Abstract
This study brings new information on major histocompatibility complex (MHC) class III sub-region genes in Old World camels and integrates current knowledge of the MHC region into a comprehensive overview for Old World camels. Out of the MHC class III genes characterized, TNFA and the LY6 gene family showed high levels of conservation, characteristic for MHC class III loci in general. For comparison, an MHC class II gene TAP1, not coding for antigen presenting molecules but functionally related to MHC antigen presenting functions was studied. TAP1 had many SNPs, even higher than the MHC class I and II genes encoding antigen presenting molecules. Based on this knowledge and using new camel genomic resources, we constructed an improved genomic map of the entire MHC region of Old World camels. The MHC class III sub-region shows a standard organization similar to that of pig or cattle. The overall genomic structure of the camel MHC is more similar to pig MHC than to cattle MHC. This conclusion is supported by differences in the organization of the MHC class II sub-region, absence of functional DY genes, different organization of MIC genes in the MHC class I sub-region, and generally closer evolutionary relationships of camel and porcine MHC gene sequences analyzed so far.
Keywords:
MHC; major histocompatibility complex; Old World camels; camels; dromedary; Bactrian camel; SNP 1. Introduction
The major histocompatibility complex (MHC) is a genomic region of critical importance for vertebrate immune functions. Class I and II MHC molecules are responsible for the presentation of peptides to T cells of intracellular and extracellular origin, respectively [1]. Different MHC molecules encoded by different alleles of multiple MHC genes can bind and present different groups of structurally similar antigenic oligopeptides. Compared to highly specific T- and B-cell receptors, MHC molecules are less specific. This phenomenon allows MHC molecules to cope with the immense diversity of protein antigens based on inherited genetic variation. Different antigen presenting molecules are encoded by different allelic variants of MHC class I and class II genes. The MHC region is characterized by many loci with immune as well as with non-immune functions. Of these, multiple loci code for antigen presenting molecules that possess many alleles and have higher heterozygosity typically than predicted by neutrality [2]. As a result, the MHC is one of the most complex and most polymorphic regions of the mammalian genome [3]. Both parasite-mediated and sexual selection shape functional MHC polymorphism in mammalian populations [4]. However, different taxonomic families, and even different species within a family, have evolved various strategies allowing them to successfully cope with the selection pressure of extremely variable pathogens and with other environmental challenges [5,6,7,8]. Therefore, further studies of mammalian species and families are needed for obtaining a comprehensive and unbiased picture of MHC function and diversity [9].
The first camelids appeared in North America, 45–40 Mya [10]. While the ancestors of New World camelids migrated to South America, the ancestors of Old World camelids migrated into Asia (around 6–7 Mya) and continued to spread further west. It is now generally accepted that the divergence between one-humped dromedary (Camelus dromedarius) and two-humped Bactrian (Camelus bactrianus) camel occurred sometime between 4 and 7 Mya [11,12]. Due to the advancements in camel evolutionary studies, it is well established that extant Old World camelids are composed of three species – Camelus dromedarius, Camelus bactrianus and Camelus ferus [11,12,13,14]. The other recent part of the Camelidae family is represented by New World camelids, which include alpaca (Vicugna pacos), vicugna (Vicugna vicugna), llama (Lama glama) and guanaco (Lama guanicoe).
Camels are an economically and socially important species for a large part of the world. Since ancient times, they have been used as animals of burden, for milk and meat production, as well as for riding and racing. They represent a sustainable livestock species with several specific features [15]. In addition to their adaptation to the arid and dusty environment of deserts, such as specialized water and fat metabolism, stress response to high temperatures, and adaptation to intense ultraviolet radiation, they are more resistant to infectious diseases when compared to other species living in the same geographical area, and they also produce milk with a high content of antimicrobial agents. [16,17,18].
While genome and transcriptome analyses reveal genes underlying the adaptations of camels to the harsh environment [11,19], another specific feature of camels, their immunogenome, has been only poorly studied so far [11,15]. Camelids are the only mammals producing heavy-chain homodimeric antibodies without a light chain and with the antigen-binding fragment reduced to a single heavy chain variable domain [20]. Similarly, camel T-cell receptors and their genes are quite unique in terms of their structure and function, especially the TR γ/δ loci. In dromedary, (Camelus dromedarius), γ/δ rearranged genes, somatic hypermutation increases repertoire diversity, very much similar to the process of affinity maturation of immunoglobulin genes [21].
In this context, it is surprising that only little attention has been paid to MHC studies not only in Old World camels but in all camelids. The first information on the existence of MHC in dromedary and alpaca genomes was provided by Antczak [22], using cytogenetic techniques. In Old and New World camelids, the MHC is located on the long arm of chromosome 20 [23,24], and the first characterization of dromedary and Bactrian MHC class II and class I genes was provided by Plasil et al. [24,25]. The most common structure of the mammalian MHC region, represented by the order MHC class II–MHC class III–MHC class I genes, was identified in camels as well [24]. The organization of the MHC genes within these regions also follows the general pattern observed in other mammalian species [25]. A low level of polymorphism of the class II genes DRA, DRB and DQA genes was observed in all three Old World camel species, as well as for class I genes [24,25]. Surprisingly, MHC class I related MR1 and MICA loci were found to be more polymorphic than a classical MHC class I locus, B-67 [25].
Considering the gaps in the so far studied general organization of the camel MHC, and based on a new dromedary genome assembly [26], the aim of this work was to characterize selected MHC class III genes, and to integrate the new and previously published knowledge into an improved overview of the MHC region with a special focus on Camelus dromedarius. For this purpose, MHC-encoded molecules with different functions were selected. TNFA was chosen as an example of a typical, well conserved MHC class III gene, while the LY6G6 genes were selected as a family of genes with a common origin and located in close physical proximity. TAP1 classified as an MHC class II gene, was selected to determine variation of gene encoding non-antigen presenting molecules, and to compare it with variation observed for the studied MHC class II [24] and MHC class III genes.
2. Materials and Methods
2.1. Analysis of Selected Non-Antigen Presenting MHC Genes
The camel DNA samples used in this study come from collections at the Research Institute of Wildlife Ecology, Vetmeduni Vienna and UPVS Brno. Bactrian camel samples were collected at three different locations in Mongolia and one sample was collected from a breeder in Austria, while dromedary samples were from Jordan, Saudi Arabia, UAE, Qatar, Sudan, Kenya, Kazakhstan and Nigeria. Numbers of analyzed camels of both species are presented in Table 1. All samples were collected commensally during veterinary procedures for previous research projects (GACR 523/09/1972; PI: P. Horin; FWF P1084-B17 and P24706-B25; PI: P. Burger).

Table 1.
List of analyzed genes and numbers of individuals of C. dromedarius and C. bactrianus.
An MHC class II gene TAP1, and MHC class III TNFA, and the LY6G6 gene family were selected for this study. All these genes were already annotated. We amplified and sequenced them as described previously for MHC class I genes [25]. Briefly, individual gene sequences were extracted from sequence resources at NCBI, as seen in Tables S1–S6. Primers were designed using either the Primer3 Webtool or NCBI PrimerBLAST in Table 2. PCR was performed according to standard protocol using either KAPA 2G Robust HS or KAPA LongRange HS (KapaBiosystems, Wilmington, DE, USA). Sequencing was performed on Illumina MiSeq platform (San Diego, CA, USA) using 500 cycles PE chemistry. Data analysis was performed as previously described in Plasil et al. [25].

Table 2.
List of primers used in amplification of selected major histocompatibility complex (MHC) genes in camels.
All SNP positions are numbered according to their respective reference sequences; for all loci, their accession numbers can be found in supplementary files.
2.2. New Annotation of the MHC Region and Phylogenetic Analyses
We used the newly available resource of the C. dromedarius genome assembly CamDro3 [26], which improves upon work published by Elbers et al. [27], for refining our previous characterization of the MHC region to produce a detailed map of all three MHC class regions [24,25]. The CamDro3 assembly is a result of upgrading the CamDro2 assembly, similarly to Elbers et al. [27], where CamDro1 assembly was upgraded to CamDro2 [28]. The CamDro2 assembly was re-scaffolded using the original Dovetail Chicago and Hi-C reads with the HiRise pipeline [29] in an attempt to fix local misassemblies. We then filled in gaps using our PacBio long-reads (SRA accession: SRP050586) [27] with PBJelly v. 15.8.24 [30] twice instead of one time, which was done for CamDro2. Instead of polishing the assembly with Pilon [31], we used a standard variant calling workflow, which increased the RNA-Seq mapping rates relative to the Pilon-polished assembly. Briefly, we first mapped, trimmed and error-corrected Illumina short-insert sequences (SRA accession: SRR2002493) [28], using BBMap v. 38.12 (https://sourceforge.net/projects/bbmap/) with the vslow and usejni settings to the PBJelly assembly. We then sorted and indexed the resulting BAM file with Sambamba v. 0.6.7 [32], and called variants with CallVariants v. 38.12 (https://sourceforge.net/projects/bbmap/). We finally used BCFtools v. 1.2 (http://samtools.github.io/bcftools/) to generate a consensus sequence, for which we filled in gaps using ABYSS Sealer v. 2.1.0 [33], using default settings except for a bloom filter size of 40 GB, and multiple K values from 90 to 20 in increments of 10.
For phylogenetic analyses of the MHC genes studied here, we included annotated sequences from other mammalian species available at NCBI, as seen in Tables S1–S6. Phylogenetic trees were constructed in MEGA7 using the maximum-likelihood (ML) approach with 1000 bootstraps and the best-fitting model was tested according to the Bayesian information criterion (BIC) [34]. I.e., the best evolutionary model for TAP1, TNFA, LY6G6E and LY6G6F followed the Tamura 3-parameter with gamma distribution (5 categories with parameters 0.5098, 0.1968, 0.5569 and 0.8816, respectively) [35]. The LY6G6C and LY6G6D trees were based on the Jukes-Cantor and Kimura 2-parameter [36,37], respectively.
We have constructed a schematic overview of the MHC regions of selected species to compare their general similarities as well as to summarize evolutionary relationships between individual genes. For this purpose, we used the above-mentioned dromedary assembly CamDro3 as well as assemblies of the following species: Cattle (ARS-UCD1.2 (GCF_002263795.1)), pig (Sscrofa11.1 (GCF_000003025.6)), dog (CanFam3.1 (GCF_000002285.3)), human (GRCh38.p12 (GCF_000001405.38)), and data from our previous studies [24,25,38].
We have also prepared a synteny plot to directly compare the MHC regions of camelids. We lifted over NCBI annotations (release 101) for the V. pacos assembly GCF_000164845.2 to the DNA Zoo V. pacos assembly (https://www.dnazoo.org/assemblies/Vicugna_pacos) with flo (https://github.com/wurmlab/flo). We then extracted the MHC region (sequences and annotations between genes RXRB and TRIM27) for the DNA Zoo V. pacos assembly and the MHC regions for C. dromedarius (CamDro3 assembly), C. bactrianus (CamBac2 assembly, GCF_000767855.1 scaffolded with CamDro3), and C. ferus genomes (CamFer2 assembly, GCF_000311805.1 scaffolded with CamDro3) [26]. Annotations for CamDro3, CamBac2, and CamFer2 were generated with MAKER v. 2.31.10 following an approach similar to Elbers et al. [27,39,40]. We generated a synteny plot of the MHC regions for V. pacos (DNA Zoo V. pacos), C. bactrianus (CamBac2), C. dromedarius (CamDro3), and C. ferus (CamFer2) with Synima [41].
3. Results
3.1. The MHC Class II Gene TAP 1
Compared to the available annotated coding sequences (CDSs) of Old World camels (XM_010994582.1:25-2265), one substitution was found in the CDS of the TAP1 gene. Out of the total of 19 SNPs within the CDS, 12 were non-synonymous and 7 were shared between Camelus bactrianus and Camelus dromedaries, as seen in Table S7, Figure S1. We detected a discrepancy between the here developed TAP1 CDS and the sequences available at NCBI, as seen in Table S1. While the alignment of the published dromedary and Bactrian camel sequences showed significant differences between sequence positions 520-585 of the CDS, we could not confirm them in our sequences as all analysed sequences bear close similarity to the sequence of C. dromedaries, as seen in Figure S2. A potential deletion between positions 265 and 306 bp was identified in the CDS of C. ferus obtained from NCBI, as seen in Table S1.
A phylogenetic analysis of the camel TAP1 sequences showed high level of conservation within Camelidae. Despite its lower bootstrap support, it suggested a relatively close evolutionary relationship with the domestic pig, Sus scrofa, as seen in Figure 1. Additional information regarding the sequence similarity of the analyzed TAP1 CDSs is available in the Table S1.
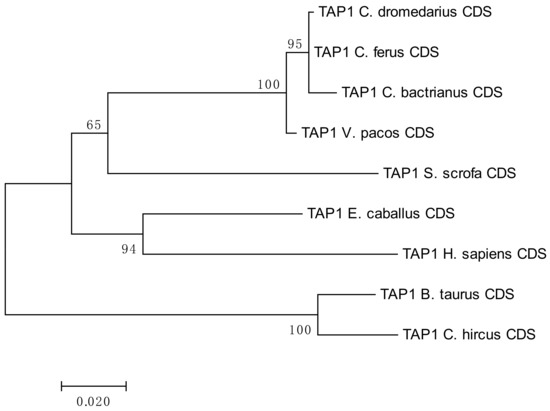
Figure 1.
Phylogenetic tree of selected TAP1 coding sequences (CDSs). The tree is based on the maximum-likelihood method using Tamura 3-parameter model with gamma distribution (five categories, parameter = 0.5098). Number of bootstraps = 1000.
3.2. MHC Class III Genes
3.2.1. The TNFA Gene
The three SNPs found within the TNFA CDS in exon 2 (1 non-synonymous) and exon 3 (1 synonymous, 1 non-synonymous) were shared between C. bactrianus and C. dromedaries, as seen in Figure S3. The haplotypes were conserved across the entire CDS, i.e., the three SNPs were either in homozygous or heterozygous constitution, and the frequency of heterozygote individuals was 0.48 and 0.27 in dromedaries and Bactrian camels, respectively. A comparison with other publicly available camelid sequences did not reveal any additional potential SNPs, as seen in Figure S4. The phylogenetic tree showed an overall high level of conservation within Camelidae, and closer evolutionary relationships with sequences from cattle and goat, as seen in Figure 2. Additional information regarding sequence similarity of the analyzed TNFA CDSs is available in the Table S2.
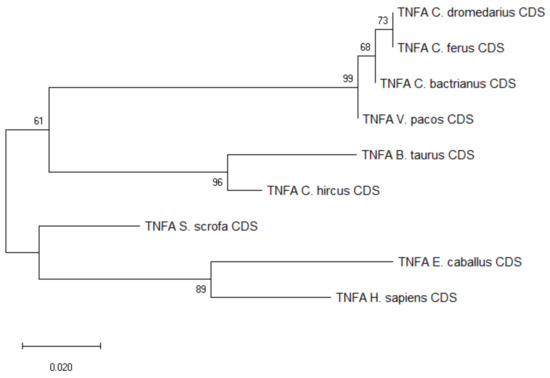
Figure 2.
Phylogenetic tree of the TNFA CDSs from selected species. The maximum-likelihood method was used based on the Tamura 3-parameter model with gamma distribution (five categories, parameter = 0.1968). The number of bootstraps was 1000.
3.2.2. The LY6G6 Gene Family
Four genes of this family, LY6G6C, LY6G6D, LY6G6E, and LY6G6F were analyzed and have been annotated in the three species of Old World camelids. While we discovered only one non-synonymous SNP within the LY6G6C CDS at position 218 (G/A) present in C. dromedaries, as seen in Figure S5, the sequences retrieved from NCBI showed another non-synonymous SNP at position 235 (C/A) as seen in Figure S6. However, this SNP was not confirmed by our dataset. The frequency of heterozygotes for the LY6G6C SNP 218 in C. dromedarius was 0.2. The CDS of the LY6G6D contained one synonymous SNP (G/T at position 405) observed only in C. bactrianus, as seen in Figure S7. The frequency of heterozygotes for this SNP was 0.2 in C. bactrianus. Comparison with other available sequences from NCBI did not reveal any additional SNPs in Old World camelids, as seen in Figure S8.
In the family Camelidae, the LY6G6E gene has been annotated as sperm acrosome membrane-associated protein 4-like. Figure S9 shows our analyses, as well as sequences retrieved from NCBI, that the LY6G6E CDSs were monomorphic in the subfamily Camelini. However, a comparison with the CDS of Vicugna pacos (Lamini) revealed two SNP positions (319 T/C and 358 G/A), where the latter was non-synonymous. Comparison with the CDS of Bos taurus showed that the CDSs compared were not of the same length.
One synonymous (A/G, at position 150) and one non-synonymous SNP (C/T, at position 659) were identified in the CDS of LY6G6F, both shared between C. bactrianus and C. dromedaries, as seen in Figure S10. The frequencies of heterozygotes in C. bactrianus and C. dromedarius, for both SNPs were 0.3 and 0.5, respectively. Individuals of both species were either heterozygous or homozygous in both SNPs, but never homozygous for one and heterozygous for the other. Comparison of the sequences available on NCBI confirmed both SNPs, as seen in Figure S11.
The results of the phylogenetic analyses summarized in Figure 3, Figure 4, Figure 5 and Figure 6 document a high level of conservation within the LY6G6 family CDSs in the Camelidae. LY6G6C showed the closest evolutionary relationship with the sequence of Sus scrofa, while LY6G6D was closer to Bos taurus and Capra hircus. LY6G6E was close to sequences of all other included Cetartyodactyla Sus scrofa, Bos taurus and Capra hircus, and LY6G6F was also closest to Sus scrofa. In humans, LY6G6E has a status of pseudogene. Sequence similarities of CDSs used for constructing the phylogenetic trees are provided in Tables S3–S6.
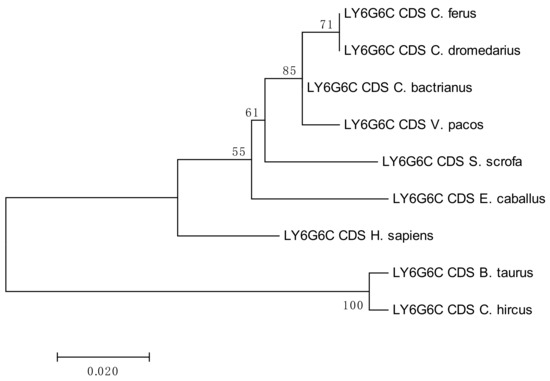
Figure 3.
Phylogenetic tree of the LY6G6C CDSs from selected species. The maximum-likelihood method was used based on the Jukes-Cantor model. The number of bootstraps was 1000.
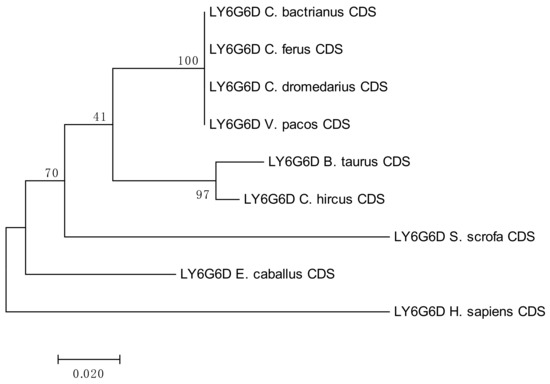
Figure 4.
Phylogenetic tree of the LY6G6D CDSs from selected species. The maximum-likelihood method was used based on the Kimura 2-parameter model. The number of bootstraps was 1000.
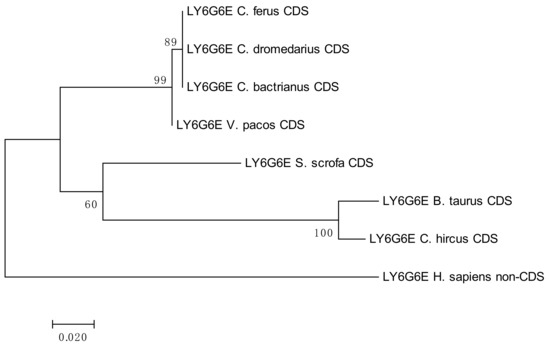
Figure 5.
Phylogenetic tree of the LY6G6E CDSs from selected species. The maximum-likelihood method was used based on the Tamura 3-parameter model with gamma distribution (five categories, parameter = 0.5569). The number of bootstraps was 1000.
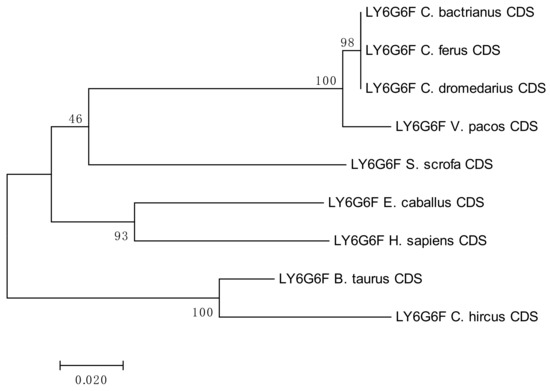
Figure 6.
Phylogenetic tree of the LY6G6F CDSs from selected species. The maximum-likelihood method was used based on the Tamura 3-parameter model with gamma distribution (five categories, parameter = 0.8816). The number of bootstraps was 1000.
3.3. New Annotation of the MHC Region in the Dromedary
A new version of the organization of the MHC region in the dromedary is shown in Figure 7. A complete annotation record will be available in Lado et al. [26]. A graphical summary of the evolutionary relationships for the genes studied is presented in Figure 8. The synteny plot for available camelid MHC regions is shown in Figure 9.
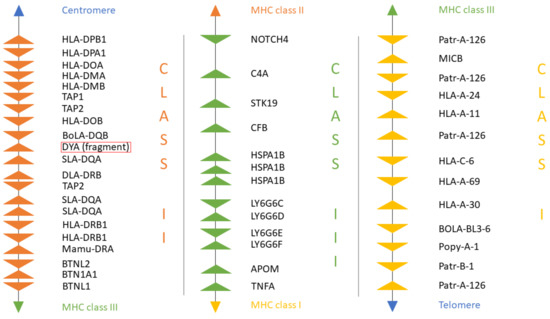
Figure 7.
The organization of the MHC region in C. dromedarius. MHC class II genes are shown in orange. MHC class III genes are shown in green. MHC class I genes are shown in yellow. Spaces between genes do not represent real distances. Details of the annotation are available in Table S8.
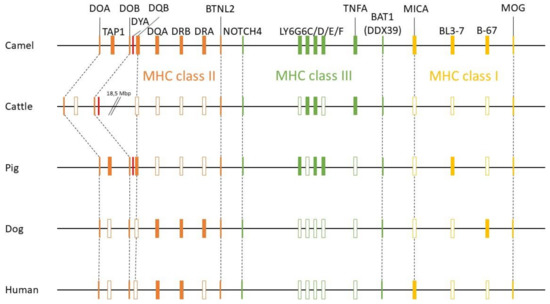
Figure 8.
Summary of evolutionary relationships of MHC genes of dromedary, cattle, pig, dog and human. Relative gene positions are presented as rectangles. The filled rectangles highlight the closest relationships observed between the dromedary and the respective species for a given locus within the groups studied.
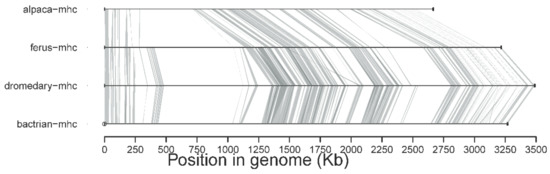
Figure 9.
The synteny plot of the camelid MHC region showing a general structural conservation among all camelids. Assemblies of the dromedary (CamDro3) [26]; Bactrian (CamBac2, GCF_000767855.1 scaffolded with CamDro3 as a reference); wild Bactrian (CamFer2, GCF_000311805.1 scaffolded with CamDro3 as a reference); and alpaca (DNA Zoo V. pacos) were used.
4. Discussion
This study brings new information on MHC class III sub-region genes in Old World camels. These data, along with new camel genomic resources, allowed us to construct an improved genomic map of the entire MHC region of Old World camels.
The MHC class III region, where no genes coding for antigen presenting molecules are located, represented a gap in our knowledge of the camel MHC. For bridging this gap in the map, we selected different types of class III genes with diverse immunological functions. Based on this, we could make a comparison of their variability with so far known genes from other MHC regions previously studied in the same groups of dromedary and Bactrian camels [24,25]. The data then primarily served for allowing us an overall characterization of the camelid MHC. As such, they do not represent a detailed analysis of the MHC class III region, which still is our pending task.
The TNFA (tumor necrosis factor alpha) is a cytokine playing a major role in the activation of both acute and chronical inflammatory processes, and contributing to the activation cascade of mother cytokines and chemokines connected with inflammation [42]. Besides this role, the TNFA also contributes to the regulation of apoptosis and carcinogenesis [43,44]. While there are no reports on the role of TNF-alpha and/or of the TNFA gene in diseases of camels, in cattle, a closely-related species, TNFA polymorphisms have been associated with multiple infectious diseases and with fertility [45,46,47,48]. The SNPs identified in this report thus represent potentially useful markers for analyzing various camel diseases. TNFA, subject to strong purifying selection [42], may be also used as a marker of a conserved part of the MHC in close physical proximity of MHC genes under positive selection pressure [48].
The LY6G6 (lymphocyte antigen 6 family, member G6) gene family is located within the MHC class III region, spanning approximately 20 kb. In humans (assembly GRCh38.p12) and mice (assembly GRCm38.p4), the family contains four individual genes, LY6G6C, LY6G6D, LY6G6E, and LY6G6F. LY6 proteins containing cysteine-rich domain are attached to the cell surface by a GPI anchor, which is involved in signal transduction. LY6 proteins might play a role in the hematopoietic differentiation [49].
This is a first report on the existence of this group of MHC class III genes in camels. Although little is currently known about their functions, they represent suitable anchor and marker loci for the MHC class III region. Sequences of all four selected genes showed high levels of conservation within Old World camels. SNPs were rather rare and were often observed in only one camel species; LY6G6E was even completely monomorphic in both species. Interestingly, while the LY6G6E sequence has the status of pseudogene in humans, we did not find any features of non-functional LY6G6E sequences in the two camel species. These findings are in agreement with those of other closely related mammals, such as Bos taurus and Sus scrofa, suggesting that this gene is probably functionally important for the Cetartiodactyla.
Having characterized selected MHC class III genes in Old World camels, we were able to build upon our previous studies of MHC class I and MHC class II regions and to update the general organization of the MHC region, describe its similarities with New World camelids, as well as with relevant mammalian species, and to point out its unique features.
The MHC class III genes studied here do not encode antigen presenting molecules and are not involved in the mechanisms of antigen presentation. For comparison, we chose TAP1, a gene not coding for antigen presenting molecules, but playing an important role in the process of antigen loading of the MHC class I molecules, located in the MHC class II region [50]. Certain TAP1 variants may have a large impact on MHC class I expression and interaction with pathogens, at least in some species [51,52]. While TNFA and the gene family LY6 showed high levels of conservation characteristic for MHC class III loci in general, the camel TAP1 was not only polymorphic, but the numbers of SNPs detected were higher than in the MHC class I and class II genes examined so far [24,25].
4.1. MHC Region Organization and Diversity in Old World Camels
The presented MHC organization is primarily based on dromedary sequences, but due to the high conservation and similarity of this region between dromedaries and Bactrian camels, we can take firm conclusions about the latter’s MHC. The overall nucleotide and amino acid sequence similarities between the two species ranged across the MHC region (delimited by the genes TRIM27 and RXRB) on chromosome 20 of the new dromedary assembly CamDro3 [26], with no major differences between the three sub-regions, as seen in Figure 9. Extensive trans-species polymorphism (TSP), i.e., allele sharing, is also a typical feature of the MHC of camelids, including llamas [24,25].
The diversity of the MHC class I, class I–related and class II genes is generally lower than expected, based on a comparative analysis of multiple vertebrate species [53]. This finding is in agreement with a lower genome-wide diversity in dromedaries than in the wild and domestic Bactrian camels [28]. We were unable to make a reliable estimate of polymorphisms of class II DQB genes. Their exon 2 encoded amino acid sequences containing the antigen-binding site are by four amino-acids longer due to a 12 bp insertion in both camel species. They contain multiple SNPs, but we were unable to identify individual loci [24]. It is possible that certain specificities in DQB sequences, such as GC rich regions and/or long repeats cause technical problems as described, e.g., for the horse DQB region [54]. Interestingly, some of the genes encoding other than antigen presenting molecules and located in the MHC class I and II regions, such as MICA and TAP1, were found to have higher polymorphism than classical MHC class I and II genes. On the other hand, the class II DRA gene, monomorphic in most mammalian species, is polymorphic both in dromedaries and Bactrian camels [24]. Recently identified MHC-linked microsatellite loci also showed unusually low diversity both in dromedaries and bactrians, often with only two or three alleles [55]. Bottlenecks experienced in the evolutionary history of the three species, and also in recent times due to domestication, might be the cause of the reduced genome-wide variability in dromedaries. However, it is not clear to what extent these processes contributed to the low diversity of the immunogenome. Additionally, the reasons for higher numbers of SNPs observed in genes coding for other than antigen presenting MHC molecules remains especially puzzling.
4.2. Cross-Species Comparisons with New World Camelids and Other Mammals
A direct comparison with alpaca sequences presented here, as well as in our previous work [24,25], showed that the studied loci are generally highly conserved in camelids. Based on the data available so far, as seen in Figure 9, it seems that the organization of the MHC genomic region in Vicugna pacos is highly similar if not nearly identical to Old World camels.
When compared to other species, the physical location and the overall organization of the MHC in camels follows the common general structure of the mammalian MHC region [24]. However, phylogenetic relationships of camel MHC genes do not always follow relationships based on other (neutral) nuclear genes [24,25,38,56], suggesting that different MHC sub-regions might have followed different evolutionary pathways. In addition, so far, we have been unable to identify a DYA locus, specific for the Bovidae, the phylogenetically closest family. Although DYA-like sequences were previously annotated in camelid genomes, as seen in Figure 7, a more focused analysis showed that they are more similar to DQA rather than to DYA sequences, and that it was impossible to identify this region within the camel MHC physical map [38]. Like in camels, the porcine MHC class II region contains a non-functional fragment of DY gene (GenBank Gene ID: 100135048), although it is annotated as DYB, while the camel sequence is annotated as DYA. Moreover, in cattle, the physical position of the TAP1 gene is different from other mammalian species, including camels, as seen in Figure 8. Overall, the organization of the camel MHC class II region is more similar to Sus scrofa rather than to Bos taurus.
Nucleotide sequences of genes located in the MHC class II sub-region are generally closer to dog and human, with the exception of DQB and TAP1, which are closely related to their porcine counterparts, as seen in Figure 7 [38]. On the other hand, the MHC class III genes and the LY6G6 gene family, are more related to the corresponding porcine sequences, with the exception of LY6G6D and TNFA, which are closer to the bovine sequences than to its porcine counterpart. Despite a probably common origin and close physical proximity of the LY6G6 gene family, the LY6G6D does not behave identically in phylogenetic trees, as seen in Figure 3, Figure 4, Figure 5 and Figure 6 [57]. It thus seems that different MHC sub-regions have not always the same evolutionary history, reflecting probably the fact that the selection pressures exerted on the immune system are different between camels and cattle.
The evolution of the MHC class I sub-region appears to be even more complex, as shown in our previous study [25]. The MHC class I locus B-67 is closely related to the canine locus DLA-88, the locus BL3-7 is related to the porcine SLA-11, while MICA is closest to the relevant human sequences. While the camel MIC genes follow the pattern of the human MICA/B genes, cattle have at least three functional MIC genes with unique sequences [25,58]. In contrast to these differences between camel and cattle, high similarities with the pig MHC class I genes were often observed. Especially, the MHC class I gene BL3-7, a locus of unclear status, highly similar to the annotated sequence BL3-6 in alpacas, is also closely related to the locus SLA-11 in pig, and is one MHC locus with unknown function and unusual structure [25,59]. Interestingly, we have also found close similarities between camels and pigs for two other complex immunogenomic regions, the natural killer complex (NKC) and the leukocyte receptor complex (LRC), encoding natural killer cell receptor molecules. Both regions differ from their cattle counterparts, resembling more the pig NKC and LRC, respectively [60]. However, in this context, it is important to note that the annotation of MHC class I loci from CamDro3, as presented in Figure 7, is based on calculations of their sequence similarities with other species. Taking into consideration that sequence similarities alone are often not informative for a correct assignment of their classical or non-classical status in non-model organisms, we do not have enough information for definitive conclusions about this status for class I genes in camels either.
5. Conclusions
This first characterization of specific MHC class III genes added to the general structure and variability of the MHC in Old World camels, allowing a synopsis of the current knowledge of the MHC region of Old World camelids. Novel phylogenetic analyses between camelids and other domestic mammals showed high conservation of this region among Old and New World camels, and a closer evolutionary relationship with pigs rather than with cattle. The knowledge of the MHCs structure, function and evolution in camels is important for understanding their immune response to diseases in their specific and extreme environments.
Supplementary Materials
The supplementary materials are available online at https://www.mdpi.com/2073-4409/8/10/1200/s1.
Author Contributions
M.P.: investigation, methodology, formal analysis, validation, visualization, writing – original draft, writing – review & editing; S.W.: investigation, formal analysis; J.P.E.: data curation, formal analysis, visualization, writing – review & editing; J.O.: formal analysis; P.A.B.: conceptualization, funding acquisition, project administration, resources, writing – review & editing; P.H.: conceptualization, funding acquisition, project administration, supervision, writing – original draft, writing – review & editing.
Funding
This work was supported by the Austrian Science Foundation (FWF) project P29623-B25 and by CEITEC - Central European Institute of Technology with research infrastructure supported by the project CZ.1.05/1.1.00/02.0068 financed from European Regional Development Fund. J.P. Elbers is funded by the FWF project P29623-B25 granted to P.A. Burger.
Acknowledgments
We thank all camel owners and veterinarian colleagues for their kind cooperation during sample collection, O. Abdelhadi, A. Abdussamad, H. Burgsteiner, B. Faye, G. Gassner, J. Juhasz, G. Konuspayeva, D. Modry, P. Nagy, A. and J. Perret, M. Qablan and R. Saleh. Open Access Funding by the Austrian Science Fund (FWF).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Janeway, C.A.; Travers, P.; Walport, M.; Shlomchik, M.J. Immunobiology: The Immune System in Health and Disease, 6th ed.; Taylor & Francis Group, Garland Science: New York, NY, USA, 2005. [Google Scholar]
- Hedrick, P.W.; Whittam, T.S.; Parham, P. Heterozygosity at individual amino acid sites: Extremely high levels for HLA-A and-B genes. Proc. Natl. Acad. Sci. USA 1991, 88, 5897–5901. [Google Scholar] [CrossRef] [PubMed]
- Kumánovics, A.; Takada, T.; Lindahl, K.F. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 2003, 21, 629–657. [Google Scholar] [CrossRef] [PubMed]
- Winternitz, J.C.; Minchey, S.G.; Garamszegi, L.Z.; Huang, S.; Stephens, P.R.; Altizer, S. Sexual selection explains more functional variation in the mammalian major histocompatibility complex than parasitism. Proc. Biol. Sci. 2013, 280, 20131605. [Google Scholar] [CrossRef] [PubMed]
- Rocha, R.G.; Magalhães, V.; López-Bao, J.V.; van der Loo, W.; Llaneza, L.; Alvares, F.; Esteves, P.J.; Godinho, R. Alternated selection mechanisms maintain adaptive diversity in different demographic scenarios of a large carnivore. BMC Evol. Biol. 2019, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Roemer, G.; Debenham, S.; Binns, M.; Garcelon, D.; Wayne, R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl. Acad. Sci. USA 2004, 101, 3490–3494. [Google Scholar] [CrossRef]
- Mikko, S.; Røed, K.; Schmutz, S.; Andersson, L. Monomorphism and polymorphism at Mhc DRB loci in domestic and wild ruminants. Immunol. Rev. 1999, 167, 169–178. [Google Scholar] [CrossRef]
- Doxiadis, G.G.; Otting, N.; de Groot, N.G.; Bontrop, R.E. Differential evolutionary MHC class II strategies in humans and rhesus macaques: Relevance for biomedical studies. Immunol. Rev. 2001, 183, 76–85. [Google Scholar] [CrossRef]
- Bernatchez, L.; Landry, C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 2003, 16, 363–377. [Google Scholar] [CrossRef]
- Burger, P.A.; Ciani, E.; Faye, B. Old World camels in a modern world—A balancing act between conservation and genetic improvement. Anim. Genet. 2019, in press. [Google Scholar] [CrossRef]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef]
- Ji, R.; Cui, P.; Ding, F.; Geng, J.; Gao, H.; Zhang, H.; Yu, J.; Hu, S.; Meng, H. Monophyletic origin of domestic bactrian camel (Camelus bactrianus) and its evolutionary relationship with the extant wild camel (Camelus bactrianus ferus). Anim. Genet. 2009, 40, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Silbermayr, K.; Orozco-terWengel, P.; Charruau, P.; Enkhbileg, D.; Walzer, C.; Vogl, C.; Schwarzenberger, F.; Kaczensky, P.; Burger, P.A. High mitochondrial differentiation levels between wild and domestic Bactrian camels: A basis for rapid detection of maternal hybridization. Anim. Genet. 2010, 41, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Sequencing, T.B.C.G.; Analysis Consortium. Genome sequences of wild and domestic bactrian camels. Nat. Commun. 2012, 3, 1202. [Google Scholar] [CrossRef]
- Burger, P.A. The history of Old World camelids in the light of molecular genetics. Trop. Anim. Health Pro. 2016, 48, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Wernery, U.; Kinne, J. Foot and mouth disease and similar virus infections in camelids: A review. Rev. Sci. Tech. Oie 2012, 31, 907–918. [Google Scholar] [CrossRef]
- Dirie, M.F.; Abdurahman, O. Observations on little known diseases of camels (Camelus dromedarius) in the Horn of Africa. Rev. Sci. Tech. Oie 2003, 22, 1043–1050. [Google Scholar] [CrossRef]
- Al Kanhal, H.A. Compositional, technological and nutritional aspects of dromedary camel milk. Int. Dairy J. 2010, 20, 811–821. [Google Scholar]
- Ali, A.; Baby, B.; Vijayan, R. Camel Genome-from Desert to Medicine. Front. Genet. 2019, 10, 17. [Google Scholar] [CrossRef]
- Muyldermans, S. Single domain camel antibodies: Current status. Rev. Mol. Biotech. 2001, 74, 277–302. [Google Scholar] [CrossRef]
- Ciccarese, S.M.; Burger, P.; Ciani, E.; Castelli, V.; Linguiti, G.; Plasil, M.; Massari, S.; Horin, P.; Antonacci, R. The camel adaptive immune receptors repertoire as a singular example of structural and functional genomics. Front. Genet. 2019. Under review. [Google Scholar] [CrossRef]
- Antczak, D. Major histocompatibility complex genes of the dromedary camel. In Proceedings of the Qatar Foundation Annual Research Conference, Doha, Qatar, 24–25, November, 2013; Hamad bin Khalifa University Press (HBKU Press): Doha, Qatar, 2013; p. BIOP015. [Google Scholar] [CrossRef]
- Avila, F.; Baily, M.P.; Perelman, P.; Das, P.J.; Pontius, J.; Chowdhary, R.; Owens, E.; Johnson, W.E.; Merriwether, D.A.; Raudsepp, T. A comprehensive whole-genome integrated cytogenetic map for the alpaca (Lama pacos). Cytogenet. Genome Res. 2014, 144, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Plasil, M.; Mohandesan, E.; Fitak, R.R.; Musilova, P.; Kubickova, S.; Burger, P.A.; Horin, P. The major histocompatibility complex in Old World camelids and low polymorphism of its class II genes. BMC Genomics 2016, 17, 167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Plasil, M.; Wijkmark, S.; Elbers, J.P.; Oppelt, J.; Burger, P.; Horin, P. The major histocompatibility complex of Old World camelids: Class I and class I-related genes. HLA 2019, 93, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Lado, S.; Elbers, J.P.; Rogers, M.F.; Perelman, P.L.; Proskuryakova, A.A.; Serdyukova, N.A.; Johnson, W.E.; Horin, P.; Corander, J.; Murphy, D.; et al. Reference-guided assembly of two Old World camel genomes and genomic diversity of Old World camelid immune response genes. Manuscript in preparation.
- Elbers, J.P.; Rogers, M.F.; Perelman, P.L.; Proskuryakova, A.A.; Serdyukova, N.A.; Johnson, W.E.; Horin, P.; Corander, J.; Murphy, D.; Burger, P.A. Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol. Ecol. Resour. 2019, 19, 1015–1026. [Google Scholar] [CrossRef]
- Fitak, R.R.; Mohandesan, E.; Corander, J.; Burger, P.A. The de novo genome assembly and annotation of a female domestic dromedary of North African origin. Mol. Ecol. Resour. 2016, 16, 314–324. [Google Scholar] [CrossRef]
- Putnam, N.H.; O’Connell, B.L.; Stites, J.C.; Rice, B.J.; Blanchette, M.; Calef, R.; Troll, C.J.; Fields, A.; Hartley, P.D.; Sugnet, C.W. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016, 26, 342–350. [Google Scholar] [CrossRef]
- English, A.C.; Richards, S.; Han, Y.; Wang, M.; Vee, V.; Qu, J.; Qin, X.; Muzny, D.M.; Reid, J.G.; Worley, K.C. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 2012, 7, e47768. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+ C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. Mammal. Prot. Metab. 1969, 3, 132. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Plasil, M. Comparative genomics of the major histocompatibility complex MHC. Ph.D. Thesis, Masaryk University, Brno, Czech Republic, 26 October 2018. [Google Scholar]
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 2011, 12, 491. [Google Scholar] [CrossRef]
- Farrer, R.A. Synima: A Synteny imaging tool for annotated genome assemblies. BMC Bioinform. 2017, 18, 507. [Google Scholar] [CrossRef]
- Chu, W.M. Tumor necrosis factor. Cancer Lett. 2013, 328, 222–225. [Google Scholar] [CrossRef]
- Pan, S.; An, P.; Zhang, R.; He, X.; Yin, G.; Min, W. Etk/Bmx as a tumor necrosis factor receptor type 2-specific kinase: Role in endothelial cell migration and angiogenesis. Mol. Cell. Biol. 2002, 22, 7512–7523. [Google Scholar] [CrossRef]
- Odbileg, R.; Konnai, S.; Ohashi, K.; Onuma, M. Molecular cloning and phylogenetic analysis of inflammatory cytokines of Camelidae (llama and camel). J. Vet. Med. Sci. 2005, 67, 921–925. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ranjan, S.; Bhushan, B.; Panigrahi, M.; Kumar, A.; Deb, R.; Kumar, P.; Sharma, D. Association and expression analysis of single nucleotide polymorphisms of partial tumor necrosis factor alpha gene with mastitis in crossbred cattle. Anim. Biotechnol. 2015, 26, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Lendez, P.A.; Passucci, J.A.; Poli, M.A.; Gutierrez, S.E.; Dolcini, G.L.; Ceriani, M.C. Association of TNF-α gene promoter region polymorphisms in bovine leukemia virus (BLV)-infected cattle with different proviral loads. Arch. Virol. 2015, 160, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Aoki, Y.; Magata, F.; Miyamoto, A.; Kawashima, C.; Hojo, T.; Okuda, K.; Shirasuna, K.; Shimizu, T. The effect of single nucleotide polymorphisms in the tumor necrosis factor-α gene on reproductive performance and immune function in dairy cattle. J. Reprod. Develop. 2014, 60, 173–178. [Google Scholar] [CrossRef]
- Seitzer, U.; Gerdes, J.; Müller-Quernheim, J. Genotyping in the MHC locus: Potential for defining predictive markers in sarcoidosis. Resp. Res. 2001, 3, 6. [Google Scholar] [CrossRef]
- Mallya, M.; Campbell, R.D.; Aguado, B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein. Sci. 2006, 15, 2244–2256. [Google Scholar] [CrossRef]
- Trowsdale, J.; Hanson, I.; Mockridge, I.; Beck, S.; Townsendt, A.; Kelly, A. Sequences encoded in the class II region of the MHC related to the ’ABC’ superfamily of transporters. Nature 1990, 348, 741. [Google Scholar] [CrossRef]
- Kaufman, J. Co-evolution with chicken class I genes. Immunol. Rev. 2015, 267, 56–71. [Google Scholar] [CrossRef]
- Praest, P.; Luteijn, R.D.; Brak-Boer, I.G.J.; Lanfermeijer, J.; Hoelen, H.; Ijgosse, L.; Costa, A.I.; Gorham, R.D.; Lebbink, R.J.; Wiertz, E. The influence of TAP1 and TAP2 gene polymorphisms on TAP function and its inhibition by viral immune evasion proteins. Mol. Immunol. 2018, 101, 55–64. [Google Scholar] [CrossRef]
- Kulski, J.K.; Shiina, T.; Anzai, T.; Kohara, S.; Inoko, H. Comparative genomic analysis of the MHC: The evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol. Rev. 2002, 190, 95–122. [Google Scholar] [CrossRef]
- Viļuma, A.; Mikko, S.; Hahn, D.; Skow, L.; Andersson, G.; Bergström, T.F. Genomic structure of the horse major histocompatibility complex class II region resolved using PacBio long-read sequencing technology. Sci. Rep. 2017, 7, 45518. [Google Scholar] [CrossRef] [PubMed]
- Wijacki, J.; (Department of Animal Morphology, Physiology and Genetics, Mendel University, Brno, Czech Republic). Personal communication, 2019.
- Wang, Q.; Yang, C. The phylogeny of the Cetartiodactyla based on complete mitochondrial genomes. Int. J. Biol. 2013, 5, 30. [Google Scholar] [CrossRef][Green Version]
- Mallya, M.; Campbell, R.D.; Aguado, B. Transcriptional analysis of a novel cluster of LY-6 family members in the human and mouse major histocompatibility complex: Five genes with many splice forms. Genomics 2002, 80, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.; Sanjuan, C.D.J.; Guzman, E.; Ellis, S.A. Genomic location and characterisation of MIC genes in cattle. Immunogenetics 2008, 60, 477–483. [Google Scholar] [CrossRef]
- Renard, C.; Vaiman, M.; Chiannilkulchai, N.; Cattolico, L.; Robert, C.; Chardon, P. Sequence of the pig major histocompatibility region containing the classical class I genes. Immunogenetics 2001, 53, 490–500. [Google Scholar] [CrossRef]
- Futas, J.; Oppelt, J.; Jelinek, A.; Elbers, J.P.; Wijacki, J.; Knoll, A.; Burger, P.A.; Horin, P. Natural killer cell receptor genes in camels: Another mammalian model. Front. Genet. 2019, 10, 620. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).