Abstract
Sugars are the major source of energy in living organisms and play important roles in osmotic regulation, cell signaling and energy storage. SWEETs (Sugars Will Eventually be Exported Transporters) are the most recent family of sugar transporters that function as uniporters, facilitating the diffusion of sugar molecules across cell membranes. In plants, SWEETs play roles in multiple physiological processes including phloem loading, senescence, pollen nutrition, grain filling, nectar secretion, abiotic (drought, heat, cold, and salinity) and biotic stress regulation. In this review, we summarized the role of SWEET transporters in plant development and abiotic stress. The gene expression dynamics of various SWEET transporters under various abiotic stresses in different plant species are also discussed. Finally, we discuss the utilization of genome editing tools (TALENs and CRISPR/Cas9) to engineer SWEET genes that can facilitate trait improvement. Overall, recent advancements on SWEETs are highlighted, which could be used for crop trait improvement and abiotic stress tolerance.
1. Introduction
Photosynthetic organisms synthesize sugars during photosynthesis, a primary source of carbon and energy in cells [1]. Synthesized sugars are assimilated, transported, and distributed from source to sink tissues through the process of carbohydrate partitioning [1]. Sucrose is the main product of photosynthetic reactions, synthesized explicitly in the cytosol and transported to the sink organs [2]. Sucrose acts as a signaling molecule to control growth and differentiation [3]. Several review articles provide a detailed account of carbon partitioning, sugar metabolism, and signaling in plants [1,2,4,5,6,7,8]. Sugars are involved in various plant growth and developmental processes by acting as the source of carbon skeletons, the substrate of respiratory reactions, intermediate metabolites in biochemical reactions, storage substances, osmolyte, and signals in biotic and abiotic stresses [9,10,11,12,13,14]. The demand for sugar increases in the shoot/root apical meristem, flower buds, and seed/fruits organs [2,15,16]. Significant increases in sugar concentrations also occur under biotic and abiotic stresses such as cold, drought, phosphorus starvation, and pathogen attack [9,10,12,17]. In contrast, sugar levels decline under reduced oxygen conditions [9,13]. Additionally, sugars play a crucial role in regulating reproductive events such as pollen germination [18]. Thus, sugar metabolites form the core of the plant metabolism in response to developmental and environmental cues.
Sugar transporters across cell membranes mediate sugar translocation. These are evolutionally conserved genes present in bacteria, fungi, archaea, and plants [5,19,20,21]. Sugar transporters are classified into the following three types in plants: monosaccharide transporters (MSTs), sucrose transporters (SUTs), and the most recent type, SWEETs (Sugars Will Eventually be Exported Transporters) [5,19]. The major facilitator superfamily (MFS) transporters contain MSTs and SUTs and are primarily involved in sugar influx into the cytosol. However, some MSTs, namely tonoplast sugar transporter (TST), and the vacuolar glucose transporter (vGTs) are involved in transporting sugars from the cytosol to vacuoles and act as H+/sugar antiporters [22]. Both MSTs and SUTs contain 12 transmembrane α-helices and mediate membrane transport of different sugars [23,24]. MSTs are localized in plasma membranes and membranes of cell organelles such as chloroplast, Golgi, and vacuoles [25,26,27,28,29,30,31].
The third type of sugar transporters is SWEET transporters containing 7-TM domains [19,32]. SWEETs play critical roles in phloem transport of sugars [33,34], pollen nutrition [35], nectar secretion [36], grain filling, and size regulation [37,38,39], floral transition [40], abiotic [17,41,42,43,44,45] and biotic stresses [46,47].
Here, we review the current state of knowledge of various biological functions of the SWEET family of sugar transporters. The role of SWEET sugar transporters in various developmental stages and abiotic stresses (i.e., drought, cold, heat and salt stress) is discussed. Additionally, how genome editing technologies such as TALENs and CRISPR/Cas9 are being utilized to engineer SWEET genes to improve agricultural traits and yield under stresses in plants.
2. Sugar Production in Plants
Sugar synthesis takes place in specialized plant-cell compartments known as chloroplast in the presence of sunlight and CO2. The produced triose-phosphate is directly transported to the cytosol or used for starch synthesis in the chloroplast. In the dark, starch degrades to hexose sugars (glucose or maltose) and gets exported to cytosol, where conversions of glucose take place [glucose to glucose-6 phosphate (G-6-P) and then G-6-P to fructose-6-phosphate]. These two products are used by sucrose-phosphate synthase to produce sucrose phosphate, which is then converted to sucrose by sucrose-phosphate phosphatase [48].
Sucrose is the primary form of sugar is gets transported over long distances in plants [48,49]. Sucrose is loaded into the phloem parenchyma via plasmodesmata (symplastic) or specialized membrane transporters (apoplastic). In the apoplastic mode of transportation, sucrose is initially transported out of mesophyll cells into apoplast via SWEETs and then imported into companion cells from the apoplast through membrane-localized sucrose/H+ symporters known as SUT1 [23,33] (For details see Figure 1). In some plants, particularly trees with higher plasmodesmata connectivity between the sieve element- companion cells complex and mesophyll cells, the sucrose transports in a concentration gradient manner and passively enters phloem [50]. The phloem loading strategies opted by plants have been discussed previously [51,52,53]. Sucrose accumulation attracts water, which generates enhanced turgidity directing the mass flow of assimilates toward sink tissues. The unloading of sucrose from phloem to sink cells takes place apoplasmically or symplasmically, followed by degradation carried out by cytoplasmic invertases (cINs) or sucrose synthase (SuSy) [54,55]. Additionally, cytosolic sucrose is taken up into vacuoles for hydrolysis mediated by vacuolar invertases (vINs) [56]. Cell wall invertases (cwINs) are involved in sucrose partitioning, plant development and cell differentiation and derive sink strength during pathogen infection [57]. The hexose produced from sucrose hydrolysis is further utilized in glycolysis and in the synthesis of sugar polymers (i.e., cellulose, fructan, and starch) [48].
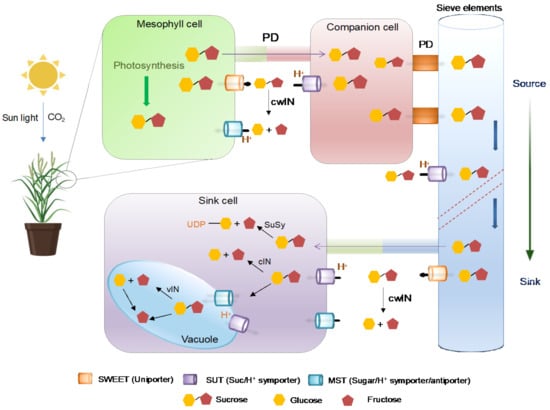
Figure 1.
Schematic diagram showing the path of sucrose transportation from source to sink. The photosynthetically synthesized sucrose is transported out from mesophyll cells via SWEETs sugar transporters. Sucrose transporters (SUTs) accumulate sucrose in the sieve element/companion cell complex for long-distance distribution throughout the plant body. PD: plasmodesmata, cwIN: cell wall invertase, cIN: cytoplasmic invertase, SuSy: sucrose synthase, vIN: vacuole invertase, Glc: glucose, Fru: fructose.
3. SWEET Gene Family in Plants
Furthermore, SUT1 plays a significant role in the phloem loading of sugars [58]; however, the mechanism by which sucrose is released into apoplastic space from leaf cells remained elusive. A new type of transporters designated as SWEETs were identified by using fluorescence resonance energy transfer-based technology [19]. SWEETs are membrane-localized uniporters that transport sugars across cell membranes [19,33]. The first two foundation members of this family were identified in Medicago truncatula (MtN3) and Drosophila melanogaster (Saliva) in the late 1990s [59,60]. Therefore, the domain present in SWEET proteins is named as MtN3/Saliva (MtN3_slv). The SWEETs consisting of transmembrane helices (TMHs) and three TMHs make a 3-TM domain. In prokaryotes, SWEETs are known as semiSWEETs, since they contain only one unit of 3-TMHs (TMH1-3). In contrast, eukaryotic SWEET genes consist of two 3-TMHs units (TMH1-3 and TMH5-7) separated by a less conserved TMH (TM4) [5]. However, some uncommon SWEET proteins have also been reported in plants and oomycetes. For example, extraSWEET contains four-five repeats of 3-TM domains attached by two single TMHs reported in Vitis vinifera and wild rice, and superSWEET with more than five to eight repeats 3-TM domain was reported in oomycetes [61]. The structures of semiSWEET, SWEET, extraSWEET, and superSWEET are presented in Figure 2. The semiSWEETs were initially thought to be uncommon in eukaryotes. However, a recent analysis of 25 plant species showed that out of 411 SWEETs, 140 SWEET genes were present in partial forms, suggesting that they are pseudogenes with truncated domains [62]. The evolution of SWEET genes in prokaryotes and higher organisms is not clear yet. Gene duplication and fusion have been proposed as key driving forces to facilitate the evolution and distribution of SWEET transporters [32].
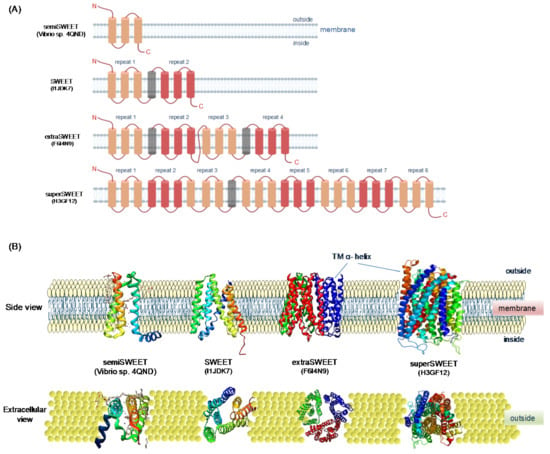
Figure 2.
Schematic representation of two-dimensional (2D) model and 3D protein structures of four types of SWEET proteins based on 1, 2, 4 and 8, 3- transmembrane helices (TMH) domains. (A) Two-D models of semiSWEET, SWEET, extraSWEET and superSWEET proteins (their UniProt/PDB IDs are shown in the corresponding models). Colored boxes indicate TMHs, and loops are marked with lines. and triangles represent functional 3-TM units. (B) Side and extracellular view of three-D protein structures of four types of SWEET proteins. Images were prepared with PYMOL.
Plant genomes contain more SWEET genes (7–68) than animals, including humans, where only one SWEET gene is present. Drosophila has two SWEET genes, and C. elegans has seven SWEET genes. SWEET genes were first reported in Arabidopsis [19], then identified in other plant species (Figure 3). A phylogenetic analysis of SWEET genes in plants classified them into four clades. Genes in clade I and II encode proteins, which transport hexose sugars such as glucose and fructose. The Clade III contains genes, which encode proteins that show preferential transport activity for sucrose over glucose. Genes in the clade IV mainly include vacuolar transporters involved in a flux of fructose across the tonoplast [19,33]. However, clade V is found in mammals, Chlamydomonas, and C. elegans [19].
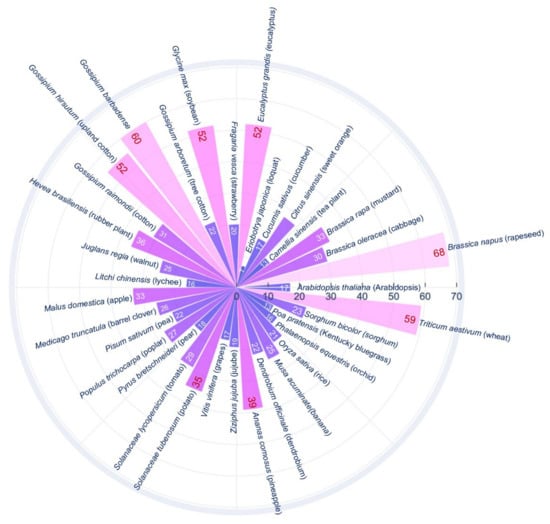
Figure 3.
Bar plot showing the SWEET genes present in different plant species. Arabidopsis thaliana [19], Brassica napus [63], Brassica oleracea [64], Brassica rapa [65,66], Camellia sinensis [44], Citrus sinensis [67,68], Cucumis sativus [69], Eriobotrya japonica [70], Eucalyptus grandis [71], Fragaria vesca [72], Glycine max [62], Gossypium arboretum [73], Gossypium barbadense [73], Gossypium hirsutum [73,74], Gossypium raimondii [73], Hevea brasiliensis [75], Juglans regia [76], Litchi chinensis [77], Malus domestica [78], Medicago truncatula [79,80], Pisum sativum [79], Populus trichocarpa [81], Pyrus bretschneideri [82], Solanaceae lycopersicum [83], Solanaceae tuberosum [84], Vitis vinifera [85], Ziziphus jujuba [86], Ananas comosus [87], Dendrobium officinale [88], Musa acuminate [43], Oryza sativa [89], Phalaenopsis equestris [88], Poa pratensis [90], Sorghum bicolor [91], Triticum aestivum [92,93]. The number depicted on the bar graphs represent number of SWEET genes in the plant species.
4. Role of SWEET Genes in Plant Development
SWEET genes are evolutionally conserved, playing a crucial role in various plant developmental processes, including phloem loading, nectar secretion, and reproductive organ development. Most of the experimental work elucidating the developmental roles of SWEETs have been performed in Arabidopsis and rice. This section describes the involvement of SWEET genes in various plant developmental processes.
4.1. Nectar Secretion
Nectar secretion is an essential and complex process to attract pollinators, which helps in pollination and maintaining genetic diversity in flowering plants. Nectar is produced in specialized organs called nectaries located inside or outside flowers. Furthermore, NEC1, an AtSWEET9 homolog, is predominantly expressed in the nectaries of Petunia hybrid and is assumed to play a role in nectar secretion [94]. A clade III member, SWEET9, is characterized as a nectary-specific sugar transporter in Arabidopsis, mustard and wild tobacco (Nicotiana attenuata) [36]. It functions in sucrose secretion from nectary parenchyma into apoplast, and mutation leads to a loss of nectar secretion. In other crops such as Hevea brasiliensis, Medicago truncatula, Pisum sativum orthologues of AtSWEET9 exhibited male flower-specific abundant expression, indicating a similar function to nectar production [75,79].
4.2. Leaf Senescence
Leaf senescence is an important trait that influences plant yield and nutritional quality. Carbohydrate offloading is mediated by SWEET (Clade II and III) and SUT (SUT1 and SUT2) in the senescing leaves [37]. The SWEET15, also called SAG29 (Senescence-Associated Gene 29), functions by remobilizing carbohydrates during senescence [41]. During senescence, SWEET15 is upregulated and can be used as a senescence marker. Overexpression of AtSWEET15 in Arabidopsis resulted in accelerated senescence that suggests its role in phloem loading during senescence [37]. The SAG gene (SAG101) encodes the membrane acyl hydrolase that regulates membrane hydrolysis in the early stages of senescence. The accumulation of hexose sugars (mainly glucose, fructose end galactose) in senescent leaves also leads to the speculation that clade II SWEETs may also function in carbon partitioning during senescence [41]. The OsSWEET5 belongs to clade II and is involved in the galactose transporter in rice. The overexpression of OsSWEET5 causes early leaf senescence, growth retardation, and change in auxin levels at the seedling stage in rice [42]. Increased clad II and III SWEET genes expression have been reported in senescing leaves of Pisum sativum and Brassica rapa [63,95]. In pear, the expression of PbSWEET4 (clade III member), a homolog of AtSWEET15, is localized in the cell membrane. The expression of the PbSWEET4 gene is potentially related to leaf development and is highly expressed in older leaves. The overexpression of PbSWEET4 in strawberry plants resulted in a reduced sugar and chlorophyll content and accelerated leaf senescence [96]. Overall, this suggests that SWEET genes can be modulated to alter leaf senescence traits in plants.
4.3. Fruit and Seed Development
Recent studies on gene expression in different plant species, including pineapple, apple, and pear, demonstrate the role of SWEETs in fruit development. For instance, in pineapple two genes, namely AnmSWEET-5 and 11 demonstrated up-regulation at the early phases of fruit development [87]. In apple, nine SWEET genes were abundantly expressed during apple fruit development. Two genes, MdSWEET9b and 15a, were associated with fruit sugar accumulation and likely to be implicated in fruit development [97]. Recently, in pear, histone acetylation-mediated regulation of SWEET genes was involved in fruit development [98]. Comparative transcriptomics of two pear varieties, ‘Nanguo’ (NG; low sucrose content) and its bud sport (BNG; high sucrose content) revealed that the PuSWEET15 gene is induced in BNG fruit. PuSWEET15 overexpression in NG fruit induced sucrose content, while silencing in BNG fruit reduced the sucrose level.
SWEET genes also play an essential role during seed development. An increase in transcript levels of several SWEET genes (ZmSWEET4c, 6b, 11, 13a, 13b, 14b and 15a) was observed during seed germination in maize [99]. These genes participate in the sucrose efflux from scutellum to embryo axis. In crops, yield is determined by the allocation of sugars from leaves to seed, which is carried out by specific SWEET transporters. In Arabidopsis, three SWEET genes (SWEET11, 12, and 15) of clad III showed spatiotemporal expression during seed development and might help to transport sucrose from seed coat to the developing embryo. Triple mutant lines of atsweet11:12:15 produce retarded embryos with reduced seed weight and low starch and lipid content, resulting in wrinkled seeds production [37]. In rice, the double knockout of ossweet11:15 accrued starch in the pericarp, whereas caryopses did not comprise a functional endosperm (Yang et al. 2018). However, the knockout of a single gene in rice (OsSWEET11) and soybean (GmSWEET15) also produces the same phenotype, i.e., decreased sucrose concentration in the embryo resulting in seed abortion [100,101]. This demonstrates that SWEET transports sucrose from seed coat to the developing embryo and plays a vital role in seed development. However, in some cases, clade II SWEET genes which transport mainly hexose, are also reported to play an essential role during seed development. In maize ZmSWEET4c, a clade II SWEET participates in the transport of hexoses across the basal endosperm transfer layer. Impaired seed filling was observed in the mutants of zmsweet4c and its rice ortholog ossweet4, suggesting that SWEET4 enhances sugar import into the endosperm in both maize and rice [38]. In Litchi chinensis, the temporal and spatial expression profiling indicated the role of LcSWEET2a and 3b in seed development [77].
4.4. Shoot Branching and Bud Outgrowth
Sugars are involved in shoot branching and bud outgrowth [102,103,104]. A SWEET gene (CmSWEET17) in Chrysanthemum morifolium displays axillary bud-specific expression after treatment with 20 mM sucrose, and the overexpression of CmSWEET17 promotes axillary bud growth [105]. Simultaneously, the CmSWEET17 overexpression lines revealed the induction of several auxin transporter genes [AUXIN RESISTANT 1 (AUX1), LIKE AUX1 2 (LAX2), PINFORMED1 (PIN1), PIN2 and PIN4], indicating that SWEET17 may be engaged in sucrose-mediated axillary bud outgrowth via the auxin transport pathway [105].
4.5. Development of Reproductive Organs
SWEET genes are expressed at different stages of pollen development. In Arabidopsis, AtSWEET8/RPG1 (Ruptured pollen grain 1) is expressed in microsporocyte and tapetum. Pollen grains of atsweet8 mutants are aborted and sterile, suggesting its involvement in anther and pollen development [106]. Furthermore, AtSWEET13/RPG2 partly restores the male fertility of atsweet8 at the late reproductive stages, which is also expressed in the anther during microsporogenesis, indicating functional redundancy among SWEETs. However, the double mutant of rpg1:rpg2 was fully sterile and was unable to restore [35]. Knockout mutants of AtSWEET11 and OsSWEET11/Os8N3/Xa13 also produced defective pollen grains and reduced male fertility in Arabidopsis and rice, respectively [107,108,109]. Some other SWEET genes such as AtSWEET1, PwSWEET1, and AtSWEET5/VEX1 are expressed at different stages of pollen development, which indicates their role in pollen development [19,110].
5. Role of SWEET Genes in Abiotic Stress
To cope with different abiotic constraints, plants tightly regulate the vacuolar storage and transport of sugars. For instance, sugar accumulation occurs in vacuoles to minimize freezing stress [44,111]. Additionally, SWEET genes are responsive to various abiotic stresses, suggesting their role in abiotic stress response (Figure 4 and Table 1). We analyzed the transcriptional dynamics of SWEET genes using Genevestigator [112] in Arabidopsis and rice under drought, heat, cold and salt stresses (Figure 4). The following sections summarize and discuss the role of SWEET genes in plant abiotic stress responses.
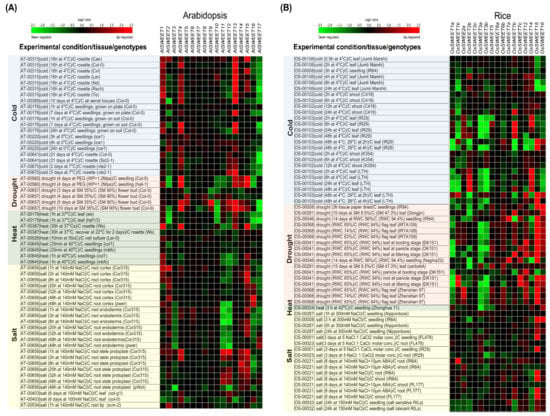
Figure 4.
Heat maps representing SWEET gene expression patterns in (A) Arabidopsis and (B) rice under abiotic stresses (cold, drought, heat and salt). Heatmap was constructed from the data obtained from the Genevestigator database containing different experiments. [Red = up-regulation and Green = down-regulation of genes].
5.1. Osmotic or Drought Stress
Prolonged drought increases the root to shoot ratio [1], which is affected by excess C assimilation in leaf that is transported to roots [113]. This suggests that sugar transporters may play a crucial role under drought stress conditions. Consistently, AtSWEET4, AtSWEET13, AtSWEET14 and AtSWEET15 were induced in Arabidopsis and OsSWEET12, OsSWEET15 and OsSWEET16 were induced in rice (Figure 4). Likewise, AtSWEET11, AtSWEET12, and AtSUC2 transcript levels were significantly induced in leaves, while AtSUC2 and AtSWEET11-15 were induced in roots of water-stressed Arabidopsis [114]. An increase in the expression of sugar transporters in both leaves and roots suggests that plants have to maintain an efficient root system under stress conditions, which is ensured by allocating more C to roots. In contrast, Durand et al. [115] reported the downregulation of AtSUC2, AtSWEET11, AtSWEET12, AtSWEET13, and AtSWEET15 and reduced sucrose transport between leaves and roots in response to poly-ethylene-glycol (PEG) treated Arabidopsis plants. This suggests that plants use distinct mechanisms to cope with drought and PEG-induced osmotic stress.

Table 1.
List of important SWEET sugar transporters genes in plants involved in different abiotic stresses.
Table 1.
List of important SWEET sugar transporters genes in plants involved in different abiotic stresses.
| Abiotic Stress/Genes | Plant Species | Experimental Results | Reference |
|---|---|---|---|
| (a) Drought Stress | |||
| AtSWEET11, 12, and 15 | Arabidopsis thaliana | Up-regulated in shoot and roots under drought stress (0.4 g water g−1 compost) | [114] |
| DsSWEET12 and 17 | Dianthus spiculifolius | DsSWEET12 overexpression (OE) Arabidopsis lines showed enhanced tolerance to osmotic stress. | [116,117] |
| GhSWEET5, 20, 49 and 55 | Gossypium hirsutum | Up-regulated under drought stress (20% PEG-6000; 1 h) | [65] |
| MdSWEET17 | Malus domestica | MdSWEET17 transgenic tomatoes showed higher drought tolerance (10% PEG-6000; 3–48 h) | [45] |
| TaSWEET14g-1A and 16a-4A | Triticum aestivum | Up-regulated under drought stress (20% PEG-8000; 6 h) | [93] |
| CaSWEET1-like, and 4 | Cicer arietinum | Up-regulated under drought stress (70% available soil water fraction) | [118] |
| GmSWEET6 and 15 | Glycine max | Up-regulated in drought stress (field water holding capacity of 35–40%) | [119] |
| StSWEET10b | Solanumtuberosum | Up-regulated under drought stress (reduced soil water content) | [120] |
| AtSWEET17 | Arabidopsis thaliana | Up-regulated in roots under drought stress (−0.5 MPa osmotic potential; PEG-8000; 6 h) | [121] |
| CsSWEET1a, 2a, 2c, 3a, 7a, 7b and 10a | Camellia sinensis | Up-regulated under drought (PEG; 72 h) | [122] |
| OsSWEET13 and 15 | Oryza sativa | Up-regulated under drought stress (20% PEG-6000) | [17] |
| (b) Heat Stress | |||
| BnSWEET9-2, 10-3, 12, 13-2 and 14 | Brassica napus | Up-regulated under heat stress (40 °C; 3–24 h) | [63] |
| GhSWEET4 and 10e | Gossypium hirsutum | Up-regulated under heat stress (40 °C; 3–10 h) | [73] |
| GhSWEET5, 49 and 55 | Gossypium hirsutum | Up-regulated under heat stress (38 °C; 6 h) | [65] |
| BrSWEET11 | Brassica rapa | Up-regulated under heat stress (38 °C; 8 h) | [66] |
| TaSWEET14g-1A, 14h-1B and 15a-7D | Triticum aestivum | Up-regulated under heat stress (42 °C; 6 h) | [93] |
| (c) Cold Stress | |||
| AtSWEET16 and AtSWEET17 | Arabidopsis thaliana | Provides higher cold tolerance (1 week; 4 °C) by transporting glucose or fructose in the tonoplasts of leaves and roots | [123,124] |
| AtSWEET11 and 12 | Arabidopsis thaliana | Up-regulated under cold stress (1 week; 4 °C) and affect vascular development | [125] |
| AtSWEET4 | Arabidopsis thaliana | AtSWEET4 OE lines have higher freezing tolerance | [126] |
| MaSWEET1, 4, and 14 | Musa acuminata | Up-regulated under cold stress (4 °C; 22 h) | [43] |
| CsSWEET16 | Camellia sinensis | Enhanced cold tolerance in CsSWEET16 OE lines | [49] |
| BoSWEET11b, 11c, 12b, 16a, and 17 | Brassica oleracea | Show variable expression pattern under cold treatments (4 °C; 3–48 h) | [64] |
| MdSWEET16 | Malus domestica | Enhanced cold tolerance in MdSWEET16 OE lines | [127] |
| CsSWEET1a, 1b, 3b, and 15c | Camellia sinensis | Show variable expression pattern under cold treatments | [122] |
| (d) Salinity Stress | |||
| AtSWEET15 | Arabidopsis thaliana | AtSWEET15/SAG29 OE plants show accelerated senescence and hypersensitivity to salinity | [41] |
| DsSWEET17 | Dianthus spiculifolius | DsSWEET17 OE Arabidopsis lines have higher slat tolerance | [117] |
| AtSWEET2, 13, 14, 16, and 17 | Arabidopsis thaliana | Show variable expression pattern under salt stress treatments (150 mM NaCl) | [128] |
| MtSWEET1a, 2a, 2b, 3c, 7, 9b, and 13 | Medicago truncatula | Show variable expression pattern under salt stress treatments (300 mM NaCl) | [80] |
| OsSWEET11 and 14 | Oryza sativa | Down-regulated under salt stress (150 mM NaCl) | [129] |
| OsSWEET13 and 15 | Oryza sativa | Up-regulated under salt stress (20 mM NaCl) | [17] |
Several other examples demonstrate the role of SWEETs in drought tolerance (Table 1). Arabidopsis seedlings overexpressing the Dianthus Spiculifolius gene DsSWEET12 have longer roots, high fructose, and glucose with a lower sucrose content and higher tolerance to osmotic and oxidative stresses compared to wild type plants [116]. Transgenic lines of tomato overexpressing Malus domestica SWEET gene MdSWEET17 showed a higher accumulation of fructose and enhanced drought tolerance [45]. It is well known that sugars act as osmoprotectants, and therefore participate in osmotic stress tolerance [130]. The total sugar content in embryos increased after PEG and NaCl treatment in sorghum, but the increase in fructose treatment was most apparent [131]. A significant reduction in the expression of SWEET10b under drought stress was also reported in potato [120].
In a recent study, another sugar transporter AtSWEET17, localized in the vacuolar membrane, was markedly upregulated during lateral root (LR) development under drought stress [121]. In another study, comparative root transcriptomics of chickpea under drought stress were carried out, and three SWEET genes (N3 (LOC101510607), SWEET1-like (LOC101515250), and SWEET4 (LOC101488443)) were reported to be upregulated in chickpea genotypes [118]. In soybean, drought stress increased leaf sucrose and soluble sugars but decreased root starch content. Consistent with this, the expression of sucrose transporters (GmSUC2, GmSWEET6, and 15) was upregulated in the leaves and roots [119]. In rice, OsSWEET13 and OsSWEET15 were induced in response to drought stress. The higher expression of these two genes was due to the binding of an ABA-responsive TF (OsbZIP72) to their promoter sequences. This modulates sucrose transport and distribution in response to drought stress, thus maintaining sugar homeostasis in response to drought stress [17]. In wheat, 13 SWEET genes showed differential expression after PEG treatment at the seedling stage. These genes include four members of clade I (TaSWEET2a1-6B, TaSWEET2a2-6D, and TaSWEET2b2-3A), four members of clade III (TaSWEET13c-6A, TaSWEET14h-6D, TaSWEET14g-1A, and TaSWEET15a-7D), and five members of clade IV (TaSWEET16c-4D, TaSWEET16a-4A, TaSWEET17a-5D, TaSWEET17c-5A, and TaSWEET17b-5B). However, these genes are expressed in a clade-specific manner, i.e., the members of clade I show downregulation, clade III show upregulation, and clade IV showed upregulation [93]. Similarly, in tea plants, seven genes (CsSWEET1a, 2a, 2c, 3a, 7a, 7b and 10a) were induced under drought stress [122]. However, further research is required to address the contrasting gene expression patterns of SWEET sugar transporters under drought and osmotic conditions.
5.2. Heat Stress
Heat stress inhibits carbon fixation while respiration increases, and heat tolerance involves the maintenance of leaf sugar content [132,133]. Heat reduces sugar export from source leaves to sink. For instance, in maize, the export rate of sugars from source leaves decreased after heat stress [134]. Heat stress caused a decline in the starch content of tomato mesophyll cells, but it increased significantly at a later time point [135]. Consistently, AtSWEET1, AtSWEET4, AtSWEET13 and AtSWEET15 were induced, and AtSWEET2, AtSWEET10 and AtSWEET17 were suppressed in Arabidopsis. In rice, OsSWEET14 and OsSWEET16 were induced, and OsSWEET3b, OsSWEET4 and OsSWEET5 were suppressed (Figure 4).
In wheat, 22 sugar transporters were up-regulated and 19 were suppressed under heat stress [136]. In Brassica napus, SWEET genes (BnSWEET9-2, 10-3, 12, 13-2 and 14) were up-regulated after heat stress [63]. In B. rapa, BrSWEET1 was expressed after 2 h of heat stress, while BrSWEET11 was expressed after 8 h of heat [66]. In cotton, GhSWEET4, 5, 10e, 49 and 55 showed induced expression under heat stress [65,73]. In wheat, 18 paralogues of nine SWEET genes (SWEET1, 2, 3, 4, 6, 14, 15, 16, and 17) showed a differential expression after 6 h of heat stress at the seedling stage. However, these genes expressed in a clade-specific manner, i.e., the members of clade II, III, and IV showed downregulation (except TaSWEET15a-7D), while the members of clade I showed both upregulation and downregulation [93]. Studies of different plant species under heat stress show that sugar levels of the source leaves decline due to decreased photosynthesis. However, functional gene studies are required to demonstrate the role of SWEET transporters in heat stress.
5.3. Cold Stress
Cold stress induces sugar accumulation in plants, and several SWEET genes such as AtSWEET15/SAG29 are up-regulated under cold stress [137]. The expression of AtSWEET1, AtSWEET2b, AtSWEET4, AtSWEET13 and AtSWEET15 was induced in Arabidopsis and OsSWEET7c and OsSWEET14 were induced in rice (Figure 4). Cold stress resulted in a higher accumulation of glucose and fructose than wild-type in Arabidopsis sweet11 and atsweet11/12 mutants, which resulted in cold stress tolerance. Enhanced tolerance observed in the double mutant may be due to the reduced number of xylem cells and smaller diameter vessels [125]. Additionally, it has been shown that the overexpression of AtSWEET16 shows freezing tolerance [123]. AtSWEET4 facilitates sugar transport in axial tissues during plant growth and development, and the transgenic plants overexpressing AtSWEET4 exhibits higher freezing tolerance [126]. In tea, CsSWEET16, a vacuolar membrane transporter, is downregulated after cold stress. The overexpression of the CsSWEET16 in Arabidopsis resulted in the compartmentation of sugars across the vacuole [44], while the overexpression of AtSWEET17 reduced the fructose content in leaves by 80% under cold stress conditions [124,138]. Furthermore, MaSWEET1, 4 and 14 expressions were upregulated in banana (Musa acuminata L.) under various stresses, indicating its role in stress tolerance to multiple stresses [43]. A genome-wide study carried out in Brassica oleracea reported the downregulation of BoSWEET11b, 11c, 12b, 16a, and 17 after chilling stress, possibly resulting in the accumulation of glucose and fructose and an enhanced chilling tolerance [64]. Similarly, several other genome-wide studies have been performed in different plants. They show differential expression of SWEET genes after cold stress (Table 1), suggesting that SWEET genes mediate cold-induced sugar-signaling responses [43,44,64,73,80,122,127]. The above studies indicate the role of SWEET genes in providing cold stress tolerance in plants; however, functional validation of these genes is still needed.
5.4. Salinity Stress
Salinity stress affects various physiological and metabolic processes, ultimately inhibiting crop productivity [139]. There are two phases of salt stress in plants; the first phase is an osmotic phase in which leaf-growth inhibition occurs, followed by the second phase of ion toxicity in which accelerated leaf senescence occurs [140]. Instead, a phase zero is also suggested, known as the transient phase, and begins quickly after salt shock, resulting in a lower turgor pressure and growth rate [141]. Sucrose also behaves similarly to osmolyte and prevents salt stress-induced damages [142]. Additionally, SWEET15 (clade III member) is mainly involved in the sucrose transportation. Our analysis showed that AtSWEET1, AtSWEET2, AtSWEET4, AtSWEET14 and AtSWEET15 were induced in Arabidopsis and OsSWEET1b, OsSWEET7c and OsSWEET15 were induced in rice (Figure 4). Consistently, AtSWEET15 is induced under osmotic stress, and AtSWEET15 overexpression leads to accelerated senescence and hypersensitivity to salt stress [41]. The transcript level of SWEET15 was observed as 64-fold higher than the control in phase 1 after salt stress, and for this property, the expression of this gene can be used as a marker to differentiate between phase 0 and phase 1 in Arabidopsis and maize [143]. The expression of MtSWEET1a, MtSWEET2b, MtSWEET7, MtSWEET9b and MtSWEET13 were upregulated under salt, while MtSWEET2a and MtSWEET3c were down-regulated [80]. However, in Arabidopsis, a lower transcript level of SWEET2, 13, 16, and 17 was observed, while a higher transcript level was observed for SWEET14 [128]. Since SWEET2, 16 and 17 transport glucose, fructose and/or sucrose across the tonoplast along the concentration gradient, their downregulation supports the hypothesis of reducing the cytosolic sugar towards storage in the vacuole [37,123]. The heterologous expression of DsSWEET17, a tonoplast sugar transporters of Dianthus spiculifolius, in A. thaliana affects sugar metabolism and tolerance to salinity, osmotic, and oxidative stresses [116]. In this study, higher fructose accumulation is observed in transgenic Arabidopsis than in the wild type, consistent with the previous study that reported a decrease in leaf fructose content in sweet17 [138].
Salt stress induces the expression of FLN2 (fructokinase-like protein2). The FLN2 knockout generated by CRISPR/Cas9 was hypersensitive to salinity stress and showed the disruption of the sugar metabolism, inhibition of Rubisco activity, and downregulation of sucrose synthesis and transportation genes. In FLN2 knockout lines, SWEET11 and SWEET14 were down-regulated after salt stress and indicated that FLN2 protein enhanced salinity tolerance in plants via influencing the sugar metabolism [129]. In rice, two SWEET genes (OsSWEET13 and 15) were also involved in the modulation of sucrose transport in response to salinity stress [17]. Several genome-wide studies using publicly available transcriptome data in different plant species, i.e., wheat, Medicago truncatula, banana, revealed differential expression of SWEET genes under salinity stress (Table 1) [80,144]. Altogether, these studies suggested that SWEET genes were also involved in salt stress tolerance in plants. However, further investigation into their functional roles in salt stress tolerance is required.
6. Additional Roles of SWEETs Other than Sugar Transportation
The SWEET members have also been demonstrated to transport hormones other than sugars. The two Arabidopsis SWEET genes (AtSWEET13 and AtSWEET14) are involved in the transportation of various forms of gibberellins (GA) [145]. The Arabidopsis sweet13;14 double mutant shows a phenotype related to the GA response, i.e., delayed anther dehiscence, and exogeneous GA application restored this phenotype. This experiment supports that SWEETs are involved in GA transport in plants. Similarly, in pea, the interaction between cytokinins, SWEET and cell wall invertase (CWIN) led to the formation of multiple shoots during pathogen infection [146]. These two studies depict the role of SWEET genes in the transportation of phytohormones and help us speculate the additional role of SWEET genes in the transport of phytohormones in addition to sugars. However, some additional research is required to confirm these extra transport functions and their vital relevance.
7. Genetic Engineering of SWEET Genes for Crop Improvement
Genome editing has modernized biology and can facilitate the targeted modifications of genomes [147]. Zinc-Finger Nucleases (ZFNs), TAL Effector Nuclease (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein-9 nuclease (CRISPR/Cas9) are the most commonly used tools. Several reports on rice and other plants (Cassava and cotton) exist, for which TALENs or CRISPR/Cas9 technologies have been used to target SWEET genes (Table 2). Most of these studies were focused on bacterial blight resistance. The bacterial transcription activator-like (TAL) effectors are involved in pathogen virulence. The TAL effectors of Xanthomonas oryzae (Xoo) transcriptionally activate rice disease-susceptibility (S) genes, including SWEET genes. Thus, genome-editing techniques could be used to enhance disease resistance by deleting effector-binding elements (EBEs) in the promoter region of S genes. For example, TALEs (AvrXa7, PthXo1, PthXo2, Tal5, and TalC) from different Xoo strains targets different SWEET genes (OsSWEET11/12/13/14) [148,149,150,151,152,153]. The EBEs in the OsSWEET14 were edited using TALENs in a susceptible rice cv. Kitake and the mutated lines were found to be resistant towards AvrXa7 and PthXo3 strains [150,152]. For the functional study of those genes for which no naturally occurring TAL effectors are present, designer TAL effectors (dTALEs) can be used. For example, in rice OsSWEET12 was induced by the infection of a Xoo strain transformed with dTALEs, hence providing susceptibility [151]. Additionally, in cassava (Manihot esculenta), by utilizing dTALE that complements TAL20Xam668 mutant phenotypes, it was shown that MeSWEET10a is the primary virulence gene that is the target of TAL20Xam668 [154].

Table 2.
Genome editing approaches utilized to target SWEET genes in plants.
Similarly, CRISPR/Cas9 technology has been implemented to edit rice SWEET genes [64,100,153,157,158,159,160,161,162,163] (Table 2). Jiang et al. [161] designed sgRNAs to edit the genes OsSWEET11 and OsSWEET14, which are involved in resistance to bacterial blight caused by Xoo. In another study, the CRISPR/Cas9 approach was utilized to edit OsSWEET13, an S-gene of the pathotype PthXo2 in rice [160]. The broad-spectrum against bacterial blight resistance was achieved in rice by disrupting the EBEs of two S genes (OsSWEET11 and 14) by CRISPR/Cas9 system. Interestingly, the mutation was introduced into the rice cultivar Kitaake, containing the recessive resistance allele of Xa25/OsSWEET13 [157,159,164]. Besides the disease-resistance dissections, CRISPR/Cas9 technology was also utilized to dissect the role of the SWEET gene during grain filling in rice. The knockout of OsSWEET11/Os8N3 showed a decreased sucrose concentration in the mutant plants’ embryo sacs, which lead to aberrant grain filling. These results suggest that OsSWEET11/Os8N3 is involved in sucrose transportation during the early phase of caryopsis development [100]. Future efforts should be carried out to target all EBE/S gene combinations and other important SWEET genes via TALEN or CRISPR/cas9 technology to confer broad-spectrum resistance, abiotic-stress resistance and plant development and growth-related traits in important crops such as rice, maize, and wheat.
8. Conclusions and Future Prospects
This review highlights the role of SWEET sugar transporters in phloem-loading, symplastic sucrose transport during, pollen nutrition, nectar secretion, grain filling, biotic and abiotic stress regulation, and transport of GAs. The functional characterization of SWEETs under various developmental and stress conditions has been well documented in Arabidopsis. However, in crop plants, functional characterization studies of SWEET transporters have just begun. Gene editing tools such as TALENs or CRISPR/Cas9 can be crucial in this context and have been used to study SWEET gene function under biotic stresses, but less functional studies exist in the case of abiotic stress regulation (Table 2). Their characterization can lead to exciting discoveries as sugar plays a central role in crop growth, development, and yield. Furthermore, SWEETs can be key targets when engineering plants with an improved abiotic stress tolerance and yield.
Author Contributions
V.G. and S.K. conceived and designed the manuscript, T.G., M.D., V.J., G.Z. wrote the manuscript, with contributions by all Authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Council of Scientific and Industrial Research (CSIR), India (Grant No. MLP-201) and Department of Science and technology for the INSPIRE faculty award.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The funding has been received from the Council of Scientific and Industrial Research (CSIR), India (Grant No. MLP-201). V.G. thanks to the Department of Science and technology for the INSPIRE faculty award. G.Z. acknowledges the support obtained from the projects funded by CSIR-FIRST (MLP-178), and DST Start-up Research Grant (SRG), SERB (GAP-294). This manuscript represents CSIR-IHBT publication number 4942.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.-L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-Sink Transport of Sugar and Regulation by Environmental Factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Horacio, P.; Martínez-Noël, G. Sucrose signaling in plants: A world yet to be explored. Plant Signal. Behav. 2013, 8, e23316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, D.M.; Wang, L.; Ruan, Y.-L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 2013, 65, 1713–1735. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of Sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [Green Version]
- Cho, L.-H.; Pasriga, R.; Yoon, J.; Jeon, J.-S.; An, G. Roles of Sugars in Controlling Flowering Time. J. Plant Biol. 2018, 61, 121–130. [Google Scholar] [CrossRef]
- Ciereszko, I. Regulatory roles of sugars in plant growth and development. Acta Soc. Bot. Pol. 2018, 87, 2. [Google Scholar] [CrossRef] [Green Version]
- Ciereszko, I. Sucrose Metabolism in Plant Tissues under Stress Conditions: Key Enzymes, Localization, and Function. In Compartmentation of Responses to Stresses in Higher Plants, True or False; Maksymiec, W., Ed.; Transworld Research Network: Kerala, India, 2009; pp. 193–218. [Google Scholar]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant. 2014, 36, 1607–1619. [Google Scholar] [CrossRef] [Green Version]
- Morkunas, I.; Borek, S.; Formela, M.; Ratajczak, L. Plant responses to sugar starvation. In Carbohydrates—Comprehensive Studies on Glycobiology and Glycotechnology; Chang, C., Ed.; IntechOpen: London, UK, 2012; Chapter 19; pp. 409–438. [Google Scholar]
- Lukaszuk, E.; Rys, M.; Możdżeń, K.; Stawoska, I.; Skoczowski, A.; Ciereszko, I. Photosynthesis and sucrose metabolism in leaves of Arabidopsis thaliana aos, ein4 and rcd1 mutants as affected by wounding. Acta Physiol. Plant. 2016, 39, 17. [Google Scholar] [CrossRef] [Green Version]
- Sami, F.; Yusuf, M.; Faizan, M.; Faraz, A.; Hayat, S. Role of sugars under abiotic stress. Plant Physiol. Biochem. 2016, 109, 54–61. [Google Scholar] [CrossRef]
- Fernandez, O.; Ishihara, H.; George, G.M.; Mengin, V.; Flis, A.; Sumner, D.; Arrivault, S.; Feil, R.; Lunn, J.E.; Zeeman, S.C.; et al. Leaf Starch Turnover Occurs in Long Days and in Falling Light at the End of the Day. Plant Physiol. 2017, 174, 2199–2212. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, C.A.; Paul, M.; Foyer, C.H. Metabolite transport and associated sugar signalling systems underpinning source/sink interactions. Biochim. Biophys. Acta 2016, 1857, 1715–1725. [Google Scholar] [CrossRef] [Green Version]
- Ko, H.-Y.; Ho, L.-H.; Neuhaus, H.E.; Guo, W.-J. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021, 187, 2230–2245. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant. 2020, 171, 620–637. [Google Scholar] [CrossRef]
- Hirsche, J.; FernáNdez, J.M.G.; Stabentheiner, E.; GroßKinsky, D.K.; Roitsch, T. Differential Effects of Carbohydrates on Arabidopsis Pollen Germination. Plant Cell Physiol. 2017, 58, 691–701. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Gerós, H.; Granell, A. Plant SWEETs: From sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021, 186, 836–852. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 Update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- Aluri, S.; Buttner, M. Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc. Natl. Acad. Sci. USA 2007, 104, 2537–2542. [Google Scholar] [CrossRef] [Green Version]
- Williams, L.E.; Lemoine, R.; Sauer, N. Sugar transporters in higher plants—A diversity of roles and complex regulation. Trends Plant Sci. 2000, 5, 283–290. [Google Scholar] [CrossRef]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 38, 151–159. [Google Scholar] [CrossRef]
- Butowt, R.; Granot, D.; Rodríguez-García, M.I. A putative plastidic glucose translocator is expressed in heterotrophic tissues that do not contain starch, during olive (Olea europea L.) fruit ripening. Plant Cell Physiol. 2003, 44, 1152–1161. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.X.; Weerasinghe, R.R.; Perdue, T.D.; Cakmakci, N.G.; Taylor, J.P.; Marzluff, W.F.; Jones, A.M. A Golgi-localized Hexose Transporter Is Involved in Heterotrimeric G Protein-mediated Early Development inArabidopsis. Mol. Biol. Cell 2006, 17, 4257–4269. [Google Scholar] [CrossRef] [Green Version]
- Wormit, A.; Trentmann, O.; Feifer, I.; Lohr, C.; Tjaden, J.; Meyer, S.; Schmidt, U.; Martinoia, E.; Neuhaus, H.E. Molecular Identification and Physiological Characterization of a Novel Monosaccharide Transporter from Arabidopsis Involved in Vacuolar Sugar Transport. Plant Cell 2006, 18, 3476–3490. [Google Scholar] [CrossRef] [Green Version]
- Büttner, M. The monosaccharide transporter(-like) gene family inArabidopsis. FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef] [Green Version]
- Rottmann, T.M.; Klebl, F.; Schneider, S.; Kischka, D.; Rüscher, D.; Sauer, N.; Stadler, R. Sugar Transporter STP7 Specificity for l-Arabinose and d-Xylose Contrasts with the Typical Hexose Transporters STP8 and STP12. Plant Physiol. 2018, 176, 2330–2350. [Google Scholar] [CrossRef] [Green Version]
- Patzke, K.; Prananingrum, P.; Klemens, P.A.; Trentmann, O.; Rodrigues, C.M.; Keller, I.; Fernie, A.R.; Geigenberger, P.; Bölter, B.; Lehmann, M.; et al. The Plastidic Sugar Transporter pSuT Influences Flowering and Affects Cold Responses. Plant Physiol. 2018, 179, 569–587. [Google Scholar] [CrossRef] [Green Version]
- Pommerrenig, B.; Müdsam, C.; Kischka, D.; Neuhaus, H.E. Treat and trick: Common regulation and manipulation of sugar transporters during sink establishment by the plant and the pathogen. J. Exp. Bot. 2020, 71, 3930–3940. [Google Scholar] [CrossRef]
- Xuan, Y.H.; Hu, Y.B.; Chen, L.-Q.; Sosso, D.; Ducat, D.C.; Hou, B.-H.; Frommer, W.B. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 2013, 110, E3685–E3694. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Doidy, J.; Grace, E.; Kühn, C.; Simon-Plas, F.; Casieri, L.; Wipf, D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012, 17, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-X.; Huang, X.-Y.; Yang, J.; Guan, Y.-F.; Yang, Z.-N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013, 26, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Sosso, D.; Luo, D.; Li, Q.-B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; Mccarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J. SWEET 11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef] [Green Version]
- Andrés, F.; Kinoshita, A.; Kalluri, N.; Fernández, V.; Falavigna, V.S.; Cruz, T.M.D.; Jang, S.; Chiba, Y.; Seo, M.; Mettler-Altmann, T.; et al. The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 53. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Park, J.-M.; Kang, S.K.; Kim, S.-G.; Park, C.-M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2010, 233, 189–200. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in Rice Causes Growth Retardation and Precocious Senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.; Sun, P.; Liu, Q.; Miao, Y.; Liu, J.; Zhang, K.; Hu, W.; Zhang, J.; Wang, J.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef]
- Lu, J.; Sun, M.-H.; Ma, Q.-J.; Kang, H.; Liu, Y.-J.; Hao, Y.-J.; You, C.-X. MdSWEET17, a sugar transporter in apple, enhances drought tolerance in tomato. J. Integr. Agric. 2019, 18, 2041–2051. [Google Scholar] [CrossRef]
- Hutin, M.; Sabot, F.; Ghesquière, A.; Koebnik, R.; Szurek, B. A Knowledge-Based Molecular Screen Uncovers a Broad-Spectrum OsSWEET14 Resistance Allele to Bacterial Blight from Wild Rice. Plant J. 2015, 84, 694–703. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gautam, T. SWEET genes and TAL effectors for disease resistance in plants: Present status and future prospects. Mol. Plant Pathol. 2021, 22, 1014–1026. [Google Scholar] [CrossRef]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Sauer, N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 2007, 581, 2309–2317. [Google Scholar] [CrossRef] [Green Version]
- Turgeon, R.; Medville, R. The absence of phloem loading in willow leaves. Proc. Natl. Acad. Sci. USA 1998, 95, 12055–12060. [Google Scholar] [CrossRef] [Green Version]
- Rennie, E.A.; Turgeon, R. A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. USA 2009, 106, 14162–14167. [Google Scholar] [CrossRef] [Green Version]
- Slewinski, T.L.; Braun, D.M. Current perspectives on the regulation of whole-plant carbohydrate partitioning. Plant Sci. 2010, 178, 341–349. [Google Scholar] [CrossRef]
- Liesche, J. Sucrose transporters and plasmodesmal regulation in passive phloem loading. J. Integr. Plant Biol. 2017, 59, 311–321. [Google Scholar] [CrossRef]
- Bieniawska, Z.; Barratt, D.H.P.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [CrossRef]
- Barratt, D.H.P.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef] [Green Version]
- Vu, D.P.; Rodrigues, C.M.; Jung, B.; Meissner, G.; Klemens, P.A.W.; Holtgräwe, D.; Fürtauer, L.; Nägele, T.; Nieberl, P.; Pommerrenig, B.; et al. Vacuolar sucrose homeostasis is critical for plant development, seed properties, and night-time survival in Arabidopsis. J. Exp. Bot. 2020, 71, 4930–4943. [Google Scholar] [CrossRef]
- Tauzin, A.S.; Giardina, T. Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. 2014, 5, 293. [Google Scholar] [CrossRef]
- Riesmeier, J.; Willmitzer, L.; Frommer, W. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 1994, 13, 1–7. [Google Scholar] [CrossRef]
- Gamas, P.; de Carvalho Niebel, F.; Lescure, N.; Cullimore, J. Use of a Subtractive Hybridization Approach to Identify New Medicago truncatula Genes Induced during Root Nodule Development. Mol. Plant Microbe Interact. 1996, 9, 233–242. [Google Scholar] [CrossRef]
- Artero, R.D.; Terol-Alcayde, J.; Paricio, N.; Ring, J.; Bargues, M.; Torres, A.; Perez-Alonso, M. Saliva, a New Drosophila Gene Expressed in the Embryonic Salivary Glands with Homologues in Plants and Vertebrates. Mech. Dev. 1998, 75, 159–162. [Google Scholar] [CrossRef]
- Jia, B.; Zhu, X.F.; Pu, Z.J.; Duan, Y.X.; Hao, L.J.; Zhang, J.; Chen, L.-Q.; Jeon, C.O.; Xuan, Y.H. Integrative View of the Diversity and Evolution of SWEET and SemiSWEET Sugar Transporters. Front. Plant Sci. 2017, 8, 2178. [Google Scholar] [CrossRef] [Green Version]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [Green Version]
- Jian, H.; Lu, K.; Yang, B.; Wang, T.; Zhang, L.; Zhang, A.; Wang, J.; Liu, L.; Qu, C.; Li, J. Genome-Wide Analysis and Expression Profiling of the SUC and SWEET Gene Families of Sucrose Transporters in Oilseed Rape (Brassica napus L.). Front. Plant Sci. 2016, 7, 1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.; Li, J. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Xuan, Y.; Jiang, J.; Wei, Y.; Piao, Z. Genome Wide Identification and Expression Profiling of SWEET Genes Family Reveals Its Role During Plasmodiophora brassicae-Induced Formation of Clubroot in Brassica rapa. Front. Plant Sci. 2018, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xiao, D.; Zhang, C.; Hou, X. The Expanded SWEET Gene Family Following Whole Genome Triplication in Brassica rapa. Genes 2019, 10, 722. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.-M.; Tang, Z.; Xu, Q.; Deng, X.-X. Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis). Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 609–624. [Google Scholar] [CrossRef]
- Yao, T.; Zhou, Y.; Hu, J.; Xiao, T.; Zhou, C. Genomic evolutionary relationship of SWEET genes and their responses to HLB disease and oxytetracycline treatment in Valencia sweet orange. Biologia 2021, 76, 1685–1689. [Google Scholar] [CrossRef]
- Hu, L.-P.; Zhang, F.; Song, S.-H.; Tang, X.-W.; Xu, H.; Liu, G.-M.; Wang, Y.; He, H.-J. Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J. Integr. Agric. 2017, 16, 1486–1501. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, Y.; Shan, Y.; Qin, Q. Characterization of SWEET family members from loquat and their responses to exogenous induction. Tree Genet. Genomes 2017, 13, 123. [Google Scholar] [CrossRef]
- Yin, Q.; Zhu, L.; Du, P.; Fan, C.; Wang, J.; Zhang, B.; Li, H. Comprehensive analysis of SWEET family genes in Eucalyptus (Eucalyptus grandis). Biotechnol. Biotechnol. Equip. 2020, 34, 595–604. [Google Scholar] [CrossRef]
- Liu, H.-T.; Lyu, W.-Y.; Tian, S.-H.; Zou, X.-H.; Zhang, L.-Q.; Gao, Q.-H.; Ni, D.-A.; Duan, K. The SWEET family genes in strawberry: Identification and expression profiling during fruit development. S. Afr. J. Bot. 2019, 125, 176–187. [Google Scholar] [CrossRef]
- Zhao, L.; Yao, J.; Chen, W.; Li, Y.; Lü, Y.; Guo, Y.; Wang, J.; Yuan, L.; Liu, Z.; Zhang, Y. A genome-wide analysis of SWEET gene family in cotton and their expressions under different stresses. J. Cotton Res. 2018, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ren, Z.; Wang, Z.; Sun, K.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Song, C.; Zhou, X.; et al. Evolution and Stress Responses of Gossypium hirsutum SWEET Genes. Int. J. Mol. Sci. 2018, 19, 769. [Google Scholar] [CrossRef] [Green Version]
- Sui, J.; Xiao, X.; Qi, J.; Fang, Y.; Tang, C. The SWEET gene family in Hevea brasiliensis—Its evolution and expression compared with four other plant species. FEBS Open Bio 2017, 7, 1943–1959. [Google Scholar] [CrossRef]
- Jiang, S.; Balan, B.; Assis, R.D.A.B.; Sagawa, C.H.D.; Wan, X.; Han, S.; Wang, L.; Zhang, L.; Zaini, P.A.; Walawage, S.L.; et al. Genome-Wide Profiling and Phylogenetic Analysis of the SWEET Sugar Transporter Gene Family in Walnut and Their Lack of Responsiveness to Xanthomonas arboricola pv. juglandis Infection. Int. J. Mol. Sci. 2020, 21, 1251. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Wang, D.; Qin, Y.; Ma, A.; Fu, J.; Qin, Y.; Hu, G.; Zhao, J. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development. BMC Plant Biol. 2019, 19, 499. [Google Scholar] [CrossRef]
- Wei, X.; Liu, F.; Chen, C.; Ma, F.; Li, M. The Malus domestica sugar transporter gene family: Identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front. Plant Sci. 2014, 5, 569. [Google Scholar] [CrossRef] [Green Version]
- Doidy, J.; Vidal, U.; Lemoine, R. Sugar transporters in Fabaceae, featuring SUT MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum. PLoS ONE 2019, 14, e0223173. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Wu, H.; Huang, W.; Song, J.; Zhou, Y.; Lin, Y. SWEET Gene Family in Medicago truncatula: Genome-Wide Identification, Expression and Substrate Specificity Analysis. Plants 2019, 8, 338. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, L.; Zhang, J.; Song, C.; Li, Y.; Li, J.; Lu, M. Expression and localization of SWEETs in Populus and the effect of SWEET7 overexpression in secondary growth. Tree Physiol. 2020, 41, 882–899. [Google Scholar] [CrossRef]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A New Insight into the Evolution and Functional Divergence of SWEET Transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef] [Green Version]
- Feng, C.-Y.; Han, J.-X.; Han, X.-X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Manck-Götzenberger, J.; Requena, N. Arbuscular Mycorrhiza Symbiosis Induces a Major Transcriptional Reprogramming of the Potato SWEET Sugar Transporter Family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Piron, M.-C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Wu, M.; Zhang, C. Sugar Transporter ZjSWEET2.2 Mediates Sugar Loading in Leaves of Ziziphus jujuba Mill. Front. Plant Sci. 2020, 11, 1081. [Google Scholar] [CrossRef]
- Guo, C.; Li, H.; Xia, X.; Liu, X.; Yang, L. Functional and evolution characterization of SWEET sugar transporters in Ananas comosus. Biochem. Biophys. Res. Commun. 2018, 496, 407–414. [Google Scholar] [CrossRef]
- Wang, T.; Song, Z.; Meng, W.L.; Li, L.B. Identification, characterization, and expression of the SWEET gene family in Phalaenopsis equestris and Dendrobium officinale. Biol. Plant. 2017, 62, 24–32. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Niu, K.; Ma, H. Identification and Expression Analysis of the SWEET Gene Family from Poa pratensis Under Abiotic Stresses. DNA Cell Biol. 2020, 39, 1606–1620. [Google Scholar] [CrossRef]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol. Biofuels 2016, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, D.P.; Zhu, X.; Li, T.Y.; Jia, B.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef]
- Gautam, T.; Saripalli, G.; Gahlaut, V.; Kumar, A.; Sharma, P.K.; Balyan, H.S.; Gupta, P.K. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2019, 46, 2327–2353. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.X.; Angenent, G.C.; Wittich, P.E.; Peters, J.; Franken, J.; Busscher, M.; Zhang, L.M.; Dahlhaus, E.; Kater, M.M.; Wullems, G.J.; et al. NEC1, a Novel Gene, Highly Expressed in Nectary Tissue of Petunia hybrida. Plant J. 2000, 24, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Ninan, A.S.; Grant, J.; Song, J.; Jameson, P.E. Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum sativum L. Plants 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Li, J.; Zhu, R.; Zhang, M.; Qi, K.; Zhang, S.; Wu, J. Overexpression of sugar transporter gene PbSWEET4 of pear causes sugar reduce and early senescence in leaves. Gene 2020, 743, 144582. [Google Scholar] [CrossRef]
- Zhen, Q.; Fang, T.; Peng, Q.; Liao, L.; Zhao, L.; Owiti, A.; Han, Y. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 2018, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Guo, W.; Li, J.; Yue, P.; Bu, H.; Jiang, J.; Liu, W.; Xu, Y.; Yuan, H.; Li, T.; et al. Histone Acetylation at the Promoter for the Transcription Factor PuWRKY31 Affects Sucrose Accumulation in Pear Fruit. Plant Physiol. 2020, 182, 2035–2046. [Google Scholar] [CrossRef] [Green Version]
- López-Coria, M.; Sánchez, S.; Martínez-Marcelo, V.H.; Aguilera-Alvarado, G.P.; Flores-Barrera, M.; King-Díaz, B.; Sánchez-Nieto, S.; Coria, L.; Marcelo, M.; Alvarado, A.; et al. SWEET Transporters for the Nourishment of Embryonic Tissues during Maize Germination. Genes 2019, 10, 780. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Zhang, D.; Miao, Q.; Yang, J.; Xuan, Y.; Hu, Y. Essential Role of Sugar Transporter OsSWEET11 During the Early Stage of Rice Grain Filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.-L.; Ma, J.F.; Shou, H. The Soybean Sugar Transporter GmSWEET15 Mediates Sucrose Export from Endosperm to Early Embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef] [Green Version]
- Evers, J.B. Sugar as a key component of the shoot branching regulation network. Plant Cell Environ. 2015, 38, 1455–1456. [Google Scholar] [CrossRef]
- Fichtner, F.; Barbier, F.; Feil, R.; Watanabe, M.; Annunziata, M.G.; Chabikwa, T.; Höfgen, R.; Stitt, M.; Beveridge, C.A.; Lunn, J.E. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 2017, 92, 611–623. [Google Scholar] [CrossRef] [Green Version]
- Schneider, A.; Godin, C.; Boudon, F.; Demotes-Mainard, S.; Sakr, S.; Bertheloot, J. Light Regulation of Axillary Bud Outgrowth Along Plant Axes: An Overview of the Roles of Sugars and Hormones. Front. Plant Sci. 2019, 10, 1296. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Peng, B.; Song, A.; Jiang, J.; Chen, F. Sugar Transporter, CmSWEET17, Promotes Bud Outgrowth in Chrysanthemum Morifolium. Genes 2019, 11, 26. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.-F.; Huang, X.-Y.; Zhu, J.; Gao, J.-F.; Zhang, H.-X.; Yang, Z.-N. RUPTURED POLLEN GRAIN1, a Member of the MtN3/saliva Gene Family, Is Crucial for Exine Pattern Formation and Cell Integrity of Microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Chu, Z.; Yuan, M.; Yao, J.; Ge, X.; Yuan, B.; Xu, C.; Li, X.; Fu, B.; Li, Z.; Bennetzen, J.L.; et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006, 20, 1250–1255. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. Pathogen-Induced Expressional Loss of Function is the Key Factor in Race-Specific Bacterial Resistance Conferred by a Recessive R Gene xa13 in Rice. Plant Cell Physiol. 2009, 50, 947–955. [Google Scholar] [CrossRef]
- Zhou, Y.; Cui, X.; Hu, A.; Miao, Y.; Zhang, L. Characterization and functional analysis of pollen-specific PwSWEET1 in Picea wilsonii. J. For. Res. 2019, 31, 1913–1922. [Google Scholar] [CrossRef] [Green Version]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 2015, 67, 461–471. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef] [Green Version]
- Hummel, I.; Pantin, F.; Sulpice, R.; Piques, M.; Rolland, G.; Dauzat, M.; Christophe, A.; Pervent, M.; Bouteillé, M.; Stitt, M.; et al. Arabidopsis Plants Acclimate to Water Deficit at Low Cost through Changes of Carbon Usage: An Integrated Perspective Using Growth, Metabolite, Enzyme, and Gene Expression Analysis. Plant Physiol. 2010, 154, 357–372. [Google Scholar] [CrossRef] [Green Version]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water Deficit Enhances C Export to the Roots in Arabidopsis thaliana Plants with Contribution of Sucrose Transporters in Both Shoot and Roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef] [Green Version]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source–sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2017, 247, 587–611. [Google Scholar] [CrossRef] [Green Version]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. DsSWEET17, a Tonoplast-Localized Sugar Transporter from Dianthus spiculifolius, Affects Sugar Metabolism and Confers Multiple Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1564. [Google Scholar] [CrossRef] [Green Version]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. A Novel Sugar Transporter from Dianthus spiculifolius, DsSWEET12, Affects Sugar Metabolism and Confers Osmotic and Oxidative Stress Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 497. [Google Scholar] [CrossRef] [Green Version]
- Bhaskarla, V.; Zinta, G.; Ford, R.; Jain, M.; Varshney, R.K.; Mantri, N. Comparative Root Transcriptomics Provide Insights into Drought Adaptation Strategies in Chickpea (Cicer arietinum L.). Int. J. Mol. Sci. 2020, 21, 1781. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, H.; Wu, J.; Xie, F. Effect of Drought Stress during Soybean R2–R6 Growth Stages on Sucrose Metabolism in Leaf and Seed. Int. J. Mol. Sci. 2020, 21, 618. [Google Scholar] [CrossRef] [Green Version]
- Aliche, E.B.; Theeuwen, T.P.; Oortwijn, M.; Visser, R.G.; van der Linden, C.G. Carbon partitioning mechanisms in POTATO under drought stress. Plant Physiol. Biochem. 2019, 146, 211–219. [Google Scholar] [CrossRef]
- Valifard, M.; Le Hir, R.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- Jiang, L.; Song, C.; Zhu, X.; Yang, J. SWEET Transporters and the Potential Functions of These Sequences in Tea (Camellia sinensis). Front. Genet. 2021, 12, 655843. [Google Scholar] [CrossRef]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the Vacuolar Sugar Carrier AtSWEET16 Modifies Germination, Growth, and Stress Tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.-J.; Nagy, R.; Chen, H.-Y.; Pfrunder, S.; Yu, Y.-C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a Facilitative Transporter, Mediates Fructose Transport across the Tonoplast of Arabidopsis Roots and Leaves. Plant Physiol. 2013, 164, 777–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Hir, R.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Géry, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Xu, H.; Zou, Q.; Zhang, J.; Jiang, S.; Fang, H.; Wang, Y.; Su, M.; Wang, N.; Chen, X. The vacuolar membrane sucrose transporter MdSWEET16 plays essential roles in the cold tolerance of apple. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 140, 129–142. [Google Scholar] [CrossRef]
- Sellami, S.; Le Hir, R.; Thorpe, M.R.; Vilaine, F.; Wolff, N.; Brini, F.; Dinant, S. Salinity Effects on Sugar Homeostasis and Vascular Anatomy in the Stem of the Arabidopsis thaliana Inflorescence. Int. J. Mol. Sci. 2019, 20, 3167. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Zhu, L.; Qian, Q. The tolerance of salinity in rice requires the presence of a functional copy of FLN2. Biomolecules 2020, 10, 17. [Google Scholar] [CrossRef] [Green Version]
- Murakeözy, É.P.; Nagy, Z.; Duhazé, C.; Bouchereau, A.; Tuba, Z. Seasonal Changes in the Levels of Compatible Osmolytes in Three Halophytic Species of Inland Saline Vegetation in Hungary. J. Plant Physiol. 2003, 160, 395–401. [Google Scholar] [CrossRef]
- Gill, P.K.; Sharma, A.D.; Singh, P.; Bhullar, S.S. Changes in Germination, Growth and Soluble Sugar Contents of Sorghum bicolor (L.) Moench Seeds under Various Abiotic Stresses. Plant Growth Regul. 2003, 40, 157–162. [Google Scholar] [CrossRef]
- Kaushal, N.; Awasthi, R.; Gupta, K.; Gaur, P.; Siddique, K.H.M.; Nayyar, H. Heat-Stress-Induced Reproductive Failures in Chickpea (Cicer arietinum) Are Associated with Impaired Sucrose Metabolism in Leaves and Anthers. Funct. Plant Biol. 2013, 40, 1334–1349. [Google Scholar] [CrossRef]
- Zhou, R.; Kjær, K.H.; Rosenqvist, E.; Yu, X.; Wu, Z.; Ottosen, C.O. Physiological Response to Heat Stress During Seedling and Anthesis Stage in Tomato Genotypes Differing in Heat Tolerance. J. Agron. Crop Sci. 2017, 203, 68–80. [Google Scholar] [CrossRef]
- Suwa, R.; Hakata, H.; Hara, H.; El-Shemy, H.A.; Adu-Gyamfi, J.J.; Nguyen, N.T.; Kanai, S.; Lightfoot, D.A.; Mohapatra, P.K.; Fujita, K. High Temperature Effects on Photosynthate Partitioning and Sugar Metabolism during Ear Expansion in Maize (Zea mays L.) Genotypes. Plant Physiol. Biochem. 2010, 48, 124–130. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, X.; Li, T.; Chang, T. Effect of Elevated Temperature Stress on the Production and Metabolism of Photosynthate in Tomato (Lycopersicon esculentum L.) Leaves. J. Hortic. Sci. Biotechnol. 2012, 87, 367–373. [Google Scholar] [CrossRef]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genom. 2008, 9, 432. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Kang, J.; Zhou, X.; Su, Z.; Qu, L.; Gu, H. Variation at the Transcriptional Level among Chinese Natural Populations of Arabidopsis thaliana in Response to Cold Stress. Chin. Sci. Bull. 2008, 53, 2989–2999. [Google Scholar] [CrossRef] [Green Version]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.W.; Spinner, L.; Clement, G.; Chietera, G.; Léran, S.; Ferrand, M.; Lacombe, B.; et al. Leaf Fructose Content Is Controlled by the Vacuolar Transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Schubert, S.; Neubert, A.; Schierholt, A.; Sümer, A.; Zörb, C. Development of Salt-Resistant Maize Hybrids: The Combination of Physiological Strategies Using Conventional Breeding Methods. Plant Sci. 2009, 177, 196–202. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant Salt-Tolerance Mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Jung, S.; Schubert, S. Transcription Profile Analysis Identifies Marker Genes to Distinguish Salt Shock and Salt Stress after Stepwise Acclimation in Arabidopsis thaliana and Zea mays. Plant Physiol. Biochem. 2019, 143, 232–245. [Google Scholar] [CrossRef]
- Qin, J.; Jiang, Y.; Lu, Y.; Zhao, P.; Wu, B.; Li, H.; Wang, Y.; Xu, S.; Sun, Q.; Liu, Z. Genome-Wide Identification and Transcriptome Profiling Reveal Great Expansion of SWEET Gene Family and Their Wide-Spread Responses to Abiotic Stress in Wheat (Triticum aestivum L.). J. Integr. Agric. 2020, 19, 1704–1720. [Google Scholar] [CrossRef]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 Regulate Gibberellin-Mediated Physiological Processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef]
- Dhandapani, P.; Song, J.; Novak, O.; Jameson, P.E. Infection by Rhodococcus fascians Maintains Cotyledons as a Sink Tissue for the Pathogen. Ann. Bot. 2017, 119, 841–852. [Google Scholar]
- Shah, T.; Andleeb, T.; Lateef, S.; Noor, M.A. Genome Editing in Plants: Advancing Crop Transformation and Overview of Tools. Plant Physiol. Biochem. 2018, 131, 12–21. [Google Scholar] [CrossRef]
- Antony, G.; Zhou, J.; Huang, S.; Li, T.; Liu, B.; White, F.; Yang, B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 2010, 22, 3864–3876. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Streubel, J.; Balzergue, S.; Champion, A.; Boch, J.; Koebnik, R.; Feng, J.; Verdier, V.; Szurek, B. Colonization of Rice Leaf Blades by an African Strain of Xanthomonas oryzae Pv. oryzae Depends on a New TAL Effector That Induces the Rice Nodulin-3 Os11N3 Gene. Mol. Plant-Microbe Interact. 2011, 24, 1102–1113. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-Efficiency TALEN-Based Gene Editing Produces Disease-Resistant Rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Streubel, J.; Pesce, C.; Hutin, M.; Koebnik, R.; Boch, J.; Szurek, B. Five Phylogenetically Close Rice SWEET Genes Confer TAL Effector-Mediated Susceptibility to Xanthomonas oryzae Pv. oryzae. New Phytol. 2013, 200, 808–819. [Google Scholar] [CrossRef]
- Blanvillain-Baufumé, S.; Reschke, M.; Solé, M.; Auguy, F.; Doucoure, H.; Szurek, B.; Meynard, D.; Portefaix, M.; Cunnac, S.; Guiderdoni, E.; et al. Targeted Promoter Editing for Rice Resistance to Xanthomonas Oryzae Pv. Oryzae Reveals Differential Activities for SWEET14-Inducing TAL Effectors. Plant Biotechnol. J. 2017, 15, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Zafar, K.; Khan, M.Z.; Amin, I.; Mukhtar, Z.; Yasmin, S.; Arif, M.; Ejaz, K.; Mansoor, S. Precise CRISPR-Cas9 Mediated Genome Editing in Super Basmati Rice for Resistance Against Bacterial Blight by Targeting the Major Susceptibility Gene. Front. Plant Sci. 2020, 11, 575. [Google Scholar] [CrossRef] [PubMed]
- Cohn, M.; Bart, R.S.; Shybut, M.; Dahlbeck, D.; Gomez, M.; Morbitzer, R.; Hou, B.-H.; Frommer, W.B.; Lahaye, T.; Staskawicz, B.J. Xanthomonas axonopodis Virulence Is Promoted by a Transcription Activator-Like Effector–Mediated Induction of a SWEET Sugar Transporter in Cassava. Mol. Plant-Microbe Interact. 2014, 27, 1186–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Huang, S.; Zhou, J.; Yang, B. Designer TAL Effectors Induce Disease Susceptibility and Resistance to Xanthomonas oryzae Pv. oryzae in Rice. Mol. Plant 2013, 6, 781–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, K.L.; Meng, F.; Wilkins, K.E.; Li, F.; Wang, P.; Booher, N.J.; Carpenter, S.C.D.; Chen, L.-Q.; Zheng, H.; Gao, X.; et al. TAL Effector Driven Induction of a SWEET Gene Confers Susceptibility to Bacterial Blight of Cotton. Nat. Commun. 2017, 8, 15588. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Luo, Y.; Vu, N.T.Q.; Shen, S.; Xia, K.; Zhang, M. CRISPR/Cas9-Mediated Mutation of OsSWEET14 in Rice Cv. Zhonghua11 Confers Resistance to Xanthomonas oryzae Pv. oryzae without Yield Penalty. BMC Plant Biol. 2020, 20, 313. [Google Scholar] [CrossRef]
- Kim, Y.A.; Moon, H.; Park, C.J. CRISPR/Cas9-Targeted Mutagenesis of Os8N3 in Rice to Confer Resistance to Xanthomonas oryzae Pv. oryzae. Rice 2019, 12, 67. [Google Scholar] [CrossRef]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.-S.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B.; et al. Gene Targeting by the TAL Effector PthXo2 Reveals Cryptic Resistance Gene for Bacterial Blight of Rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/SgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Veley, K.M.; Okwuonu, I.; Jensen, G.; Yoder, M.; Taylor, N.J.; Meyers, B.C.; Bart, R.S. Gene Tagging via CRISPR-Mediated Homology-Directed Repair in Cassava. G3 Genes Genomes Genet. 2021, 11, jkab028. [Google Scholar] [CrossRef]
- Eom, J.-S.; Luo, D.; Atienza-Grande, G.; Yang, J.; Ji, C.; Luu, V.T.; Huguet-Tapia, J.C.; Char, S.N.; Liu, B.; Nguyen, H.; et al. Diagnostic kit for rice blight resistance. Nat. Biotechnol. 2019, 37, 1372–1379. [Google Scholar] [CrossRef] [Green Version]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.-S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-Spectrum Resistance to Bacterial Blight in Rice Using Genome Editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).