Exome Study of Single Nucleotide Variations in Patients with Syndromic and Non-Syndromic Autism Reveals Potential Candidate Genes for Diagnostics and Novel Single Nucleotide Variants
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
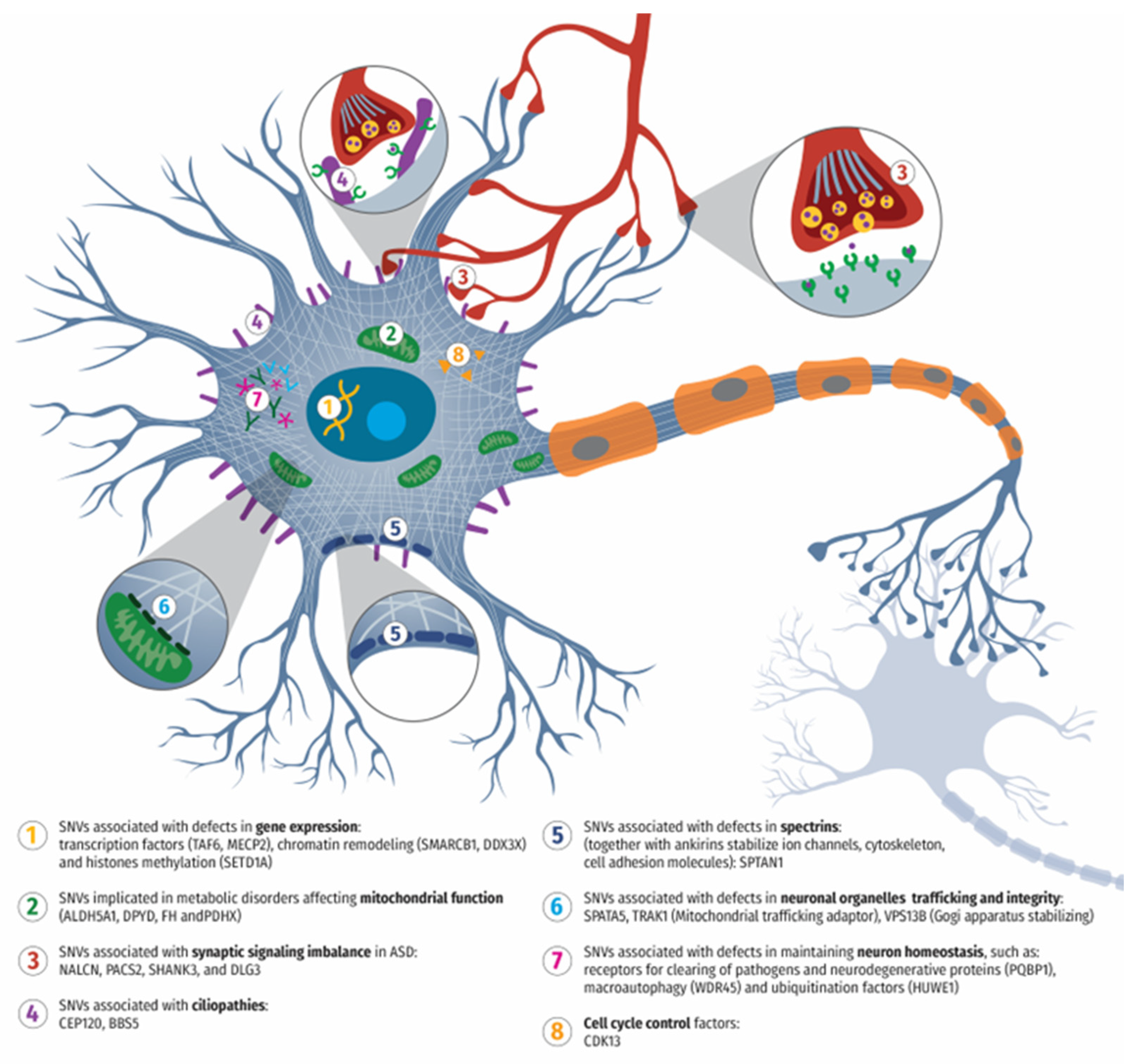
4.1. SNVs Associated with Synaptic Structure, Function, and Signaling Imbalance of Neurons in ASD
4.2. SNVs Implicated in Mitochondrial Dysfunction and ASD
4.3. SNVs Associated with ASD and Defects in Gene Expression: Transcription Factors, Chromatin Remodeling, and Histone Methylation
4.4. SNVs Associated with ASD and Cell Cycle Regulation, Ciliopathies, and Spectrin Function
4.5. SNVs Associated with ASD and Affecting Neuronal Organelle Trafficking and Homeostasis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hirota, T.; King, B.H. Autism Spectrum Disorder: A Review. JAMA 2023, 329, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Hodges, H.; Fealko, C.; Soares, N. Autism spectrum disorder: Definition, epidemiology, causes, and clinical evaluation. Transl. Pediatr. 2020, 9, S55–S65. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Brugha, T.S.; Charman, T.; Cusack, J.; Dumas, G.; Frazier, T.; Jones, E.J.H.; Jones, R.M.; Pickles, A.; State, M.W.; et al. Autism spectrum disorder. Nat. Rev. Dis. Primers 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Genovese, A.; Butler, M.G. The Autism Spectrum: Behavioral, Psychiatric and Genetic Associations. Genes 2023, 14, 677. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders; American Psychiatric Association: Arlington, TX, USA, 2022. [Google Scholar]
- Durkin, M.S. Increasing Prevalence of Developmental Disabilities Among Children in the US: A Sign of Progress? Pediatrics 2019, 144, e20192005. [Google Scholar] [CrossRef]
- Casanova, M.F.; Casanova, E.L.; Frye, R.E.; Baeza-Velasco, C.; LaSalle, J.M.; Hagerman, R.J.; Scherer, S.W.; Natowicz, M.R. Editorial: Secondary vs. Idiopathic Autism. Front. Psychiatry 2020, 11, 297. [Google Scholar] [CrossRef]
- Horecka-Lewitowicz, A.; Lewitowicz, W.; Wawszczak-Kasza, M.; Lim, H.; Lewitowicz, P. Autism Spectrum Disorder Pathogenesis-A Cross-Sectional Literature Review Emphasizing Molecular Aspects. Int. J. Mol. Sci. 2024, 25, 11283. [Google Scholar] [CrossRef]
- Havdahl, A.; Niarchou, M.; Starnawska, A.; Uddin, M.; van der Merwe, C.; Warrier, V. Genetic contributions to autism spectrum disorder. Psychol. Med. 2021, 51, 2260–2273. [Google Scholar] [CrossRef]
- Zafeiriou, D.I.; Ververi, A.; Vargiami, E. Childhood autism and associated comorbidities. Brain Dev. 2007, 29, 257–272. [Google Scholar] [CrossRef]
- Weuring, W.; Geerligs, J.; Koeleman, B.P.C. Gene Therapies for Monogenic Autism Spectrum Disorders. Genes 2021, 12, 1667. [Google Scholar] [CrossRef]
- Sztainberg, Y.; Zoghbi, H.Y. Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci. 2016, 19, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Moss, J.; Howlin, P. Autism spectrum disorders in genetic syndromes: Implications for diagnosis, intervention and understanding the wider autism spectrum disorder population. J. Intellect. Disabil. Res. 2009, 53, 852–873. [Google Scholar] [CrossRef] [PubMed]
- The Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015, 519, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Pourtavakoli, A.; Hussen, B.M.; Taheri, M.; Ayatollahi, S.A. A Review on the Role of Genetic Mutations in the Autism Spectrum Disorder. Mol. Neurobiol. 2023, 60, 5256–5272. [Google Scholar] [CrossRef]
- Wright, A.F. Genetic Variation: Polymorphisms and Mutations. eLS 2001. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Kereszturi, E. Diversity and Classification of Genetic Variations in Autism Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 16768. [Google Scholar] [CrossRef]
- Yap, C.X.; Alvares, G.A.; Henders, A.K.; Lin, T.; Wallace, L.; Farrelly, A.; McLaren, T.; Berry, J.; Vinkhuyzen, A.A.E.; Trzaskowski, M.; et al. Analysis of common genetic variation and rare CNVs in the Australian Autism Biobank. Mol. Autism 2021, 12, 12. [Google Scholar] [CrossRef]
- Apte, M.; Kumar, A. Correlation of mutated gene and signalling pathways in ASD. IBRO Neurosci. Rep. 2023, 14, 384–392. [Google Scholar] [CrossRef]
- Qiu, S.; Qiu, Y.; Li, Y.; Cong, X. Genetics of autism spectrum disorder: An umbrella review of systematic reviews and meta-analyses. Transl. Psychiatry 2022, 12, 249. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Alsharshani, M. New Horizons for Molecular Genetics Diagnostic and Research in Autism Spectrum Disorder. Adv. Neurobiol. 2020, 24, 43–81. [Google Scholar] [CrossRef] [PubMed]
- Vicari, S.; Napoli, E.; Cordeddu, V.; Menghini, D.; Alesi, V.; Loddo, S.; Novelli, A.; Tartaglia, M. Copy number variants in autism spectrum disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 92, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, C.P.; Betancur, C.; Yuen, R.K.C.; Parr, J.R.; Skuse, D.H.; Gallagher, L.; Bernier, R.A.; Buchanan, J.A.; Buxbaum, J.D.; Chen, C.A.; et al. A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat. Rev. Genet. 2020, 21, 367–376. [Google Scholar] [CrossRef]
- Viggiano, M.; Ceroni, F.; Visconti, P.; Posar, A.; Scaduto, M.C.; Sandoni, L.; Baravelli, I.; Cameli, C.; Rochat, M.J.; Maresca, A.; et al. Genomic analysis of 116 autism families strengthens known risk genes and highlights promising candidates. NPJ Genom. Med. 2024, 9, 21. [Google Scholar] [CrossRef]
- Al-Beltagi, M.; Saeed, N.K.; Bediwy, A.S.; Elbeltagi, R. Metabolomic changes in children with autism. World J. Clin. Pediatr. 2024, 13, 92737. [Google Scholar] [CrossRef]
- Acuna-Hidalgo, R.; Bo, T.; Kwint, M.P.; van de Vorst, M.; Pinelli, M.; Veltman, J.A.; Hoischen, A.; Vissers, L.E.; Gilissen, C. Post-zygotic Point Mutations Are an Underrecognized Source of De Novo Genomic Variation. Am. J. Hum. Genet. 2015, 97, 67–74. [Google Scholar] [CrossRef]
- Lord, C.; Rutter, M.; Le Couteur, A. Autism Diagnostic Interview-Revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994, 24, 659–685. [Google Scholar] [CrossRef]
- Lord, C.; Risi, S.; Lambrecht, L.; Cook, E.H., Jr.; Leventhal, B.L.; DiLavore, P.C.; Pickles, A.; Rutter, M. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000, 30, 205–223. [Google Scholar] [CrossRef]
- Van Wijngaarden-Cremers, P.J.; van Eeten, E.; Groen, W.B.; Van Deurzen, P.A.; Oosterling, I.J.; Van der Gaag, R.J. Gender and age differences in the core triad of impairments in autism spectrum disorders: A systematic review and meta-analysis. J. Autism Dev. Disord. 2014, 44, 627–635. [Google Scholar] [CrossRef]
- Kschonsak, M.; Chua, H.C.; Noland, C.L.; Weidling, C.; Clairfeuille, T.; Bahlke, O.O.; Ameen, A.O.; Li, Z.R.; Arthur, C.P.; Ciferri, C.; et al. Structure of the human sodium leak channel NALCN. Nature. 2020, 587, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Liu, H.; Zhao, Q.; Wu, J.; Yan, Z. Architecture of the human NALCN channelosome. Cell Discov. 2022, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Cochet-Bissuel, M.; Lory, P.; Monteil, A. The sodium leak channel, NALCN, in health and disease. Front Cell Neurosci. 2014, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Kottgen, M.; Benzing, T.; Simmen, T.; Tauber, R.; Buchholz, B.; Feliciangeli, S.; Huber, T.B.; Schermer, B.; Kramer-Zucker, A.; Hopker, K.; et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 2005, 24, 705–716. [Google Scholar] [CrossRef]
- Olson, H.E.; Jean-Marcais, N.; Yang, E.; Heron, D.; Tatton-Brown, K.; van der Zwaag, P.A.; Bijlsma, E.K.; Krock, B.L.; Backer, E.; Kamsteeg, E.J.; et al. A Recurrent De Novo PACS2 Heterozygous Missense Variant Causes Neonatal-Onset Developmental Epileptic Encephalopathy, Facial Dysmorphism, and Cerebellar Dysgenesis. Am. J. Hum. Genet. 2018, 102, 995–1007. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, K.; Wang, S.; Zhang, Y.H.; Yang, Z.X.; Wu, Y.; Jiang, Y.W. PACS gene family-related neurological diseases: Limited genotypes and diverse phenotypes. World J. Pediatr. 2024, 20, 82–91. [Google Scholar] [CrossRef]
- Sheng, M.; Kim, E. The Shank family of scaffold proteins. J. Cell Sci. 2000, 113 Pt 11, 1851–1856. [Google Scholar] [CrossRef]
- Uchino, S.; Waga, C. SHANK3 as an autism spectrum disorder-associated gene. Brain Dev. 2013, 35, 106–110. [Google Scholar] [CrossRef]
- Nemirovsky, S.I.; Cordoba, M.; Zaiat, J.J.; Completa, S.P.; Vega, P.A.; Gonzalez-Moron, D.; Medina, N.M.; Fabbro, M.; Romero, S.; Brun, B.; et al. Whole genome sequencing reveals a de novo SHANK3 mutation in familial autism spectrum disorder. PLoS ONE 2015, 10, e0116358. [Google Scholar] [CrossRef]
- Huang, G.; Chen, S.; Chen, X.; Zheng, J.; Xu, Z.; Doostparast Torshizi, A.; Gong, S.; Chen, Q.; Ma, X.; Yu, J.; et al. Uncovering the Functional Link Between SHANK3 Deletions and Deficiency in Neurodevelopment Using iPSC-Derived Human Neurons. Front. Neuroanat. 2019, 13, 23. [Google Scholar] [CrossRef]
- Wu, S.; Wang, J.; Zhang, Z.; Jin, X.; Xu, Y.; Si, Y.; Liang, Y.; Ge, Y.; Zhan, H.; Peng, L.; et al. Shank3 deficiency elicits autistic-like behaviors by activating p38alpha in hypothalamic AgRP neurons. Mol. Autism 2024, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, P.C.; Stanford, L.E.; Coba, M.P.; Ainge, J.A.; Fink, A.E.; Opazo, P.; Delgado, J.Y.; Komiyama, N.H.; O’Dell, T.J.; Grant, S.G. Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J. Neurosci. 2007, 27, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Trobiani, L.; Meringolo, M.; Diamanti, T.; Bourne, Y.; Marchot, P.; Martella, G.; Dini, L.; Pisani, A.; De Jaco, A.; Bonsi, P. The neuroligins and the synaptic pathway in Autism Spectrum Disorder. Neurosci. Biobehav. Rev. 2020, 119, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Sergi, C.; Parayil Sankaran, B. Succinic Semialdehyde Dehydrogenase Deficiency; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Didiasova, M.; Banning, A.; Brennenstuhl, H.; Jung-Klawitter, S.; Cinquemani, C.; Opladen, T.; Tikkanen, R. Succinic Semialdehyde Dehydrogenase Deficiency: An Update. Cells 2020, 9, 477. [Google Scholar] [CrossRef]
- Julia-Palacios, N.A.; Kuseyri Hubschmann, O.; Olivella, M.; Pons, R.; Horvath, G.; Lucke, T.; Fung, C.W.; Wong, S.N.; Cortes-Saladelafont, E.; Rovira-Remisa, M.M.; et al. The continuously evolving phenotype of succinic semialdehyde dehydrogenase deficiency. J. Inherit. Metab. Dis. 2024, 47, 447–462. [Google Scholar] [CrossRef]
- Frye, R.E. Succinic semialdehyde dehydrogenase deficiency: A model of neurocircuit imbalances in autism and potential insight into new biomarkers. Dev. Med. Child. Neurol. 2023, 65, 1544–1545. [Google Scholar] [CrossRef]
- Gogou, M.; Spilioti, M.; Tramma, D.; Papadopoulou-Alataki, E.; Evangeliou, A. Succinic Semialdehyde Dehydrogenase Deficiency Presenting as Autism Spectrum Disorder. Indian J. Pediatr. 2016, 83, 1036–1037. [Google Scholar] [CrossRef]
- Chung, T.; Na, J.; Kim, Y.I.; Chang, D.Y.; Kim, Y.I.; Kim, H.; Moon, H.E.; Kang, K.W.; Lee, D.S.; Chung, J.K.; et al. Dihydropyrimidine Dehydrogenase Is a Prognostic Marker for Mesenchymal Stem Cell-Mediated Cytosine Deaminase Gene and 5-Fluorocytosine Prodrug Therapy for the Treatment of Recurrent Gliomas. Theranostics 2016, 6, 1477–1490. [Google Scholar] [CrossRef]
- van Kuilenburg, A.B.P.; Meijer, J.; Meinsma, R.; Perez-Duenas, B.; Alders, M.; Bhuiyan, Z.A.; Artuch, R.; Hennekam, R.C.M. Dihydropyrimidine Dehydrogenase Deficiency: Homozygosity for an Extremely Rare Variant in DPYD due to Uniparental Isodisomy of Chromosome 1. JIMD Rep. 2019, 45, 65–69. [Google Scholar] [CrossRef]
- Fleger, M.; Willomitzer, J.; Meinsma, R.; Alders, M.; Meijer, J.; Hennekam, R.C.M.; Huemer, M.; van Kuilenburg, A.B.P. Dihydropyrimidine Dehydrogenase Deficiency: Metabolic Disease or Biochemical Phenotype? JIMD Rep. 2017, 37, 49–54. [Google Scholar] [CrossRef]
- Carter, M.T.; Nikkel, S.M.; Fernandez, B.A.; Marshall, C.R.; Noor, A.; Lionel, A.C.; Prasad, A.; Pinto, D.; Joseph-George, A.M.; Noakes, C.; et al. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin. Genet. 2011, 80, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Kerrigan, J.F.; Aleck, K.A.; Tarby, T.J.; Bird, C.R.; Heidenreich, R.A. Fumaric aciduria: Clinical and imaging features. Ann. Neurol. 2000, 47, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.P.; O’Brien, T.W.; Subramony, S.H.; Shuster, J.; Stacpoole, P.W. The spectrum of pyruvate dehydrogenase complex deficiency: Clinical, biochemical and genetic features in 371 patients. Mol. Genet. Metab. 2012, 106, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Bhandary, S.; Aguan, K. Pyruvate dehydrogenase complex deficiency and its relationship with epilepsy frequency—An overview. Epilepsy Res. 2015, 116, 40–52. [Google Scholar] [CrossRef]
- Gibson, G.E.; Jope, R.; Blass, J.P. Decreased synthesis of acetylcholine accompanying impaired oxidation of pyruvic acid in rat brain minces. Biochem. J. 1975, 148, 17–23. [Google Scholar] [CrossRef]
- Tuc, E.; Bengur, F.B.; Aykut, A.; Sahin, O.; Alanay, Y. The third family with TAF6-related phenotype: Alazami-Yuan syndrome. Clin. Genet. 2020, 97, 795–796. [Google Scholar] [CrossRef]
- Selicorni, A.; Mariani, M.; Lettieri, A.; Massa, V. Cornelia de Lange Syndrome: From a Disease to a Broader Spectrum. Genes 2021, 12, 1075. [Google Scholar] [CrossRef]
- Vuu, Y.M.; Roberts, C.T.; Rastegar, M. MeCP2 Is an Epigenetic Factor That Links DNA Methylation with Brain Metabolism. Int. J. Mol. Sci. 2023, 24, 4218. [Google Scholar] [CrossRef]
- Liu, Y.; Flamier, A.; Bell, G.W.; Diao, A.J.; Whitfield, T.W.; Wang, H.C.; Wu, Y.; Schulte, F.; Friesen, M.; Guo, R.; et al. MECP2 directly interacts with RNA polymerase II to modulate transcription in human neurons. Neuron 2024, 112, 1943–1958.e10. [Google Scholar] [CrossRef]
- Zachariah, R.M.; Rastegar, M. Linking epigenetics to human disease and Rett syndrome: The emerging novel and challenging concepts in MeCP2 research. Neural Plast. 2012, 2012, 415825. [Google Scholar] [CrossRef]
- Weissmiller, A.M.; Wang, J.; Lorey, S.L.; Howard, G.C.; Martinez, E.; Liu, Q.; Tansey, W.P. Inhibition of MYC by the SMARCB1 tumor suppressor. Nat. Commun. 2019, 10, 2014. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Kleefstra, T.; Brunner, H.G.; Amiel, J.; Oudakker, A.R.; Nillesen, W.M.; Magee, A.; Genevieve, D.; Cormier-Daire, V.; van Esch, H.; Fryns, J.P.; et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 2006, 79, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Selten, M.; Mossink, B.; Keller, J.M.; Linda, K.; Moerschen, R.; Qu, J.; Koerner, P.; Jansen, S.; Oudakker, A.; et al. Distinct Pathogenic Genes Causing Intellectual Disability and Autism Exhibit a Common Neuronal Network Hyperactivity Phenotype. Cell Rep. 2020, 30, 173–186. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef]
- Venkataramanan, S.; Gadek, M.; Calviello, L.; Wilkins, K.; Floor, S.N. DDX3X and DDX3Y are redundant in protein synthesis. RNA 2021, 27, 1577–1588. [Google Scholar] [CrossRef]
- Nicola, P.; Blackburn, P.R.; Rasmussen, K.J.; Bertsch, N.L.; Klee, E.W.; Hasadsri, L.; Pichurin, P.N.; Rankin, J.; Raymond, F.L.; Study, D.D.D.; et al. De novo DDX3X missense variants in males appear viable and contribute to syndromic intellectual disability. Am. J. Med. Genet. A 2019, 179, 570–578. [Google Scholar] [CrossRef]
- Kusch, T. Histone H3 lysine 4 methylation revisited. Transcription 2012, 3, 310–314. [Google Scholar] [CrossRef][Green Version]
- Wang, S.; Bleeck, A.; Nadif Kasri, N.; Kleefstra, T.; van Rhijn, J.R.; Schubert, D. SETD1A Mediated H3K4 Methylation and Its Role in Neurodevelopmental and Neuropsychiatric Disorders. Front. Mol. Neurosci. 2021, 14, 772000. [Google Scholar] [CrossRef]
- Yu, X.; Yang, L.; Li, J.; Li, W.; Li, D.; Wang, R.; Wu, K.; Chen, W.; Zhang, Y.; Qiu, Z.; et al. De Novo and Inherited SETD1A Variants in Early-onset Epilepsy. Neurosci. Bull. 2019, 35, 1045–1057. [Google Scholar] [CrossRef]
- C Yuen, R.K.; Merico, D.; Bookman, M.; L Howe, J.; Thiruvahindrapuram, B.; Patel, R.V.; Whitney, J.; Deflaux, N.; Bingham, J.; Wang, Z.; et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017, 20, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Harlow, E.; Hunt, T.; Hunter, T.; Lahti, J.M.; Manning, G.; Morgan, D.O.; Tsai, L.H.; Wolgemuth, D.J. Cyclin-dependent kinases: A family portrait. Nat. Cell Biol. 2009, 11, 1275–1276. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.R.; Lin, G.T.; Huang, C.K.; Fann, M.J. Cdk12 and Cdk13 regulate axonal elongation through a common signaling pathway that modulates Cdk5 expression. Exp. Neurol. 2014, 261, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Sifrim, A.; Hitz, M.P.; Wilsdon, A.; Breckpot, J.; Turki, S.H.; Thienpont, B.; McRae, J.; Fitzgerald, T.W.; Singh, T.; Swaminathan, G.J.; et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 2016, 48, 1060–1065. [Google Scholar] [CrossRef]
- Bostwick, B.L.; McLean, S.; Posey, J.E.; Streff, H.E.; Gripp, K.W.; Blesson, A.; Powell-Hamilton, N.; Tusi, J.; Stevenson, D.A.; Farrelly, E.; et al. Phenotypic and molecular characterisation of CDK13-related congenital heart defects, dysmorphic facial features and intellectual developmental disorders. Genome Med. 2017, 9, 73. [Google Scholar] [CrossRef]
- Chang, C.H.; Chen, T.Y.; Lu, I.L.; Li, R.B.; Tsai, J.J.; Lin, P.Y.; Tang, T.K. CEP120-mediated KIAA0753 recruitment onto centrioles is required for timely neuronal differentiation and germinal zone exit in the developing cerebellum. Genes Dev. 2021, 35, 1445–1460. [Google Scholar] [CrossRef]
- Meka, D.P.; Kobler, O.; Hong, S.; Friedrich, C.M.; Wuesthoff, S.; Henis, M.; Schwanke, B.; Krisp, C.; Schmuelling, N.; Rueter, R.; et al. Centrosome-dependent microtubule modifications set the conditions for axon formation. Cell Rep. 2022, 39, 110686. [Google Scholar] [CrossRef]
- Karam, A.; Delvallee, C.; Estrada-Cuzcano, A.; Geoffroy, V.; Lamouche, J.B.; Leuvrey, A.S.; Nourisson, E.; Tarabeux, J.; Stoetzel, C.; Scheidecker, S.; et al. WGS Revealed Novel BBS5 Pathogenic Variants, Missed by WES, Causing Ciliary Structure and Function Defects. Int. J. Mol. Sci. 2023, 24, 8729. [Google Scholar] [CrossRef]
- Wingfield, J.L.; Lechtreck, K.F.; Lorentzen, E. Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 2018, 62, 753–763. [Google Scholar] [CrossRef]
- Forsythe, E.; Beales, P.L. Bardet-Biedl syndrome. Eur. J. Hum. Genet. 2013, 21, 8–13. [Google Scholar] [CrossRef]
- Shamseldin, H.E.; Shaheen, R.; Ewida, N.; Bubshait, D.K.; Alkuraya, H.; Almardawi, E.; Howaidi, A.; Sabr, Y.; Abdalla, E.M.; Alfaifi, A.Y.; et al. The morbid genome of ciliopathies: An update. Genet. Med. 2020, 22, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, D.N.; Edwards, R.J.; Slavutsky, A.L. Spectrins: Molecular organizers and targets of neurological disorders. Nat. Rev. Neurosci. 2023, 24, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, D.N. Cargo hold and delivery: Ankyrins, spectrins, and their functional patterning of neurons. Cytoskeleton (Hoboken) 2020, 77, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Black, J.; Kisiel, N.; Kulesz-Martin, M.F. SPAF, a new AAA-protein specific to early spermatogenesis and malignant conversion. Oncogene 2000, 19, 1579–1588. [Google Scholar] [CrossRef]
- Puusepp, S.; Kovacs-Nagy, R.; Alhaddad, B.; Braunisch, M.; Hoffmann, G.F.; Kotzaeridou, U.; Lichvarova, L.; Liiv, M.; Makowski, C.; Mandel, M.; et al. Compound heterozygous SPATA5 variants in four families and functional studies of SPATA5 deficiency. Eur. J. Hum. Genet. 2018, 26, 407–419. [Google Scholar] [CrossRef]
- Tanaka, A.J.; Cho, M.T.; Millan, F.; Juusola, J.; Retterer, K.; Joshi, C.; Niyazov, D.; Garnica, A.; Gratz, E.; Deardorff, M.; et al. Mutations in SPATA5 Are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am. J. Hum. Genet. 2015, 97, 457–464. [Google Scholar] [CrossRef]
- Stancheva, M.A., T.; Todorov, T.; Atemin, S.; Pavlova, Z.; Turturikov, I.; Kadiyska, T.; Marinova, E.; Popova, D.; Alanay, Y. Clinical Presentation and Genetic Correlations in Bulgarian Patients with SPATA5 Gene Mutations. Rare Dis. Orphan Drugs 2021, 11, 19–23. [Google Scholar] [CrossRef]
- van Spronsen, M.; Mikhaylova, M.; Lipka, J.; Schlager, M.A.; van den Heuvel, D.J.; Kuijpers, M.; Wulf, P.S.; Keijzer, N.; Demmers, J.; Kapitein, L.C.; et al. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron 2013, 77, 485–502. [Google Scholar] [CrossRef]
- Seifert, W.; Kuhnisch, J.; Maritzen, T.; Horn, D.; Haucke, V.; Hennies, H.C. Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem. 2011, 286, 37665–37675. [Google Scholar] [CrossRef]
- Tanaka, H.; Okazawa, H. PQBP1: The Key to Intellectual Disability, Neurodegenerative Diseases, and Innate Immunity. Int. J. Mol. Sci. 2022, 23, 6227. [Google Scholar] [CrossRef]
- Wan, H.; Wang, Q.; Chen, X.; Zeng, Q.; Shao, Y.; Fang, H.; Liao, X.; Li, H.S.; Liu, M.G.; Xu, T.L.; et al. WDR45 contributes to neurodegeneration through regulation of ER homeostasis and neuronal death. Autophagy 2020, 16, 531–547. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; D’Arca, D.; Lim, W.K.; Brahmachary, M.; Carro, M.S.; Ludwig, T.; Cardo, C.C.; Guillemot, F.; Aldape, K.; Califano, A.; et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev. Cell 2009, 17, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Reichow, B.; George-Puskar, A.; Lutz, T.; Smith, I.C.; Volkmar, F.R. Brief Report: Systematic Review of Rett Syndrome in Males. J. Autism Dev. Disord. 2015, 45, 3377–3383. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Cheng, T.-L.; Li, G.-z.; Sun, S.-B.; Yu, S.-Y.; Zhang, Y.; Du, Y.-S.; Qiu, Z. Identification of autism-related MECP2 mutations by whole-exome sequencing and functional validation. Mol. Autism 2017, 8, 43. [Google Scholar] [CrossRef]
| Case | Gene ID | Sex | Age of Testing (Years) | Syndromic/Non-Syndromic Phenotype | Variant (GRCh37) | Variant Type | Zygosity | Inheritance | Pathogenicity * |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MECP2 | Male | 2 | Syndromic | chrX:g.153296071dup, NM_004992.3: c.1208dup, p.(Glu404Ter) | Nonsense | Hemizygous | Maternal | Likely pathogenic |
| 2 | TAF6 | Female | 15 | Syndromic | chr7: g.99711522A>G, NM_001190415.1: c.323T>C, p.(Ile108Thr) | Missense | Homozygous | Biparental | Likely pathogenic |
| 3 | SMARCB1 | Male | 6 | Syndromic | chr22: g.24145549C>T, NM_003073.3: c.568C>T, p.(Arg190Trp) | Missense | Heterozygous | de novo | Likely pathogenic |
| 4 | PACS2 | Male | 5 | Syndromic | chr14: g.105834449G>A, NM_001100913.3: c.625G>A, p.(Glu209Lys) | Missense | Heterozygous | de novo | Pathogenic |
| 5 | WDR45 | Female | 4 | Syndromic | chrX:g.48933330del, NM_007075.3:c.601_602del, p.(Leu201LysfsTer21) | Frameshift | Heterozygous | de novo | Likely pathogenic |
| 6 | PQBP1 | Male | 34 | Syndromic | chrX:g.48760017C>T, NM_001032381.1:c.586C>T, p.(Arg196Ter) | Nonsense | Hemizygous | Maternal | Pathogenic |
| 7 | SPATA5 | Male | 13 | Syndromic | chr4: g.123855300G>A, NM_145207.2: c. 554G>A, p.(Gly185Glu) p. | Missense | Heterozygous | Maternal | VUS |
| chr4: g.123900503C>T, NM_145207.2: c.1831C>T, (Pro611Ser) | Missense | Heterozygous | Paternal | VUS | |||||
| 8 | NALCN | Female | 7 | Syndromic | chr13: g.101944423A>G, NM_052867.2: c.965T>C, p.(Ile322Thr) | Missense | Heterozygous | de novo | Likely pathogenic |
| 9 | FH | Female | 8 | Syndromic | chr1: g.241667402G>A, NM_000143.4: c.1048C>T, p.(Arg350Trp) | Missense | Homozygous | Biparental | Likely pathogenic |
| 10 | CEP120 | Male | 3 | Syndromic | chr5: g.122758670A>C, NM_153223.3: c.23T>G, p.(Leu8Trp) | Missense | Heterozygous | Paternal | VUS |
| chr5: g.122700222G>C NM_153223.3: c.2548C>G, p.(Arg850Gly) | Missense | Heterozygous | Maternal | VUS | |||||
| 11 | BBS5 | Male | 12 | Syndromic | chr2:g.170343603G>A, NM_152384.3:c.167G>A, p.(Arg56Lys) | Missense | Heterozygous | Maternal | Likely pathogenic |
| chr2:170354136G>C, NM_152384.3: c.619-1G>C | Splice site | Heterozygous | Paternal | Pathogenic | |||||
| 12 | SPTAN1 | Male | 6 | Syndromic | chr9: g.131394565C>T, NM_001130438.3: c.6922C>T, p.(Arg2308Cys) | Missense | Heterozygous | de novo | Likely pathogenic |
| 13 | VPS13B | Female | 10 | Syndromic | chr8:g.100844840_100844849delinsAC, NM_152564.5: c.9574_9583delinsAC, p.(Val3192ThrfsTer33) | Frameshift | Heterozygous | Paternal | Likely pathogenic |
| chr8: g.100733139C>T, NM_152564.5: c.6914C>T, p.(Thr2305Ile) | Missense | Heterozygous | Maternal | VUS | |||||
| 14 | SHANK3 | Male | 17 | Non-syndromic | chr22: g.51153476G>A, NM_001372044.2: c.2490+1G>A | Splice site | Heterozygous | de novo | Pathogenic |
| DLG3 | chrX: g.69712394G>A, NM_021120.4:c.1721G>A, p.(Arg574Gln) | Missense | Hemizygous | Maternal | VUS | ||||
| 15 | CDK13 | Male | 10 | Syndromic | chr7: g.40085606A>T NM_003718.5: , c.2525A>T, p.(Asn842Ile) | Missense | Heterozygous | de novo | Pathogenic |
| 16 | PDHX | Female | 3 | Syndromic | chr11: g.35016549C>T, NM_003477.3: c.1336C>T, p.(Arg446Ter) | Nonsence | Homozygous | Biparental | Pathogenic |
| 17 | SETD1A | Male | 15 | Syndromic | chr16: g.30995020delG, NM_014712.3: c.4879del, p.(Val1627TrpfsTer41) | Frameshift | Heterozygous | de novo | Pathogenic |
| 18 | TRAK1 | Female | 5 | Non-syndromic | chr3:g.42240742T>A, NM_001042646.3:c.1187T>A, p.(Ile396Asn) | Missense | Heterozygous | de novo | VUS |
| 19 | ALDH5A1 | Female | 3 | Syndromic | chr6:g.24515433dup NM_170740.1:c.804dup, p.(Val269fsTer19) | Frameshift | Heterozygous | Paternal | Pathogenic |
| chr6:g.24528277G>A NM_170740.1:c.1265G>A, p.(Gly422Asp) | Missense | Heterozygous | Maternal | Likely pathogenic | |||||
| 20 | DPYD | Male | 6 | Syndromic | chr1:g.97915614C>T, NM_000110.3: c.1905+1G>A | Splice site | Homozygous | Biparental | Likely pathogenic |
| 21 | DDX3X | Female | 2 | Syndromic | chrX:g.41203374C>A, NM_001356.3:c.857C>A, p.(Ala286Asp) | Missense | Heterozygous | de novo | VUS |
| 22 | HUWE1 | Male | 8 | Syndromic | chrX:g.53578038C>T, NM_031407.7:c.9209G>A, p.(Arg3070His) | Missense | Hemizygous | de novo | Pathogenic |
| Case | Gene ID | Sex | Age of Testing (Years) | Clinical and Neuropsychological Profile of the Patient |
|---|---|---|---|---|
| 1 | MECP2 | Male | 2 | West syndrome: abnormal EEG, chaotic brain waves (hypsarrhythmia), specific infantile spasms with twitching of the head, arms, body tremors, stereotyped movements, and epileptic seizures, combined with axial muscular hypotony and motor developmental delay, mild fascial dysmorphism and smaller left auricle, clinodactyly of second left toe, and normal metabolic screening; communication deficits, lack of speech, and responding to commands, stereotyped movements. |
| 2 | TAF6 | Female | 15 | Congenital cerebellar hypoplasia, hypotrophy (underrepresented subcutaneous fat tissue), ataxic gait, mild muscular hypotony, discretely impaired fine motor skills, mild mental retardation, defects in sound pronunciation, speech delay, mood disorders, insufficient concentration, and anxiety. |
| 3 | SMARCB1 | Male | 6 | Epileptic seizures, focal epileptiform changes, and generalized paroxysmal manifestations of myocytic type, facial dysmorphism, delay in speech and neuropsychiatric development, deterioration in communication, infrequent eye contact, and stereotyped movements. |
| 4 | PACS2 | Male | 5 | Microcephaly, facial dysmorphism, discrete facial symmetry, antimongoloid eye slits, epicanthus, hypertelorism, ocular coloboma, facial dysmorphism, backward rotated dysplastic auricles, muscular hypotony, epileptic seizures, and delay in speech and psychomotor development. |
| 5 | WDR45 | Female | 4 | Epileptic encephalopathy with late epileptic spasms and focal seizures, moderate mental retardation, communication deficits. |
| 6 | PQBP1 | Male | 34 | Confined atrophy of the brain, mental retardation since early childhood, behavioral stereotypes, tics, communication deficits. |
| 7 | SPATA5 | Male | 13 | EEG abnormality, delayed onset of psychomotor development, speech delay, stereotypic movements. |
| 8 | NALCN | Female | 7 | Delay in speech, neuropsychiatric and psychomotor development, generalized muscular hypotension and hyporeflexia in the neonatal period, ulnar deviation of the fingers and hip dysplasia, speech and communication deterioration; lack of organic pathological changes in the examined intracranial anatomical components according to MRT of CNS and MR spectroscopy. |
| 9 | FH | Female | 8 | Microcephaly and unspecified encephalopathy; transfontanel ultrasound analyses shows mild diffuse dilatation of subarachnoid space and of the lateral ventricles; seizures, generalized muscular hypotension, delay in speech, and neuropsychiatric development. |
| 10 | CEP120 | Male | 3 | Delay in speech and neuropsychiatric development, epileptic seizures, dolichocephaly, and hyperprolinemia type I. |
| 11 | BBS5 | Male | 12 | Polydactyly, undeveloped expressive speech, communication deterioration, anxiety, psychomotor and sensory deterioration, and repetitive behavioral patterns. |
| 12 | SPTAN1 | Male | 6 | Febrile seizures, behavior deterioration, and communication difficulties. |
| 13 | VPS13B | Female | 10 | Abnormal EEG, paralysis cerebralis, divergent strabismus, hydrocephaly, large fontanelle in infancy, planovagus deformity of the feet, pronounced scoliosis, severe neurodevelopmental disorder, speech delay, and stereotyped movements. |
| 14 | SHANK3 | Male | 17 | Loss of previous skills over the years, loss of speech, and deterioration in communication. |
| DLG3 | ||||
| 15 | CDK13 | Male | 10 | Moderate mental retardation, significant behavioral disorder requiring care and treatment; mental and physical developmental delay, lack of speech, stereotyped movements and behavior, lack of attention, and communication deficits. |
| 16 | PDHX | Female | 3 | Cerebral cortex atrophy, demyelination, mental retardation, significant behavioral disorder, alalia, central quadriplegia, microcephaly, blindness, and leukodystrophy. |
| 17 | SETD1A | Male | 15 | Neurobehavioral retardation, facial dysmorphism, muscular hypotony, enterocolitis, hepatosplenomegaly, and hemangioma parietis thoracis since birth. |
| 18 | TRAK1 | Female | 5 | Speech delay and communication and behavioral deteriorations. |
| 19 | ALDH5A1 | Female | 3 | Global developmental delay, including speech and behavioral disorders, hypotonia, coordination problems, hyporeflexia, movement disorders, and epilepsy. |
| 20 | DPYD | Male | 6 | Microcephaly, severe developmental delay, hypotonia, seizures, speech delay, and communication difficulties. |
| 21 | DDX3X | Female | 2 | Facial dysmorphism, delay in speech, and psychomotor development. |
| 22 | HUWE1 | Male | 8 | Microcephaly, epilepsy, severe mental retardation, significant behavioral deterioration, lack of speech, and delay in motor development. |
| Gene (SNV) | Protein Name | Role in Neuronal Structure or Function | SFARI Classification |
|---|---|---|---|
| NALCN | Sodium Leak Channel, Non-Selective | Regulates resting membrane potential and excitability [32,33,34] | Strong candidate |
| PACS2 | Phosphofurin Acidic Cluster Sorting Protein 2 | Synaptic signaling, organelle communication, calcium signaling, and mitochondrial function [35,36,37] | Syndromic |
| SHANK3 | ProSAP SH3 and multiple ankyrin repeat domain protein 3 | Synapse formation and maintenance [38,39,40,41,42] | High confidence |
| DLG3 | disks large membrane-associated guanylate kinase scaffold protein 3, synapse-associated protein 102 (SAP-102) | Synaptic signaling involved in N-methyl-D-aspartate receptor clustering at excitatory synapses; synaptic plasticity [43,44] | Not listed |
| ALDH5A1 | Aldehyde Dehydrogenase 5 Family Member A1 | GABA metabolism, mitochondrial function [45,46,47,48,49] | High confidence, syndromic |
| DPYD | Dihydropyrimidine Dehydrogenase | Mitochondrial enzyme [50] and pyrimidine degradation [50,51,52,53] | Strong candidate |
| FH | Fumarate hydratase | Mitochondrial function, Krebs cycle enzyme; maintaining levels of neurotransmitters like glutamate, aspartate, and GABA [54] | Not listed |
| PDHX | pyruvate dehydrogenase X | Mitochondrial function; links glycolysis to tricarboxy acid cycle; neurotransmitter balance conversion of pyruvate to acetyl-CoA, maintaining levels of neurotransmitters like glutamate, aspartate, and GABA [42,55,56,57] | Not listed |
| TAF6 | TATA-Box Binding Protein Associated Factor 6 | Transcription initiation complex component [58,59] | Strong candidate |
| MECP2 | Methyl-CpG Binding Protein 2 | Transcription regulation [60,61,62] | High confidence, syndromic |
| SMARCB1 | SWI/SNF-related, matrix- associated, actin-dependent regulator of chromatin subfamily B member 1 | Chromatin remodeling complex subunit [63,64,65,66] | Not listed |
| DDX3X | DEAD-Box Helicase 3 X-Linked | RNA metabolism, translation initiation [67,68,69] | High confidence, syndromic |
| SETD1A | SET domain containing protein 1A or histone methyltransferase | Histone methylation and transcription regulation [70,71,72,73] | High confidence, syndromic |
| CDK13 | cyclin-dependent kinase 13 | Cell cycle control factors; transcriptional regulation and RNA splicing [74,75,76,77] | Syndromic |
| CEP120 | centrosomal protein 120 | Ciloigenesis, axonal growth, and cerebellar development [78,79] | Not listed |
| BBS5 | BBSome | Cilia function and intracellular transport [80,81,82,83] | Not listed |
| SPTAN1 | αII spectrin subunit | Membrane structure; synaptic support [84,85] | Not listed |
| SPATA5 | spermatogenesis-associated protein 5 | Mitochondrial dynamics; ATP production in neurons [86,87,88,89] | Not listed |
| TRAK1 | trafficking kinesin binding protein 1 | Mitochondrial transport in neurons; [90] | Not listed |
| VPS13B | vacuolar sorting protein 13 | Golgi integrity; vesicle trafficking in neurons [91] | High confidence, strong candidate |
| PQBP1 | Polyglutamine binding protein-1 | Neuron homeostasis; clearance of neurotoxic proteins [92] | Not listed |
| WDR45 | WD repeat-containing protein 45 | Macroautophagy; removal of damaged organelles [93] | Not listed |
| HUWE1 | HECT, UBA, and WWE Domain Containing E3 Ubiquitin Protein Ligase 1 | Protein degradation and cortical development [94] | Syndromic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belenska-Todorova, L.; Zamfirov, M.; Todorov, T.; Atemin, S.; Sleptsova, M.; Pavlova, Z.; Kadiyska, T.; Maver, A.; Peterlin, B.; Todorova, A. Exome Study of Single Nucleotide Variations in Patients with Syndromic and Non-Syndromic Autism Reveals Potential Candidate Genes for Diagnostics and Novel Single Nucleotide Variants. Cells 2025, 14, 915. https://doi.org/10.3390/cells14120915
Belenska-Todorova L, Zamfirov M, Todorov T, Atemin S, Sleptsova M, Pavlova Z, Kadiyska T, Maver A, Peterlin B, Todorova A. Exome Study of Single Nucleotide Variations in Patients with Syndromic and Non-Syndromic Autism Reveals Potential Candidate Genes for Diagnostics and Novel Single Nucleotide Variants. Cells. 2025; 14(12):915. https://doi.org/10.3390/cells14120915
Chicago/Turabian StyleBelenska-Todorova, Lyudmila, Milen Zamfirov, Tihomir Todorov, Slavena Atemin, Mila Sleptsova, Zornitsa Pavlova, Tanya Kadiyska, Ales Maver, Borut Peterlin, and Albena Todorova. 2025. "Exome Study of Single Nucleotide Variations in Patients with Syndromic and Non-Syndromic Autism Reveals Potential Candidate Genes for Diagnostics and Novel Single Nucleotide Variants" Cells 14, no. 12: 915. https://doi.org/10.3390/cells14120915
APA StyleBelenska-Todorova, L., Zamfirov, M., Todorov, T., Atemin, S., Sleptsova, M., Pavlova, Z., Kadiyska, T., Maver, A., Peterlin, B., & Todorova, A. (2025). Exome Study of Single Nucleotide Variations in Patients with Syndromic and Non-Syndromic Autism Reveals Potential Candidate Genes for Diagnostics and Novel Single Nucleotide Variants. Cells, 14(12), 915. https://doi.org/10.3390/cells14120915









