The Endoplasmic Reticulum Is a Key Battleground between Phytoplasma Aggression and Host Plant Defense
Abstract
1. Introduction
2. Materials and Methods
2.1. Phytoplasma Strains and Graft Inoculation
2.2. DNA and RNA Extraction
2.3. Production of Polyclonal Antibodies against the Immunodominant Membrane Protein (IDP) of PPT Phytoplasma
2.4. Western Blot Analysis
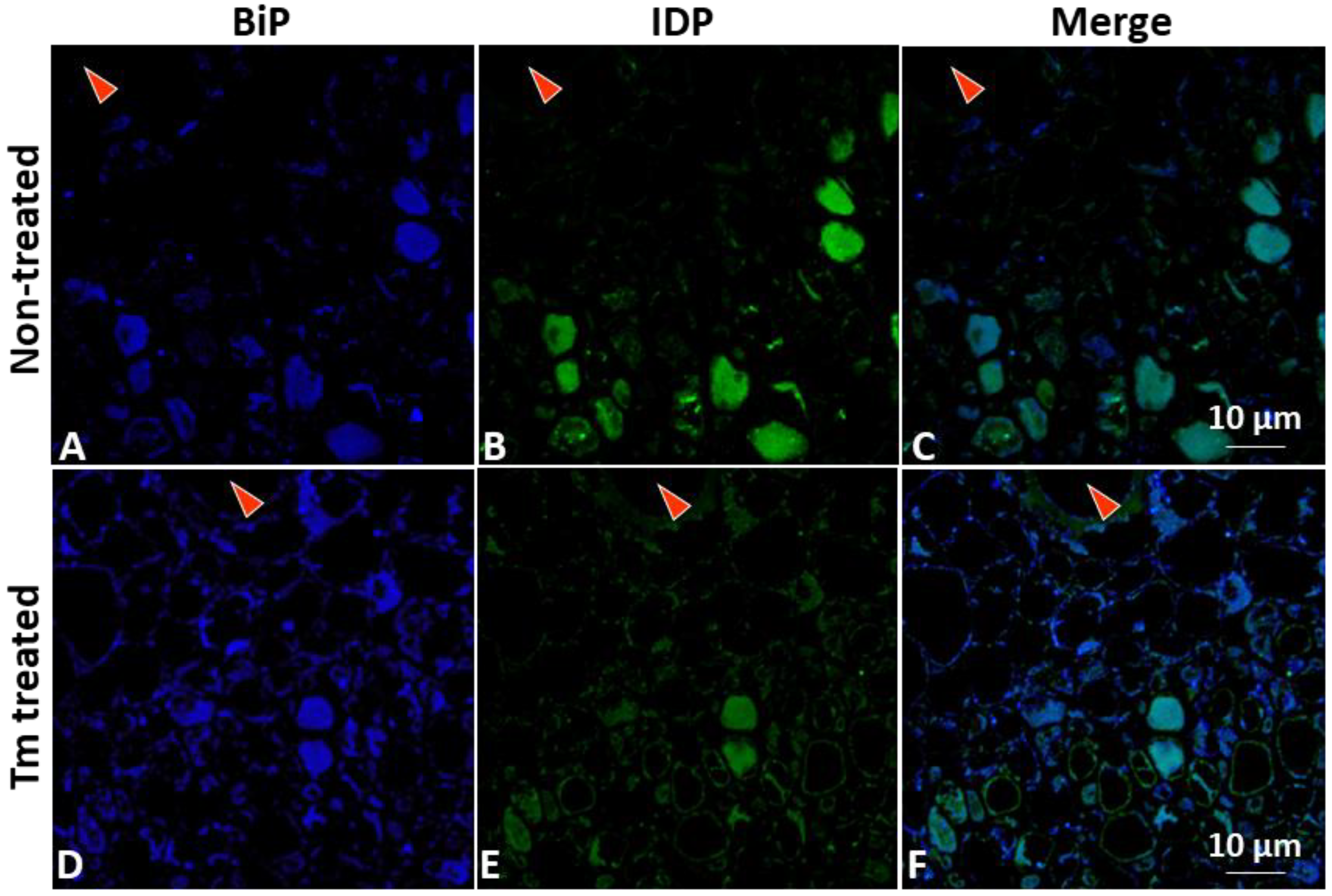
2.5. Visualization and Localization of ER-Resident Proteins and Phytoplasmas in Plants by Immunostaining
2.6. Transmission Electron Microscopy (TEM)
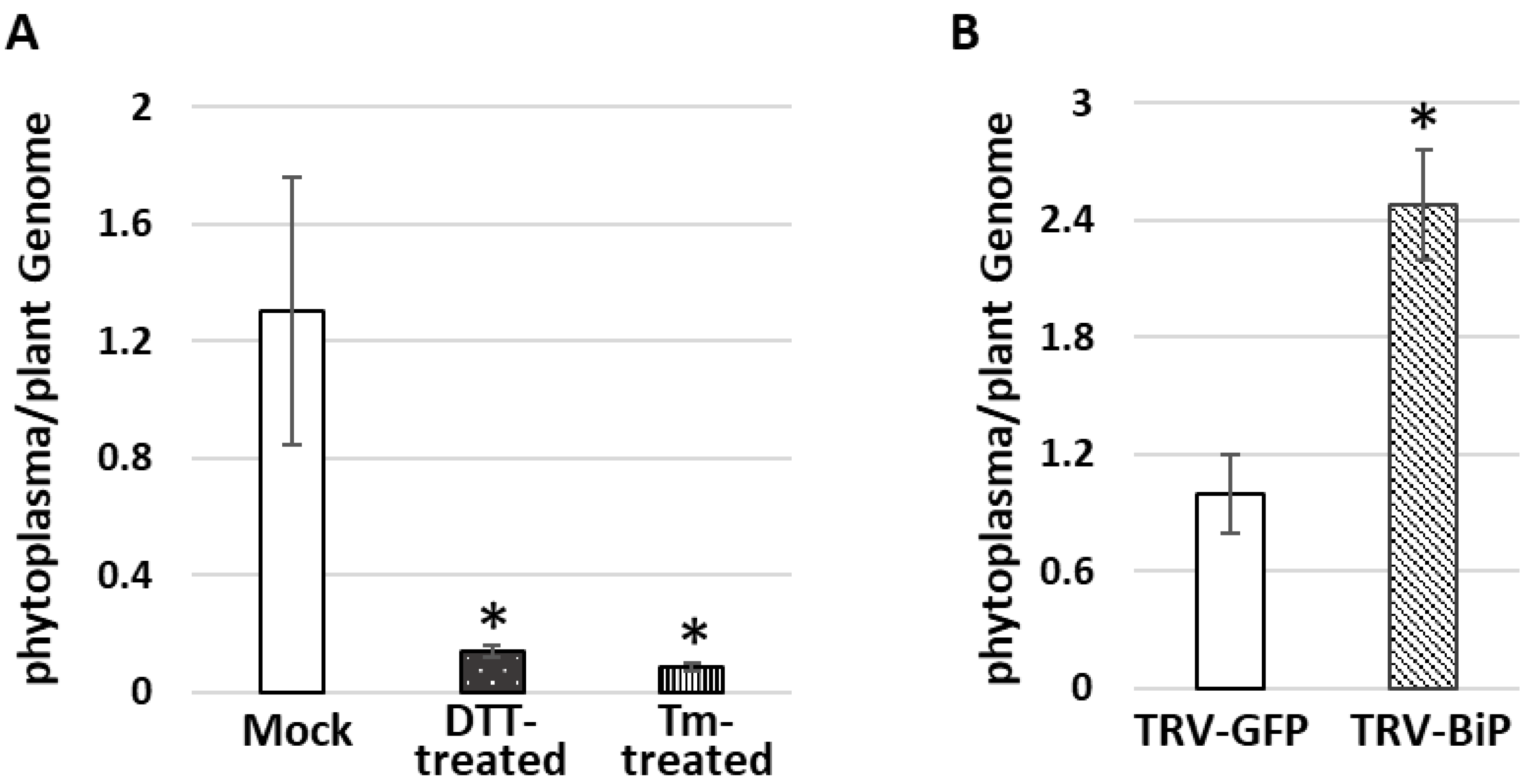
2.7. Application of Unfolded Protein Response (UPR) Chemical Inducers to Plants
2.8. Assessment of the Expression Levels of Targeted UPR and SA Marker Genes by Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
2.9. Measurement of Phytoplasma Titer by Quantitative PCR (qPCR)
2.10. Gene Knockdown in Tomato Plants by Tobacco Rattle Virus (TRV) Vector
3. Results
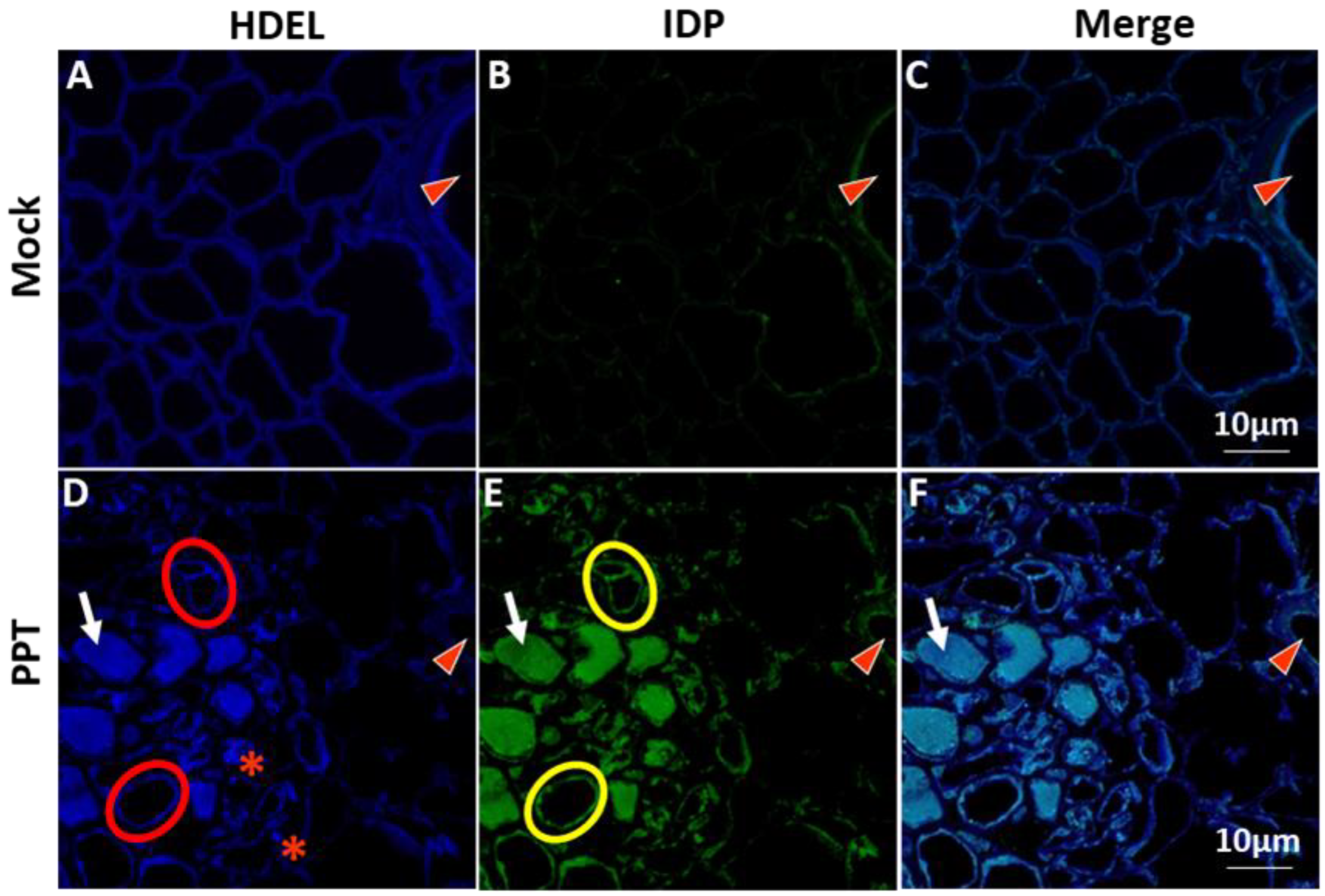
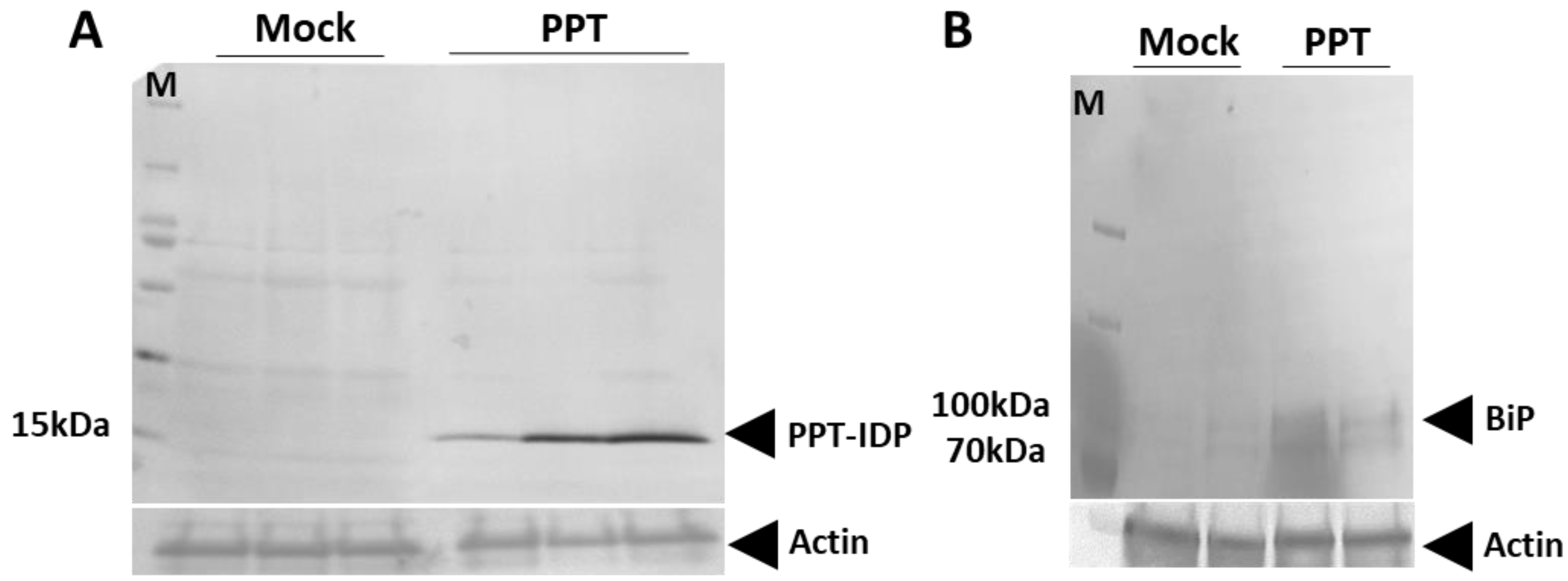
3.1. PPT Phytoplasma Infection Induced Excessive Accumulation of ER-Resident Proteins and Caused ER Stress in Tomato Plants
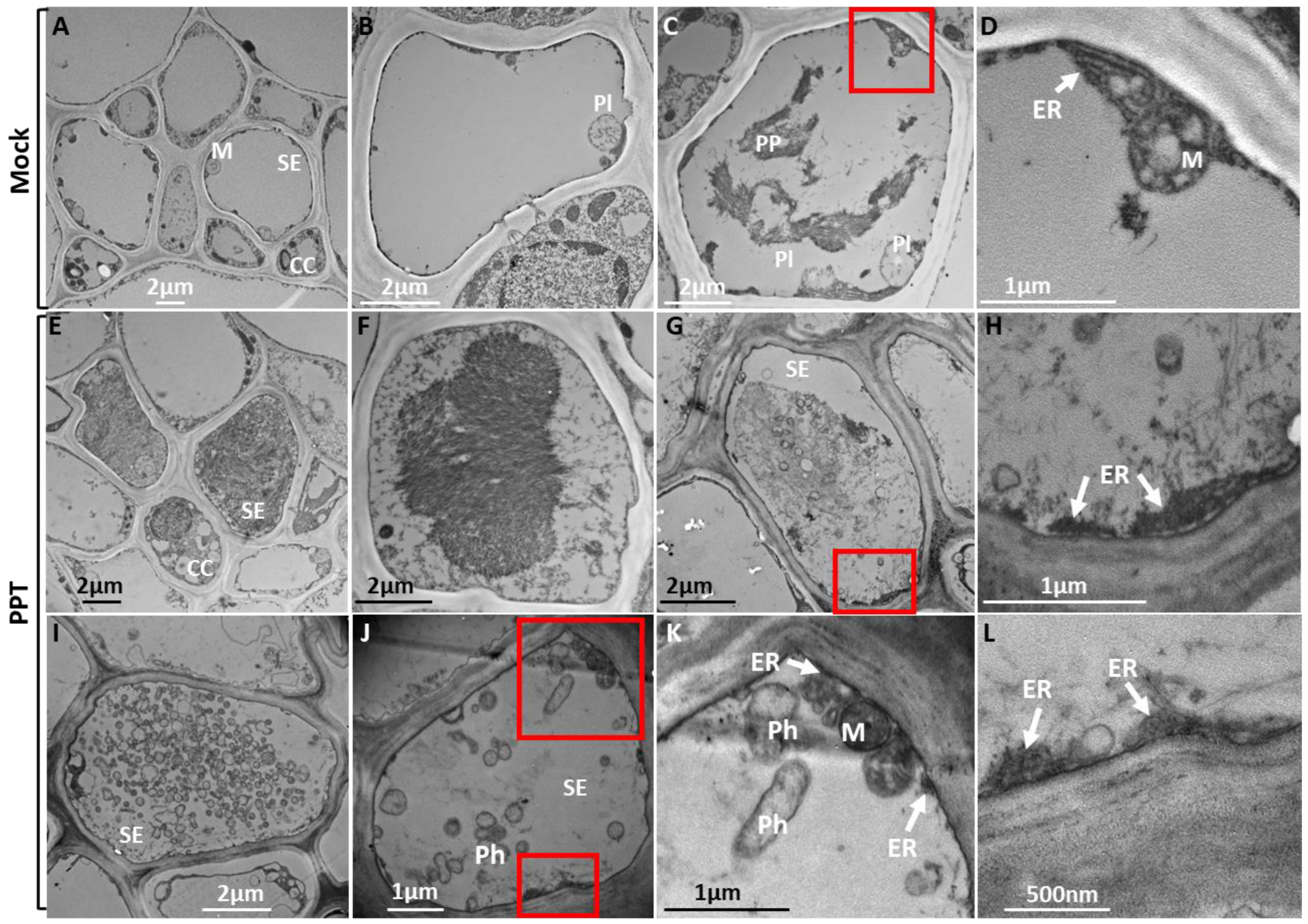
3.2. PPT Phytoplasma Induced Abnormal ER Morphology in Phloem
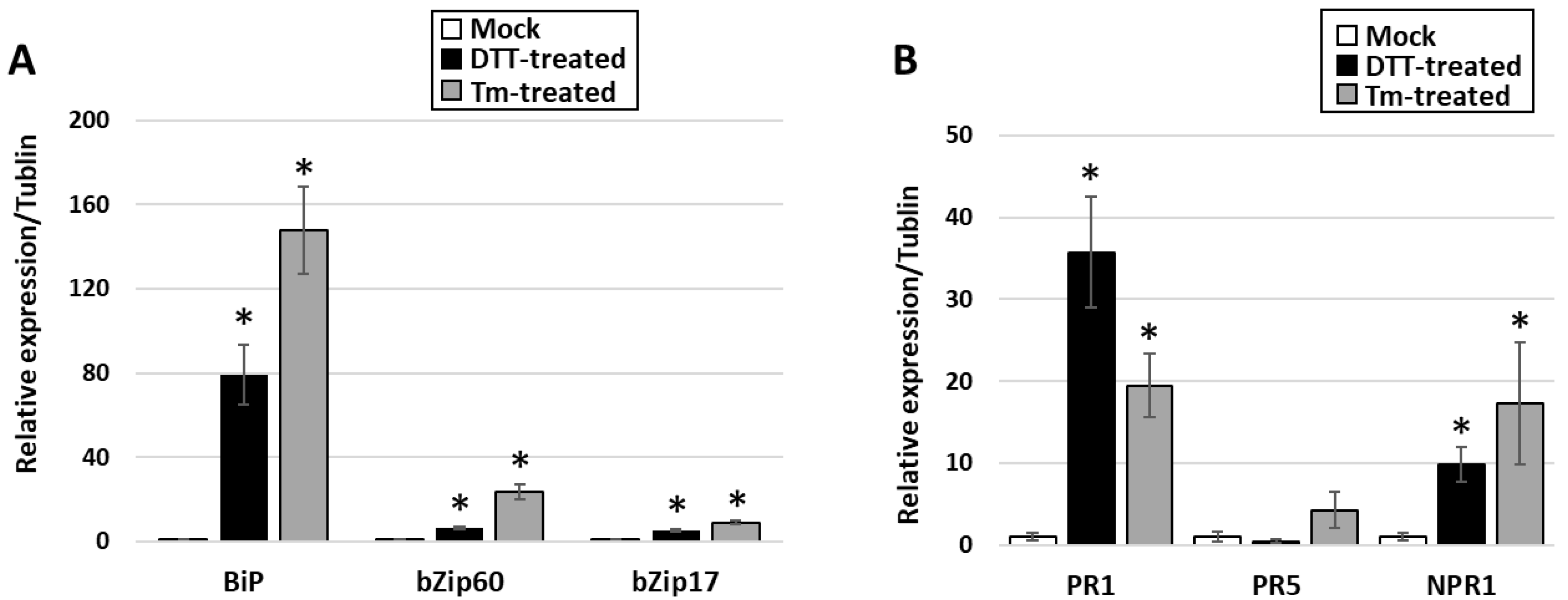
3.3. Activation of UPR in PPT-Phytoplasma-Infected Tomato Plants
3.4. Do Phytoplasmas Utilize ER and Its Resident Proteins for Their Own Infection and Multiplication?
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kube, M.; Mitrovic, J.; Duduk, B.; Rabus, R.; Seemüller, E. Current view on phytoplasma genomes and encoded metabolism. Sci. World J. 2012, 2012, 185942. [Google Scholar] [CrossRef]
- Wei, W.; Zhao, Y. Phytoplasma taxonomy: Nomenclature, classification, and identification. Biology 2022, 11, 1119. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Oshima, K.; AMMAR, E.D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Inaba, J.; Zhao, Y.; Mowery, J.D.; Hammond, R. Phytoplasma infection blocks starch breakdown and triggers chloroplast degradation, leading to premature leaf senescence, sucrose reallocation, and spatiotemporal redistribution of phytohormones. Int. J. Mol. Sci. 2022, 23, 1810. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Beanland, L. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef]
- Oshima, K.; Kakizawa, S.; Nishigawa, H.; Jung, H.-Y.; Wei, W.; Suzuki, S.; Arashida, R.; Nakata, D.; Miyata, S.-i.; Ugaki, M. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 2004, 36, 27–29. [Google Scholar] [CrossRef]
- Tan, Y.; Li, Q.; Zhao, Y.; Wei, H.; Wang, J.; Baker, C.J.; Liu, Q.; Wei, W. Integration of metabolomics and existing omics data reveals new insights into phytoplasma-induced metabolic reprogramming in host plants. PLoS ONE 2021, 16, e0246203. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef]
- Sun, J.-L.; Li, J.-Y.; Wang, M.-J.; Song, Z.-T.; Liu, J.-X. Protein quality control in plant organelles: Current progress and future perspectives. Mol. Plant 2021, 14, 95–114. [Google Scholar] [CrossRef]
- Park, C.-J.; Seo, Y.-S. Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015, 31, 323. [Google Scholar] [CrossRef]
- Vitale, A.; Boston, R.S. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 2008, 9, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Verchot, J. Plant virus infection and the ubiquitin proteasome machinery: Arms race along the endoplasmic reticulum. Viruses 2016, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Endoplasmic reticulum: The favorite intracellular niche for viral replication and assembly. Viruses 2016, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Toruño, T.Y.; Elowsky, C.G.; Zhang, C.; Steinbrenner, J.; Beynon, J.; Alfano, J.R. The Pseudomonas syringae type III effector H op D 1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL 9. New Phytol. 2014, 201, 1358–1370. [Google Scholar] [CrossRef]
- Park, C.-J.; Bart, R.; Chern, M.; Canlas, P.E.; Bai, W.; Ronald, P.C. Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS ONE 2010, 5, e9262. [Google Scholar] [CrossRef]
- Caplan, J.L.; Zhu, X.; Mamillapalli, P.; Marathe, R.; Anandalakshmi, R.; Dinesh-Kumar, S.P. Induced ER chaperones regulate a receptor-like kinase to mediate antiviral innate immune response in plants. Cell Host Microbe 2009, 6, 457–469. [Google Scholar] [CrossRef]
- Ye, C.; Dickman, M.B.; Whitham, S.A.; Payton, M.; Verchot, J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 2011, 156, 741–755. [Google Scholar] [CrossRef]
- Jing, M.; Wang, Y. Plant pathogens utilize effectors to hijack the host endoplasmic reticulum as part of their infection strategy. Engineering 2020, 6, 500–504. [Google Scholar] [CrossRef]
- Buxa, S.V.; Degola, F.; Polizzotto, R.; De Marco, F.; Loschi, A.; Kogel, K.-H.; di Toppi, L.S.; van Bel, A.J.; Musetti, R. Phytoplasma infection in tomato is associated with re-organization of plasma membrane, ER stacks, and actin filaments in sieve elements. Front. Plant Sci. 2015, 6, 650. [Google Scholar] [CrossRef]
- Musetti, R.; Pagliari, L.; Buxa, S.V.; Degola, F.; De Marco, F.; Loschi, A.; Kogel, K.-H.; van Bel, A.J. OHMS**: Phytoplasmas dictate changes in sieve-element ultrastructure to accommodate their requirements for nutrition, multiplication and translocation. Plant Signal. Behav. 2016, 11, e1138191. [Google Scholar] [CrossRef][Green Version]
- Musetti, R.; Pagliari, L.; Mian, G.; De Oliveira Cantao, F.R.; Bernardini, C.; Santi, S.; van Bel, A.J. The sieve-element endoplasmic reticulum: A focal point of phytoplasma-host plant interaction? Front. Microbiol. 2023, 14, 1030414. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, S.; Oshima, K.; Nishigawa, H.; Jung, H.Y.; Wei, W.; Suzuki, S.; Tanaka, M.; Miyata, S.I.; Ugaki, M.; Namba, S. Secretion of immunodominant membrane protein from onion yellows phytoplasma through the Sec protein-translocation system in Escherichia coli. Microbiology 2004, 150, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Oshima, K.; Kakizawa, S.; Arashida, R.; Jung, H.Y.; Yamaji, Y.; Nishigawa, H.; Ugaki, M.; Namba, S. Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 4252–4257. [Google Scholar] [CrossRef]
- Ozawa, M. Cloning of a human homologue of mouse reticulocalbin reveals conservation of structural domains in the novel endoplasmic reticulum resident Ca2+-binding protein with multiple EF-hand motifs. J. Biochem. 1995, 117, 1113–1119. [Google Scholar] [CrossRef]
- Sturbois-Balcerzak, B.; Vincent, P.; Maneta-Peyret, L.; Duvert, M.; Satiat-Jeunemaitre, B.; Cassagne, C.; Moreau, P. ATP-dependent formation of phosphatidylserine-rich vesicles from the endoplasmic reticulum of leek cells. Plant Physiol. 1999, 120, 245–256. [Google Scholar] [CrossRef]
- Wei, W.; Davis, R.E.; Nuss, D.L.; Zhao, Y. Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc. Natl. Acad. Sci. USA 2013, 110, 19149–19154. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Kakizawa, S.; Suzuki, S.; Jung, H.-Y.; Nishigawa, H.; Miyata, S.-i.; Oshima, K.; Ugaki, M.; Hibi, T.; Namba, S. In planta dynamic analysis of onion yellows phytoplasma using localized inoculation by insect transmission. Phytopathology 2004, 94, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Shao, J.; Bottner-Parker, K.D.; Zhao, Y. Draft Genome Sequence Resource of CBPPT1, a ‘Candidatus Phytoplasma trifolii’-Related Strain Associated with Potato Purple Top Disease in the Columbia Basin, USA. Plant Dis. 2023, 107, 922–925. [Google Scholar] [CrossRef]
- Inaba, J.-i.; Nagy, P.D. Tombusvirus RNA replication depends on the TOR pathway in yeast and plants. Virology 2018, 519, 207–222. [Google Scholar] [CrossRef]
- Wu, W.; Ding, Y.; Wei, W.; Davis, R.; Lee, I.M.; Hammond, R.; Zhao, Y. Salicylic acid-mediated elicitation of tomato defence against infection by potato purple top phytoplasma. Ann. Appl. Biol. 2012, 161, 36–45. [Google Scholar] [CrossRef]
- Sjolund, R.D. The phloem sieve element: A river runs through it. Plant Cell 1997, 9, 1137. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, S.-i.; Brodsky, J.L.; Nakatsukasa, K. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J. Biochem. 2005, 137, 551–555. [Google Scholar] [CrossRef]
- Iwata, Y.; Koizumi, N. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. 2012, 17, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Howell, S.H. The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 2017, 8, 344. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, M.; Achor, D.; Ghosh, S.; Kontsedalov, S.; Lebedev, G.; Levy, A. ‘Candidatus Liberibacter asiaticus’ accumulates inside endoplasmic reticulum associated vacuoles in the gut cells of Diaphorina citri. Sci. Rep. 2017, 7, 16945. [Google Scholar] [CrossRef]
- Steele-Mortimer, O. The Salmonella-containing vacuole—Moving with the times. Curr. Opin. Microbiol. 2008, 11, 38–45. [Google Scholar] [CrossRef]
- Machner, M.P.; Isberg, R.R. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 2006, 11, 47–56. [Google Scholar] [CrossRef]
- Mehlitz, A.; Karunakaran, K.; Herweg, J.A.; Krohne, G.; van de Linde, S.; Rieck, E.; Sauer, M.; Rudel, T. The chlamydial organism S imkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cell. Microbiol. 2014, 16, 1224–1243. [Google Scholar] [CrossRef]
- Derré, I. Chlamydiae interaction with the endoplasmic reticulum: Contact, function and consequences. Cell. Microbiol. 2015, 17, 959–966. [Google Scholar] [CrossRef]
- Saraste, J.; Prydz, K. Assembly and cellular exit of coronaviruses: Hijacking an unconventional secretory pathway from the pre-Golgi intermediate compartment via the Golgi ribbon to the extracellular space. Cells 2021, 10, 503. [Google Scholar] [CrossRef] [PubMed]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberté, J.F.; Zheng, H. Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Egan, A.J.; Vollmer, W. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 2013, 1277, 8–28. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inaba, J.; Kim, B.M.; Zhao, Y.; Jansen, A.M.; Wei, W. The Endoplasmic Reticulum Is a Key Battleground between Phytoplasma Aggression and Host Plant Defense. Cells 2023, 12, 2110. https://doi.org/10.3390/cells12162110
Inaba J, Kim BM, Zhao Y, Jansen AM, Wei W. The Endoplasmic Reticulum Is a Key Battleground between Phytoplasma Aggression and Host Plant Defense. Cells. 2023; 12(16):2110. https://doi.org/10.3390/cells12162110
Chicago/Turabian StyleInaba, Junichi, Bo Min Kim, Yan Zhao, Andrew M. Jansen, and Wei Wei. 2023. "The Endoplasmic Reticulum Is a Key Battleground between Phytoplasma Aggression and Host Plant Defense" Cells 12, no. 16: 2110. https://doi.org/10.3390/cells12162110
APA StyleInaba, J., Kim, B. M., Zhao, Y., Jansen, A. M., & Wei, W. (2023). The Endoplasmic Reticulum Is a Key Battleground between Phytoplasma Aggression and Host Plant Defense. Cells, 12(16), 2110. https://doi.org/10.3390/cells12162110






