]
- Laopichienpong, N.; Tawichasri, P.; Chanhome, L.; Phatcharakullawarawat, R.; Singchat, W.; Kantachumpoo, A.; Muangmai, N.; Suntrarachun, S.; Matsubara, K.; Srikulnath, K.; et al. A novel method of caenophidian snake sex identification using molecular markers based on two gametologous genes. Ecol. Evol. 2017, 7, 4661–4669. [Google Scholar] [CrossRef]
- Tawichasri, P.; Laopichienpong, N.; Chanhome, L.; Phatcharakullawarawat, R.; Singchat, W.; Koomgun, T.; Prasongmaneerut, T.; Rerkamnuaychokee, W.; Sillapaprayoona, S.; Muangmai, N.; et al. Using blood and non-invasive shed skin samples to identify sex of caenophidian snakes based on multiplex PCR assay. Zool. Anz. 2017, 271, 6–14. [Google Scholar] [CrossRef]
- Augstenová, B.; Johnson, P.M.; Altmanova, M.; Frynta, D.; Rovatsos, M.; Kratochvil, L. ZW, XY, and yet ZW: Sex chromosome evolution in snakes even more complicated. Evolution 2018. [Google Scholar] [CrossRef] [PubMed]
- Pennell, M.W.; Mank, J.E.; Peichel, C.L. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 2018, 27, 3950–3963. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Potter, S.; O’Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.B.; Fukui, K.; Marshall Graves, J.A.; Griffin, D.; Grutzner, F.; et al. Chromosomics: Bridging the Gap between Genomes and Chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef]
- Deakin, J.E.; Ezaz, T. Understanding the evolution of reptile chromosomes through happlications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef]
- Rovatsos, M.; Farkačová, K.; Altmanová, M.; Johnson, P.M.; Kratochvíl, L. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 2019, 28, 3042–3052. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Sex chromosome and sex-linked genes Springer. Chromosoma 1967, 23, 1–9. [Google Scholar] [CrossRef]
- Janzen, F.J.; Paukstis, G.L. Environmental sex determination in reptiles: Ecology, evolution, and experimental design. Q. Rev. Biol. 1991, 66, 149–179. [Google Scholar] [CrossRef]
- Pokorná, M.; Kratochvil, L. Phylogeny of sex-determining mechanisms in squamate reptiles: Are sex chromosomes an evolutionary trap. Zool. J Linn. Soc. 2009, 156, 168–183. [Google Scholar] [CrossRef]
- Pokorná, M.J.; Kratochvíl, L. What was the ancestral sex-determining mechanism in amniote vertebrates. Biol. Rev. Camb. Philos. Soc. 2016, 91, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Janes, D.E.; Organ, C.L.; Fujita, M.K.; Shedlock, A.M.; Edwards, S.V. Genome evolution in reptilia, the sister group of mammals. Annu. Rev. Genom. Hum. Genet. 2010, 11, 239–264. [Google Scholar] [CrossRef]
- O′Meally, D.; Ezaz, T.; Georges, A.; Sarre, S.D.; Graves, J.A. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 2012, 20, 7–19. [Google Scholar] [CrossRef]
- Matsubara, K.; O’Meally, D.; Sarre, S.D.; Georges, A.; Srikulnath, K.; Ezaz, T. ZW sex chromosomes in Australian dragon lizards (Agamidae) originated from a combination of duplication and translocation in the nucleolar organising region. Genes 2019, 10, 861. [Google Scholar] [CrossRef]
- Tachibana, M. Epigenetics of sex determination in mammals. Reprod. Med. Biol. 2015, 15, 59–67. [Google Scholar] [CrossRef]
- Suryamohan, K.; Krishnankutty, S.P.; Guillory, J.; Jevit, M.; Schröder, M.S.; Wu, M.; Kuriakose, B.; Mathew, O.K.; Perumal, R.C.; Koludarov, I.; et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020, 52, 106–117. [Google Scholar] [CrossRef]
- Furman, B.; Metzger, D.; Darolti, I.; Wright, A.E.; Sandkam, B.A.; Almeida, P.; Shu, J.J.; Mank, J.E. Sex Chromosome Evolution: So Many Exceptions to the Rules. Genome Biol. Evol. 2020, 12, 750–763. [Google Scholar] [CrossRef]
- Organ, C.L.; Janes, D.E. Evolution of sex chromosomes in Sauropsida. Integr. Comp. Biol. 2008, 48, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Quinn, A.E.; Sarre, S.D.; Ezaz, T.; Graves, J.A.M.; Georges, A. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol. Lett. 2011, 443–448. [Google Scholar] [CrossRef]
- Sarre, S.D.; Ezaz, T.; Georges, A. Transitions between sex-determining systems in reptiles and amphibians. Annu. Rev. Genom. Hum. Genet. 2011, 12, 391–406. [Google Scholar] [CrossRef]
- Holleley, C.E.; O’Meally, D.; Sarre, S.D.; Graves, J.A.M.; Ezaz, T.; Matsubara, K.; Azad, B.; Zhang, X.; Georges, A. Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 2015, 523, 79–82. [Google Scholar] [CrossRef]
- Graves, J.A.M. Did sex chromosome turnover promote divergence of the major mammal groups?: De novo sex chromosomes and drastic rearrangements may have posed reproductive barriers between monotremes, marsupials and placental mammals. BioEssays 2016, 38, 734–743. [Google Scholar] [CrossRef]
- Montiel, E.E.; Badenhorst, D.; Tamplin, J.; Burke, R.L.; Valenzuela, N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 2016, 126, 105–113. [Google Scholar] [CrossRef]
- Bista, B.; Valenzuela, N. Turtle Insights into the Evolution of the Reptilian Karyotype and the Genomic Architecture of Sex Determination. Genes 2020, 11, 416. [Google Scholar] [CrossRef]
- Beukeboom, L.W.; Perrin, N. The Evolution of Sex Determination; Oxford University Press: Oxford, UK, 2014; p. 222. [Google Scholar]
- Landeen, E.L.; Presgraves, D.C. Evolution: From Autosomes to Sex Chromosomes—and Back. Curr. Biol. 2013, 23, 848–850. [Google Scholar] [CrossRef] [PubMed]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Mawaribuchi, S.; Yoshimoto, S.; Ohashi, S.; Takamatsu, N.; Ito, M. Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 2012, 20, 139–151. [Google Scholar] [CrossRef]
- Vicoso, B. Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat. Ecol. Evol. 2019, 3, 1632–1641. [Google Scholar] [CrossRef]
- Bull, J.J.; Charnov, E.L. Changes in the heterogametic mechanism of sex determination. Heredity 1977, 39, 1–14. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, G.S.; Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 2007, 449, 909–912. [Google Scholar] [CrossRef]
- van Doorn, G.S.; Kirkpatrick, M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 2010, 186, 629–645. [Google Scholar] [CrossRef]
- Blaser, O.; Grossen, C.; Neuenschwander, S.; Perrin, N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 2013, 67, 635–645. [Google Scholar] [CrossRef]
- Blaser, O.; Neuenschwander, S.; Perrin, N. Sex-chromosome turnovers: The hot-potato model. Am. Nat. 2014, 183, 140–146. [Google Scholar] [CrossRef]
- Veller, C.; Muralidhar, P.; Constable, G.W.A.; Nowak, M.A. Drift-Induced selection between male and female heterogamety. Genetics 2017, 207, 711–727. [Google Scholar] [CrossRef]
- Palmer, D.H.; Rogers, T.F.; Dean, R.; Wright, A.E. How to identify sex chromosomes and their turnover. Mol. Ecol. 2019, 28, 4709–4724. [Google Scholar] [CrossRef] [PubMed]
- Kitano, J.; Ross, J.A.; Mori, S.; Kume, M.; Jones, F.C.; Chan, Y.F.; Absher, D.M.; Grimwood, J.; Schmutz, J.; Myers, R.M.; et al. A role for a neo-sex chromosome in stickleback speciation. Nature 2009, 461, 1079–1083. [Google Scholar] [CrossRef]
- Natri, H.M.; Shikano, T.; Merilä, J. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol. Biol. Evol. 2013, 30, 1131–1144. [Google Scholar] [CrossRef]
- Jetybayev, I.E.; Bugrov, A.G.; Ünal, M.; Buleu, O.G.; Rubtsov, N.B. Molecular cytogenetic analysis reveals the existence of two independent neo-XY sex chromosome systems in Anatolian Pamphagidae grasshoppers. BMC Evol. Biol. 2017, 17, 20. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 1980, 35, 205–214. [Google Scholar] [CrossRef]
- Rice, W.R. Evolution of the Y sex chromosome in animals. Biosciences 1996, 46, 331–343. [Google Scholar] [CrossRef]
- Roberts, R.B.; Ser, J.R.; Kocher, T.D. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 2009, 326, 998–1001. [Google Scholar] [CrossRef]
- Wang, J.; Na, J.K.; Yu, Q.; Gschwend, A.R.; Han, J.; Zeng, F.; Aryal, R.; VanBuren, R.; Murray, J.E.; Zhang, W.; et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13710–13715. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Bachtrog, D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 2012, 337, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Bergero, R.; Charlesworth, D. Testing for the footprint of sexually antagonistic polymorphisms in the pseudoautosomal region of a plant sex chromosome pair. Genetics 2013, 194, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Mank, J.E.; Vicoso, B.; Berlin, S.; Charlesworth, B. Effective population size and the Faster-X effect: Empirical results and their interpretation. Evolution 2010, 64, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Saunders, P.A.; Neuenschwander, S.; Perrin, N. Sex chromosome turnovers and genetic drift: A simulation study. J. Evol. Biol. 2018, 31, 1413–1419. [Google Scholar] [CrossRef]
- Ezaz, T.; Deakin, J.E. Repetitive sequence and sex chromosome evolution in vertebrates. Adv. Evol. Biol. 2014, 9. [Google Scholar] [CrossRef]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef]
- Mittwoch, U. Sex determination and sex reversal: Genotype, phenotype, dogma and semantics. Hum. Genet. 1992, 89, 467–479. [Google Scholar] [CrossRef]
- Li, H.; Holleley, C.E.; Elphick, M.; Georges, A.; Shine, R. The behavioural consequences of sex reversal in dragons. Proc. R. Soc. B. 2016, 283, 20160217. [Google Scholar] [CrossRef]
- Perrin, N. Sex reversal: A fountain of youth for sex chromosomes? Evolution 2009, 63, 3043–3049. [Google Scholar] [CrossRef]
- Dufresnes, C.; Borzée, A.; Horn, A.; Stöck, M.; Ostini, M.; Sermier, R.; Wassef, J.; Litvinchuck, S.N.; Kosch, T.A.; Waldman, B.; et al. Sex-chromosome homomorphy in Palearctic tree frogs results from both turnovers and X-Y recombination. Mol. Biol. Evol. 2015, 32, 2328–2337. [Google Scholar] [CrossRef]
- Rodrigues, N.; Studer, T.; Dufresnes, C.; Perrin, N. Sex-Chromosome Recombination in Common Frogs Brings Water to the Fountain-of-Youth. Mol. Biol. Evol. 2018, 35, 942–948. [Google Scholar] [CrossRef]
- Rovatsos, M.; Pokorná, M.; Altmanová, M.; Kratochvíl, L. Cretaceous park of sex determination: Sex chromosomes are conserved across iguanas. Biol. Lett. 2014, 10, 20131093. [Google Scholar] [CrossRef]
- Acosta, A.; Martínez-Pacheco, M.L.; Díaz-Barba, K.; Porras, N.; Gutiérrez-Mariscal, M.; Cortez, D. Deciphering ancestral sex chromosome turnovers based on analysis of male mutation bias. Genome Biol. Evol. 2019, 11, 3054–3067. [Google Scholar] [CrossRef]
- Nielsen, S.V.; Guzmán-Méndez, I.A.; Gamble, T.; Blumer, M.; Pinto, B.J.; Kratochvíl, L.; Rovatsos, M. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol. Lett. 2019, 15, 20190498. [Google Scholar] [CrossRef]
- Matsuda, Y.; Nishida-Umehara, C.; Tarui, H.; Kuroiwa, A.; Yamada, K.; Isobe, T.; Ando, J.; Fujiwara, A.; Hirao, Y.; Nishimura, O.; et al. Highly conserved linkage homology between birds and turtles: Bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 2005, 13, 601–615. [Google Scholar] [CrossRef]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y.; Nishida, C. Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet. Genome Res. 2009, 125, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Nishida, C.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y. Karyotypic evolution in squamate reptiles: Comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res. 2009, 17, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Matsuda, Y. Karyotype evolution in monitor lizards: Cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res. 2013, 21, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Nishida, C.; Olsson, M.; Matsuda, Y. Identification of the linkage group of the Z sex chromosomes of the sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 2014, 123, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Ota, H.; Matsuda, Y. Karyotype reorganization in the Hokou gecko (Gekko hokouensis, Gekkonidae): The process of microchromosome disappearance in Gekkota. PLoS ONE 2015, 10, e0134829. [Google Scholar] [CrossRef]
- Alföldi, J.; Palma, F.D.; Grabherr, M. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587. [Google Scholar] [CrossRef]
- Ezaz, T.; Azad, B.; O’Meally, D.; Young, M.J.; Matsubara, K.; Edwards, M.J.; Zhang, X.; Holleley, C.E.; Deakin, J.E.; Graves, J.A.M.; et al. Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R-spondin 1. BMC Genom. 2013, 14, 899. [Google Scholar] [CrossRef]
- Damas, J.; O’Connor, R.; Farré, M.; Lenis, V.; Martell, H.J.; Mandawala, A.; Fowler, K.; Joseph, S.; Swain, M.T.; Griffin, D.K.; et al. Upgrading short-read animal genome assemblies to chromosome level using comparative genomics and a universal probe set. Genome Res. 2017, 27, 875–884. [Google Scholar] [CrossRef]
- O’Connor, R.E.; Romanov, M.N.; Kiazim, L.G.; Barrett, P.M.; Farré, M.; Damas, J.; Ferguson-Smith, M.; Valenzuela, N.; Larkin, D.M.; Griffin, D.K. Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat. Commun. 2018, 9, 1883. [Google Scholar] [CrossRef]
- O’Connor, R.E.; Farré, M.; Joseph, S.; Damas, J.; Kiazim, L.; Jennings, R.; Bennett, S.; Slack, E.A.; Allanson, E.; Larkin, D.M.; et al. Chromosome-level assembly reveals extensive rearrangement in saker falcon and budgerigar, but not ostrich, genomes. Genome Biol. 2018, 19, 171. [Google Scholar] [CrossRef]
- Pasquesi, G.I.M.; Adams, R.H.; Card, D.C.; Schield, D.R.; Corbin, A.B.; Perry, B.W.; Reyes-Velasco, J.; Ruggiero, R.P.; Vandewege, M.W.; Shortt, J.A.; et al. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat. Commun. 2018, 9, 2774. [Google Scholar] [CrossRef]
- Gemmell, N.J.; Rutherford, K.; Prost, S.; Tollis, M.; Winter, D.; Macey, J.R.; Adelson, D.L.; Suh, A.; Bertozzi, T.; Grau, J.H.; et al. The tuatara genome reveals ancient features of amniote evolution. Nature 2020, 584, 403–409. [Google Scholar] [CrossRef]
- Srikulnath, K.; Azad, B.; Singchat, W.; Ezaz, T. Distribution and amplification of interstitial telomeric sequences (ITSs) in Australian dragon lizards support frequent chromosome fusions in Iguania. PLoS ONE 2019, 14, e0212683. [Google Scholar] [CrossRef]
- Charlesworth, D. Plant sex chromosomes. Annu. Rev. Plant Biol. 2016, 67, 397–420. [Google Scholar] [CrossRef]
- Rovatsos, M.; Vukić, J.; Kratochvíl, L. Mammalian X homolog acts as sex chromosome in lacertid lizards. Heredity 2016, 117, 8–13. [Google Scholar] [CrossRef]
- Rice, W.R. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution 1987, 41, 911–914. [Google Scholar] [CrossRef] [PubMed]
- Manolakou, P.; Lavranos, G.; Angelopoulou, R. Molecular patterns of sex determination in the animal kingdom: A comparative study of the biology of reproduction. Reprod. Biol. Endocrinol. 2006, 4, 59. [Google Scholar] [CrossRef][Green Version]
- Cline, T.W. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 1993, 9, 385–390. [Google Scholar] [CrossRef]
- Pomiankowski, A.; Nöthiger, R.; Wilkins, A. The evolution of the Drosophila sex-determination pathway. Genetics 2004, 166, 1761–1773. [Google Scholar] [CrossRef]
- Erickson, J.W.; Quintero, J.J. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007, 5, e332. [Google Scholar] [CrossRef]
- Fritz, A.J.; Barutcu, A.R.; Martin-Buley, L.; van Wijnen, A.J.; Zaidi, S.K.; Imbalzano, A.N.; Lian, J.B.; Stein, J.L.; Stein, G.S. Chromosomes at Work: Organization of Chromosome Territories in the Interphase Nucleus. J. Cell Biochem. 2016, 117, 9–19. [Google Scholar] [CrossRef]
- Tanabe, S. Contamination and toxic effects of persistent endocrine disrupters in marine mammals and birds. Mar. Pollut. Bull. 2002, 45, 69–77. [Google Scholar] [CrossRef]
- Gilbert, N.; Gilchrist, S.; Bickmore, W.A. Chromatin organization in the mammalian nucleus. Int. Rev. Cytol. 2005, 242, 283–336. [Google Scholar] [CrossRef]
- Skinner, B.M.; Robertson, L.B.; Tempest, H.G.; Langley, E.J.; Ioannou, D.; Fowler, K.E.; Crooijmans, R.P.; Hall, A.D.; Griffin, D.K.; Völker, M. Comparative genomics in chicken and Pekin duck using FISH mapping and microarray analysis. BMC Genom. 2009, 10, 357. [Google Scholar] [CrossRef]
- Greaves, I.K.; Rens, W.; Fergurson-Smith, M.A.; Griffin, D.K.; Graves, J.A.M. Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res. 2003, 11, 503–512. [Google Scholar] [CrossRef]
- Mahy, N.L.; Perry, P.E.; Bickmore, W.A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 2002, 159, 753–763. [Google Scholar] [CrossRef]
- Bellott, D.W.; Skaletsky, H.; Pyntikova, T.; Mardis, E.R.; Graves, T.; Kremitzki, C.; Brown, L.G.; Rozen, S.; Warren, W.C.; Wilson, R.K.; et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 2010, 466, 612–616. [Google Scholar] [CrossRef]
- Vallender, E.J.; Lahn, B.T. How mammalian sex chromosomes acquired their peculiar gene content. Bioessays 2004, 26, 159–169. [Google Scholar] [CrossRef]
- Mank, J.E. Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet. 2013, 29, 677–683. [Google Scholar] [CrossRef]
- Alam, S.; Altmanová, M.; Prasongmaneerut, T.; Georges, A.; Sarre, S.D.; Nielsen, S.V.; Gamble, T.; Srikulnath, K.; Rovatsos, M.; Kratochvíl, L.; et al. Cross-Species BAC Mapping Highlights Conservation of Chromosome Synteny across Dragon Lizards (Squamata: Agamidae). Genes 2020, 11, 698. [Google Scholar] [CrossRef]
- Waters, P.D.; Delbridge, M.L.; Deakin, J.E.; El-Mogharbel, N.; Kirby, P.J.; Carvalho-Silva, D.R.; Graves, J.A. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus): Implications for mammalian sex chromosome evolution. Chromosome Res. 2005, 13, 401–410. [Google Scholar] [CrossRef]
- Veyrunes, F.; Waters, P.D.; Miethke, P.; Rens, W.; McMillan, D.; Alsop, A.E.; Grützner, F.; Deakin, J.E.; Whittington, C.M.; Schatzkamer, K.; et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008, 18, 965–973. [Google Scholar] [CrossRef]
- Autuoro, J.M.; Pirnie, S.P.; Carmichael, G.G. Long noncoding RNAs in imprinting and X chromosome inactivation. Biomolecules 2014, 4, 76–100. [Google Scholar] [CrossRef]
- Kawagoshi, T.; Nishida, C.; Matsuda, Y. The origin and differentiation process of X and Y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines). Chromosome Res. 2012, 20, 95–110. [Google Scholar] [CrossRef][Green Version]
- Kawagoshi, T.; Uno, Y.; Nishida, C.; Matsuda, Y. The Staurotypus turtles and aves share the same origin of sex chromosomes but evolved different types of heterogametic sex determination. PLoS ONE 2014, 9, e105315. [Google Scholar] [CrossRef][Green Version]
- Srikulnath, K.; Uno, Y.; Matsubara, K.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Nishida, C.; Matsuda, Y. Chromosomal localization of the 18S–28S and 5s rRNA genes and (TTAGGG)n sequences of butterfly lizards (Leiolepis belliana belliana and Leiolepis boehmei, Agamidae, Squamata). Genet. Mol. Biol. 2011, 34, 582–586. [Google Scholar] [CrossRef]
- Porter, C.A.; Haiduk, M.W.; de Queiroz, K. Evolution and phylogenetic significance of ribosomal gene location in chromosomes of squamate reptiles. Copeia 1994, 1994, 302. [Google Scholar] [CrossRef]
- Pellegrino, K.C.M.; Bertolotto, C.E.V.; Yonenaga-Yassuda, Y.; Rodrigues, M.T. Banding patterns, heteromorphic sex chromosomes and Agstained NORs after pachytene stage in the meiosis of the Brazilian lizard Urostrophus vautieri (Squamata, Polychrotidae). Caryologia 1999, 52, 21–26. [Google Scholar] [CrossRef]
- Young, M.J.; O’Meally, D.; Sarre, S.D.; Georges, A.; Ezaz, T. Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 2013, 21, 361–374. [Google Scholar] [CrossRef]
- Acosta, M.J.; Marchal, J.A.; Fernández-Espartero, C.; Romero-Fernández, I.; Rovatsos, M.T.; Giagia-Athanasopoulou, E.B.; Gornung, E.; Castiglia, R.; Sánchez, A. Characterization of the satellite DNA Msat-160 from species of Terricola (Microtus) and Arvicola (Rodentia, Arvicolinae). Genetica 2010, 138, 1085–1098. [Google Scholar] [CrossRef]
- Hughes, J.F.; Skaletsky, H.; Pyntikova, T.; Graves, T.A.; van Daalen, S.K.; Minx, P.J.; Fulton, R.S.; McGrath, S.D.; Locke, D.P.; Friedman, C.; et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 2010, 463, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.; Geneva, A.J.; Glor, R.E.; Zarkower, D. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 2014, 68, 1027–1041. [Google Scholar] [CrossRef]
- Rovatsos, M.; Altmanová, M.; Pokorná, M.; Kratochvíl, L. Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution 2014, 68, 2079–2085. [Google Scholar] [CrossRef]
- Soh, Y.Q.; Alföldi, J.; Pyntikova, T.; Brown, L.G.; Graves, T.; Minx, P.J.; Fulton, R.S.; Kremitzki, C.; Koutseva, N.; Mueller, J.L.; et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 2014, 159, 800–813. [Google Scholar] [CrossRef]
- Altmanová, M.; Rovatsos, M.; Kratochvíl, L.; Johnson Pokorná, M. Minute Y chromosomes and karyotype evolution in Madagascan iguanas (Squamata: Iguania: Opluridae). Biol. J. Linn. Soc. 2016, 118, 618–633. [Google Scholar] [CrossRef]
- Morgan, A.P.; Pardo-Manuel de Villena, F. Sequence and Structural Diversity of Mouse Y Chromosomes. Mol. Biol. Evol. 2017, 34, 3186–3204. [Google Scholar] [CrossRef] [PubMed]
- Iannucci, A.; Altmanová, M.; Ciofi, C.; Ferguson-Smith, M.; Milan, M.; Pereira, J.C.; Pether, J.; Rehák, I.; Rovatsos, M.; Stanyon, R.; et al. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity 2019, 123, 215–227. [Google Scholar] [CrossRef]
- Rovatsos, M.; Rehák, I.; Velenský, P.; Kratochvíl, L. Shared Ancient Sex Chromosomes in Varanids, Beaded Lizards, and Alligator Lizards. Mol. Biol. Evol. 2019, 36, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Graphodatsky, A.S.; Trifonov, V.A.; Stanyon, R. The genome diversity and karyotype evolution of mammals. Mol. Cytogenet. 2011, 4, 22. [Google Scholar] [CrossRef]
- Olmo, E. Chromorep: A Reptile Chromosomes Database. 2013. Available online: http://chromorep.univpm.it/ (accessed on 15 May 2020).
- Vicoso, B.; Emerson, J.J.; Zektser, Y.; Mahajan, S.; Bachtrog, D. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013, 11, e1001643. [Google Scholar] [CrossRef]
- Matsubara, K.; Sarre, S.D.; Georges, A.; Matsuda, Y.; Marshall Graves, J.A.; Ezaz, T. Highly differentiated ZW sex microchromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS ONE 2014, 9, e95226. [Google Scholar] [CrossRef]
- O’Neill, M.J.; O’Neill, R.J. Sex chromosome repeats tip the balance towards speciation. Mol. Ecol. 2018, 27, 3783–3798. [Google Scholar] [CrossRef]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of Bkm sequences (GATA repeats): Predominant association with sex chromosomes and potential role in higher order chromatin organization and function. Bioinformatics 2003, 19, 681–685. [Google Scholar] [CrossRef]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.; Meller, V.H. Sex chromosome evolution: Life, death and repetitive DNA. Fly 2014, 8, 197–199. [Google Scholar] [CrossRef][Green Version]
- Iwase, M.; Satta, Y.; Hirai, Y.; Hirai, H.; Imai, H.; Takahata, N. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc. Natl. Acad. Sci. USA 2003, 100, 5258–5263. [Google Scholar] [CrossRef]
- Xu, L.; Auer, G.; Peona, V.; Suh, A.; Deng, Y.; Feng, S.; Zhang, G.; Blom, M.; Christidis, L.; Prost, S.; et al. Dynamic evolutionary history and gene content of sex chromosomes across diverse songbirds. Nat. Ecol. Evol. 2019, 3, 834–844. [Google Scholar] [CrossRef]
- Kent, T.; Uzunovic, J. Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160458. [Google Scholar] [CrossRef]
- Bonchev, G.; Willi, Y. Accumulation of transposable elements in selfing populations of Arabidopsis lyrata supports the ectopic recombination model of transposon evolution. New Phytol. 2018, 219, 767–778. [Google Scholar] [CrossRef]
- Charlesworth, B.; Sniegowsky, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Plohl, M.; Mestrovic, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [CrossRef]
- Nakayama, I.; Foresti, F.; Tewari, R.; Schartl, M.; Chourrout, D.; National, I. Sex chromosome polymorphism and heterogametic males revealed by two cloned DNA probes in the ZW/ZZ fish Leporinus elongatus. Chromosoma 1994, 103, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Hobza, R.; Lengerova, M.; Svoboda, J.; Kubekova, H.; Kejnovsky, E.; Vyskot, B. An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma 2006, 115, 376–382. [Google Scholar] [CrossRef]
- Mariotti, B.; Manzano, S.; Kejnovsky, E.; Vyskot, B.; Jamilena, M. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Mol. Genet. Genom. 2009, 281, 249–259. [Google Scholar] [CrossRef]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Seki, R.; Nishida, C.; Matsuda, Y. Molecular cloning and characterization of satellite DNA sequences from constitutive heterochromatin of the habu snake (Protobothrops flavoviridis, Viperidae) and the Burmese python (Python bivittatus, Pythonidae). Chromosoma 2015, 124, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Pokorná, M.; Kratochvíl, L.; Kejnovský, E. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 2011, 12. [Google Scholar] [CrossRef]
- Matsubara, K.; Uno, Y.; Srikulnath, K.; Matsuda, Y.; Miller, E.; Olsson, M. No interstitial telomeres on autosomes but remarkable amplification of telomeric repeats on the W sex chromosome in the sand lizard (Lacerta agilis). J. Hered. 2015, 106, 753–757. [Google Scholar] [CrossRef]
- Lisachov, A.P.; Borodin, P.M. Microchromosome polymorphism in the sand lizard, Lacerta agilis Linnaeus, 1758 (Reptilia, Squamata). Comp. Cytogenet. 2016, 10, 387. [Google Scholar] [CrossRef][Green Version]
- Lisachov, A.P.; Giovannotti, M.; Pereira, J.C.; Andreyushkova, D.A.; Romanenko, S.A.; Ferguson-Smith, M.A.; Borodin, P.M.; Trifonov, V.A. Chromosome painting does not support a sex chromosome turnover in Lacerta agilis Linnaeus, 1758. Cytogenet. Genome Res. 2020, 160, 134–140. [Google Scholar] [CrossRef]
- Giovannotti, M.; Cerioni, P.N.; Slimani, T.; Splendiani, A.; Paoletti, A.; Fawzi, A.; Barucchi, V.C. Cytogenetic characterization of a population of Acanthodactylus lineomaculatus Duméril and Bibron, 1839 (Reptilia, Lacertidae), from Southwestern Morocco and insights into sex chromosome evolution. Cytogenet. Genome Res. 2017, 153, 86–95. [Google Scholar] [CrossRef]
- Giovannotti, M.; Nisi Cerioni, P.; Rojo, V.; Olmo, E.; Slimani, T.; Splendiani, A.; Caputo Barucchi, V. Characterization of a satellite DNA in the genera Lacerta and Timon (Reptilia, Lacertidae) and its role in the differentiation of the W chromosome. J. Exp. Zool. Part B Mol. Dev. Evol. 2018, 330, 83–95. [Google Scholar] [CrossRef]
- Singh, R.J.; Röbbelen, G.; Okamoto, M. Somatic association at interphase studied by Giemsa banding technique. Chromosoma 1976, 56, 265–273. [Google Scholar] [CrossRef]
- Singh, L.; Purdom, I.F.; Jones, K.W. Sex chromosome associated satellite DNA: Evolution and conservation. Chromosoma 1980, 79, 137–157. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B.; Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 2005, 95, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Schmid, M.; Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 2016, 125, 553–571. [Google Scholar] [CrossRef]
- Singchat, W.; Areesirisuk, P.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Suntrarachun, S.; Chanhome, L.; Peyachoknagul, S.; Srikulnath, K. Complete mitochondrial genome of Siamese cobra (Naja kaouthia) determined using next-generation sequencing. Mitochondrial DNA B 2019, 4, 577–578. [Google Scholar] [CrossRef]
- Smeds, L.; Warmuth, V.; Bolivar, P.; Uebbing, S.; Burri, R.; Suh, A.; Nater, A.; Bureš, S.; Garamszegi, L.Z.; Hogner, S.; et al. Evolutionary analysis of the female-specific avian W chromosome. Nat. Commun. 2015, 6, 7330. [Google Scholar] [CrossRef] [PubMed]
- Viana, P.F.; Ezaz, T.; de Bello Cioffi, M.; Almeida, B.J.; Feldberg, E. Evolutionary Insights of the ZW Sex Chromosomes in Snakes: A New Chapter Added by the Amazonian Puffing Snakes of the Genus Spilotes. Genes 2019, 10, 288. [Google Scholar] [CrossRef]
- Heise, P.J.; Maxson, L.R.; Dowling, H.G.; Hedges, S.B. Higher-level snake phylogeny inferred from mitochondrial DNA sequences of 12S rRNA and 16S rRNA genes. Mol. Biol. Evol. 1995, 12, 259–265. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef]
- Harrington, S.M.; Reeder, T.W. Phylogenetic inference and divergence dating of snakes using molecules, morphology and fossils: New insights into convergent evolution of feeding morphology and limb reduction. Biol. J. Linn. Soc. 2017, 121, 379–394. [Google Scholar] [CrossRef][Green Version]
- Uetz, P.; Freed, P.; Hošek, J. The Reptile Database. 2019. Available online: http://www.reptile-database.org (accessed on 25 August 2020).
- Vidal, N.; Hedges, S.B. Higher-level relationships of caenophidian snakes inferred from four nuclear and mitochondrial genes. Comptes Rendus Biol. 2002, 325, 987–995. [Google Scholar] [CrossRef]
- Lawson, R.; Slowinski, J.B.; Crother, B.I.; Burbrink, F.T. Phylogeny of the Colubroidea (Serpentes): New evidence from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 2005, 37, 581–601. [Google Scholar] [CrossRef]
- Oguiura, N.; Ferrarezzi, H.; Batistic, R.F. Cytogenetics and molecular data in snakes: A phylogenetic approach. Cytogenet. Genome Res. 2009, 127, 128–142. [Google Scholar] [CrossRef]
- Booth, W.; Johnson, D.H.; Moore, S.; Schal, C.; Vargo, E.L. Evidence for viable, non-clonal but fatherless boa constrictors. Biol. Lett. 2011, 7, 257–260. [Google Scholar] [CrossRef]
- Booth, W.; Million, L.; Reynolds, R.G.; Burghardt, G.M.; Vargo, E.L.; Schal, C.; Tzika, A.C.; Schuett, G.W. Consecutive virgin births in the New World boid snake, the Colombian rainbow boa, Epicrates maurus. J. Hered. 2011, 102, 759–763. [Google Scholar] [CrossRef][Green Version]
- Booth, W.; Schuett, G.W.; Ridgway, A.; Buxton, D.W.; Castoe, T.A.; Bastone, G.; Bennett, C.; McMahan, W. Facultative parthenogenesis in pythons. Biol. J. Linn. Soc. Lond. 2014, 112, 461–468. [Google Scholar] [CrossRef]
- Kinney, M.E.; Wack, R.F.; Grahn, R.A.; Lyons, L. Parthenogenesis in a Brazilian rainbow boa (Epicrates cenchria cenchria). Zoo Biol. 2012, 32, 172–176. [Google Scholar] [CrossRef]
- Mallery, C.S., Jr.; Carrillo, M.M. A case study of sex-linkage in Python regius (Serpentes: Boidae), with new insights into sex determination in Henophidia. Phyllomedusa 2016, 15, 29–42. [Google Scholar] [CrossRef]
- Mengden, G.A.; Stock, A.D. Chromosomal evolution in Serpentes; a comparison of G and C chromosome banding patterns of some colubrid and boid genera. Chromosoma 1980, 79, 53–64. [Google Scholar] [CrossRef]
- Viana, P.F.; Ribeiro, L.B.; Souza, G.M.; Chalkidis, H.d.M.; Gross, M.C.; Feldberg, E. Is the Karyotype of Neotropical Boid Snakes Really Conserved? Cytotaxonomy, Chromosomal Rearrangements and Karyotype Organization in the Boidae Family. PLoS ONE. 2016, 1, e0160274. [Google Scholar] [CrossRef][Green Version]
- Augstenová, B.; Mazzoleni, S.; Kostmann, A.; Altmanová, M.; Frynta, D.; Kratochvíl, L.; Rovatsos, M. Cytogenetic Analysis Did Not Reveal Differentiated Sex Chromosomes in Ten Species of Boas and Pythons (Reptilia: Serpentes). Genes 2019, 10, 934. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Kumazawa, Y.; Ota, H.; Nishida, C.; Matsuda, Y. Karyotype Analysis of Four Blind Snake Species (Reptilia: Squamata: Scolecophidia) and Karyotypic Changes in Serpentes. Cytogenet. Genome Res. 2019, 157, 98–106. [Google Scholar] [CrossRef]
- Rovatsos, M.; Vukić, J.; Lymberakis, P.; Kratochvíl, L. Evolutionary stability of sex chromosomes in snakes. Proc. Biol. Sci. 2015, 282, 20151992. [Google Scholar] [CrossRef]
- Matsubara, K.; Nishida, C.; Matsuda, Y.; Kumazawa, Y. Sex chromosome evolution in snakes inferred from divergence patterns of two gametologous genes and chromosome distribution of sex chromosome-linked repetitive sequences. Zool. Lett. 2016, 2, 19. [Google Scholar] [CrossRef]
- Viana, P.F.; Ezaz, T.; de Bello Cioffi, M.; Liehr, T.; Al-Rikabi, A.; Goll, L.G.; Rocha, A.M.; Feldberg, E. Landscape of snake’ sex chromosomes evolution spanning 85 MYR reveals ancestry of sequences despite distinct evolutionary trajectories. Sci. Rep. 2020, 10, 12499. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R.; Nicol, J.A.; Tamm, H.; Kullman, B.; Kullman, K.; Leitch, I.J.; Murray, B.G.; Kapraun, D.F.; Greilhuber, J.; Bennett, M.D. Eukaryotic genome size databases. Nucleic Acids Res. 2007, 35, 332–338. [Google Scholar] [CrossRef]
- Castoe, T.A.; Hall, K.T.; Guibotsy Mboulas, M.L.; Gu, W.; Jason De Koning, A.P.; Fox, S.E.; Poole, A.W.; Vemulapalli, V.; Daza, J.M.; Mockler, T.; et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol. Evol. 2011, 3, 641–653. [Google Scholar] [CrossRef]
- Anderson, J.L.; Rodríguez Marí, A.; Braasch, I.; Amores, A.; Hohenlohe, P.; Batzel, P.; Postlethwait, J.H. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS ONE 2012, 7, e40701. [Google Scholar] [CrossRef]
- Pan, Q.; Anderson, J.; Bertho, S.; Herpin, A.; Wilson, C.; Postlethwait, J.H.; Schartl, M.; Guiguen, Y. Vertebrate sex-determining genes play musical chairs. Comptes Rendus Biol. 2016, 339, 258–262. [Google Scholar] [CrossRef]
- English, A.C.; Salerno, W.J.; Hampton, O.A.; Gonzaga-Jauregui, C.; Ambreth, S.; Ritter, D.I.; Beck, C.R.; Davis, C.F.; Dahdouli, M.; Ma, S.; et al. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genom. 2015, 16, 286. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef]
- Schield, D.R.; Card, D.C.; Hales, N.R.; Perry, B.W.; Pasquesi, G.M.; Blackmon, H.; Adams, R.H.; Corbin, A.B.; Smith, C.F.; Ramesh, B.; et al. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019, 29, 590–601. [Google Scholar] [CrossRef]
- Lewin, H.A.; Larkin, D.M.; Pontius, J.; O’Brien, S.J. Every genome sequence needs a good map. Genome Res. 2009, 19, 1925–1928. [Google Scholar] [CrossRef] [PubMed]
- Seehausen, O.; Butlin, R.K.; Keller, I.; Wagner, C.E.; Boughman, J.W.; Hohenlohe, P.A.; Peichel, C.L.; Saetre, G.P.; Bank, C.; Brännström, Å.; et al. Genomics and the origin of species. Nat. Rev. Genet. 2014, 15, 176–192. [Google Scholar] [CrossRef]
- Chapus, C.; Edwards, S.V. Genome evolution in Reptilia: In silico chicken mapping of 12,000 BAC-end sequences from two reptiles and a basal bird. BMC Genom. 2009, 10, S8. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Fritz, A.J.; Zaidi, S.K.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Nickerson, J.A.; Imbalzano, A.N.; Stein, G.S. C-ing the Genome: A Compendium of Chromosome Conformation Capture Methods to Study Higher-Order Chromatin Organization. J. Cell Physiol. 2016, 231, 31–35. [Google Scholar] [CrossRef]
- Deschamps, S.; Zhang, Y.; Llaca, V.; Ye, L.; Sanyal, A.; King, M.; May, G.; Lin, H. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef]
- Bidon, T.; Schreck, N.; Hailer, F.; Nilsson, M.A.; Janke, A. Genome-wide search identifies 1.9 Mb from the polar bear Y chromosome for evolutionary analyses. Genome Biol. Evol. 2015, 7, 2010–2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Low, W.Y.; Tearle, R.; Koren, S.; Ghurye, J.; Rhie, A.; Phillippy, A.M.; Rosen, B.D.; Bickhart, D.M.; Smith, T.; et al. New insights into mammalian sex chromosome structure and evolution using high-quality sequences from bovine X and Y chromosomes. BMC Genom. 2019, 20, 1000. [Google Scholar] [CrossRef]
- Stevens, N.M. Studies in spermatogenesis with especial reference to the “accessory chromosome”. Carnegie Inst. Wash. Publ. 1905, 36, 1–33. [Google Scholar]
- Smit, A.; Hubley, R.; Grenn, P. RepeatMasker Open-4.0. 2015. Available online: http://www.repeatmasker.org (accessed on 20 August 2020).
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, 745–751. [Google Scholar] [CrossRef]
- Brionne, A.; Juanchich, A.; Hennequet-Antier, C. ViSEAGO: A Bioconductor package for clustering biological functions using Gene Ontology and semantic similarity. BioData Min. 2019, 12, 16. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).