Abstract
Asteraceae, as the largest angiosperm family, has an architecturally complex capitulum (inflorescences) composed of heteromorphic florets with distinct morphologies and functions. AGAMOUS (AG) MADS-box transcription factors act as key regulators in flower development and are essential for the formation of the characteristic capitulum and florets. To explore the potential functions of the AG genes in Asteraceae, we conducted a genome-wide identification and analysis of 52 AG-like genes across 22 species within this family. Additionally, we studied the functions of the Tagetes erecta class C genes TeAG1 and TeAG2 by introducing these genes into T. erecta and Nicotiana tabacum. Gene structure and phylogenomic analyses indicated that AG-like genes may have conserved and specific biological functions in Asteraceae plants. Phenotypic analyses revealed that the T. erecta class C genes TeAG1 and TeAG2 played a conserved and redundant role in regulating stamen and carpel development. The simultaneous downregulation of TeAG1 and TeAG2 led to the homeotic transformation of both stamens and carpels into corolla-like structures. However, silencing TeAG1 or TeAG2 individually in T. erecta did not affect any floral organ development. Furthermore, the ectopic expression of TeAG1 and TeAG2 in N. tabacum resulted in the transformation of sepals into pistils and corollas into stamens, respectively. Additionally, qRT-PCR analyses revealed that TeAG1 and TeAG2 repressed the expression of class A genes. Our findings expand our understanding of the function of class C genes within Asteraceae and provide strategies for breeding double-flower cultivars.
1. Introduction
The emergence of flowering plants has profoundly driven the evolutionary diversification of the plant kingdom. As the reproductive structures of angiosperms, flowers are regarded as the most significant morphological innovation in plant evolution [1]. Since the early 1990s, researchers have intensively investigated floral initiation and development [2]. Studies of floral homeotic mutants in the model species led to the formulation of the ABC model of floral development [2,3,4], and the knowledge of this model has also been adapted for, in part, ornamental plants [5,6,7]. According to this model, the identity of floral organs is specified by the combinatorial activity of three classes of homeotic genes: class A genes alone determine sepal identity; class A and B genes jointly specify petal identity; class B and C genes control stamen development; and class C genes alone govern carpel formation [2,8,9]. Molecular characterization reveals that the class A, B, and C genes largely encode MADS-box transcription factors, except for the A-class gene APETALA2 (AP2), which is a member of the AP2/ERF superfamily.
AGAMOUS (AG), a class C gene, primarily promotes floral meristem determinacy and regulates stamen and carpel development [10,11,12]. In an Arabidopsis thaliana ag mutant, stamens are converted to petals and carpels to sepals [13], whereas the ectopic expression of A. thaliana AG induces the transformation of sepals into stigmatic tissues and petals into stamens [14]. Orthologous class C genes in dicotyledons and even in monocotyledons exhibit similar functions to those observed in A. thaliana. For example, in Zea mays, mutation in the AG gene ZMM2 disrupts stamen and carpel development [15]. In Chrysanthemum × morifolium, silencing CAG results in petaloid stamens and arrested stigma development in ray florets [16]. However, the number of AG genes is varied across species, and these genes often exhibit various degrees of functional redundancy or subfunctionalization. For example, three AG orthologs (OsMADS3, OsMADS13, and OsMADS58) have been identified in the monocot Oryza sativa. OsMADS3 and OsMADS58 specify stamen and carpel identity, and together with OsMADS13, they are important for floral meristem determinacy [17]. Additionally, in eudicot Eschscholzia californica, two AG homologs determine stamen and carpel identity and repress B-class genes [18].
Asteraceae, also known as Compositae or the sunflower plant family, is one of the largest angiosperm families, comprising approximately 1700 genera and 35,000 species distributed worldwide [19,20]. This family comprises vital crops, ornamentals, medicinal species, and even weeds, collectively endowing it with exceptional economic, ornamental, and medicinal value [21]. The capitulum is a typical characteristic of Asteraceae plants, and the evolutionary success of this family is largely attributed to this unique inflorescence [22]. Each capitulum is a densely packed aggregation of hundreds of morphologically and functionally distinct florets [23]. This highly condensed and specialized capitulum consists of peripheral sterile ray florets with showy zygomorphic corollas and central fertile disk florets exhibiting actinomorphic symmetry. The Asteraceae family is divided into 13 subfamilies, among which Asteroideae, Carduoideae, and Cichorioideae collectively constitute the core of its diversity [19]. Asteroideae comprises over 17,000 species characterized by two types of florets, including many well-known ornamental Asteraceae plants such as C. morifolium and Gerbera hybrida [19]. Carduoideae is one of the largest subfamilies within Asteraceae, comprising approximately 2400 species across 73 genera, all of which contain only disk florets [24]. In contrast, species of Cichorioideae are characterized by the possession of only sterile ray florets.
Understanding capitulum development in Asteraceae focuses critically on inflorescence development and floral organ identity [25,26]. These have been identified and studied in G. hybrida [27], C. morifolium [16], and Helianthus annuus [28,29]. Tagetes erecta, a member of the same family, similarly has a complex capitulum. Compared with the model Asteraceae species G. hybrida, the T. erecta capitulum is composed of two sharply distinct floret types: the outermost whorl consists of zygomorphic, pistillate ray florets, whereas the inner whorls comprise actinomorphic, bisexual disk florets. In contrast to the sterile ray florets of H. annuus, those of T. erecta retain a fertile pistil. Moreover, T. erecta occupies a phylogenetically derived position within Asteraceae [19] and exhibits a short life cycle. Collectively, these attributes render T. erecta an excellent system for investigating floral organ development in the Asteraceae.
To elucidate the characteristics of MADS-box AG genes in Asteraceae, this study systematically analyzed the physicochemical properties, gene structures, and phylogenetic relationship of AG genes derived from diverse members of Asteraceae. Additionally, to dissect the functions of T. erecta AG genes, we constructed both RNA interference and overexpression vectors targeting these genes and introduced them, via Agrobacterium-mediated transformation, into T. erecta and Nicotiana tabacum, respectively. These findings provide pivotal genetic resources for the breeding of novel double-flower cultivars of T. erecta and substantially expand our understanding of AG gene function within Asteraceae.
2. Materials and Methods
2.1. Plant Materials
The T. erecta cultivar ‘Milestone’ used for plant transformation was obtained from Chifeng Sainuo Horticulture Co., Ltd. (Chifeng, China). Kanamycin-resistant transgenic lines were transplanted to soil and cultivated in a growth chamber under long-day conditions (16 h light/8 h dark) at 25 °C. Wild-type N. tabacum was used for plant transformation. Transgenic N. tabacum plants were cultivated in a controlled-environment room (16 h light/8 h dark, 25 °C).
2.2. Identification of AG Gene in Asteraceae Plants
Given that the Asteroideae, Cichorioideae, and Carduoideae subfamilies represent 95% of Asteraceae species and the core of its diversity, this study analyzed the AG gene characteristics in these three subfamilies. The genome data of Asteraceae plants mainly from the subfamilies Asteroideae, Cichorioideae, and Carduoideae were mainly downloaded from Asteraceae multi-omics information resource (https://yanglab.hzau.edu.cn/AMIR/home/, accessed on 3 June 2025) (Table S1). First, the identified AG protein sequences from T. erecta and H. annuus were used to conduct Blast searches in the proteome databases of other Compositae plants to obtain homologous sequences. Then, all AG motif domain sequences of the species were downloaded from the Pfam database (http://pfam.xfam.org/, accessed on 8 May 2025), and a hidden Markov model was constructed using Hmmer2.3 (http://hmmer.janelia.org/, accessed on 8 May 2025) to search for sequences containing the AG motif domain in the Compositae protein database. Finally, the obtained AG proteins were analyzed using the online tool NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd, accessed on 13 July 2025) to identify the domain structure, and sequences without the AG domain were removed to obtain the final AG protein dataset (Supplementary Table S1).
2.3. Bioinformatic Analysis of AG Gene in Asteraceae
The molecular weight and isoelectric point of these AG proteins in the Asteraceae plants were predicted using the ExPASy Proteomics Server (http://www.expasy.org/proteomics, accessed on 19 July 2025). The subcellular localization of these AG proteins was predicted using CELLO v.2.5 via online software MBC (http://cello.life.nctu.edu.tw, accessed on 19 July 2025). The MUSCLE program in MEGA7 was used to perform multiple sequence alignment of the identified AG protein sequences, and a phylogenetic tree was constructed using the Neighbor-Joining (NJ) method (bootstrap = 1000). The motifs of each subfamily member were identified using the online tool MEME Suite5.5.0 (http://meme-suite.org/, accessed on 21 July 2025). The above results were analyzed and visualized using TBtools software v2.0 and the online tool iTOL v7 (https://itol.embl.de/, accessed on 21 July 2025).
2.4. Vector Construction and Plant Transformation
The sequences of AG genes (TeAG1 and TeAG2) in T. erecta have been reported previously [5]. To construct the TeAG1, TeAG2, and TeAG1/TeAG2 double RNAi constructs, the Gateway binary vector pK7GWIWG2D(II) [30] was used for plant transformation. Gene-specific primers (V097-TeAG1-F, V097-TeAG2-F, V097-TeAG1-R, and V097-TeAG2-R) containing BamHI and EcoRI restriction sites (Table S2) were designed based on the sequences of the two T. erecta C-class genes TeAG1 (Acc. No. MT452648) and TeAG2 (Acc. No. MT452649). PCR products were linked to entry vector V097. To construct the TeAG1/TeAG2 double RNAi constructs, we inserted the SspBI-digested fragment containing the TeAG2 insert into the SspBI site at the attL2 recombination site of the TeAG1 entry vector. The RNAi vectors of RNAi-TeAG1, RNAi-TeAG2, and RNAi-TeAG1-TeAG2 were generated via an LR reaction between recombinant entry vectors and gateway binary vector pK7GWIWG2D(II). These three RNAi recombinant vectors were introduced into Agrobacterium tumefaciens strain EHA105 and then transformed into T. erecta cultivar ‘Milestone’. The Agrobacterium-mediated transformation of T. erecta followed the protocol described by Yu et al. (2023) [31]. Transgenic plant phenotypes were observed and subsequently subjected to detailed characterization and analysis.
To further explore the functions of TeAG1 and TeAG2, overexpression vectors were constructed. The full-length coding sequences of TeAG1 and TeAG2 were amplified using specific primers (Table S2) and then cloned into a P2300 vector digested by BamHI. The recombinant vectors were named 35S:TeAG1 and 35S:TeAG2. These vectors were then introduced into Escherichia coli DH5a. Positive cells were tested by PCR, and 2 or 3 positive clones were randomly selected for sequencing by the Sangon company in Shanghai. After the recombinant constructs were successfully sequenced, they were then introduced into the Agrobacterium tumefaciens strain GV3101. Agrobacterium-mediated N. tabacum transformation was performed via leaf disk inoculation according to the protocol of Gallois and Marinho (1995) [32]. The phenotypic changes in transgenic plants were recorded and analyzed.
2.5. Total RNA Extraction and Expression Analysis
To confirm that the phenotypic alterations in transgenic plants correlate with transgene expressions, quantitative real-time PCR (qRT-PCR) was used to analyze the expression levels of TeAG1 and TeAG2 in T0-generation transgenic and wild-type T. erecta plants. The expression levels of TeAG1 and TeAG2 were analyzed by semi-quantitative RT-PCR (semi-qRT-PCR) in wild-type N. tabacum plants and in three randomly selected T1 transgenic plants exhibiting strong, medium, or weak phenotypes. Additionally, to further analyze the mechanism of floral organs’ phenotype changes in transgenic plants, the expression levels of the class A, B, and C genes of T. erecta and N. tabacum were detected by qRT-PCR. The primers were listed in Supplementary Table S2.
Total RNA was isolated from blooming disk florets in the center from transgenic T. erecta RNAi-TeAG1, RNAi-TeAG2, and RNAi-TeAG1/TeAG2 and wild-type plants using the RN33-PLANTpure kit with gDNA Eraser (Aidlab, Beijing, China) and then reversed-transcribed by TRUEscript RT kit (Aidlab, Beijing, China). Similarly, total RNA was also extracted from the flowers, sepals, corollas, stamens, and pistils of wild-type N. tabacum and transgenic N. tabacum lines. N. tabacum EF1a (NtEF1a, GenBank accession: D63396.1) was used to reference genes. The qRT-PCR experiment was carried out with the SYBR Green kit (Aidlab, Beijing, China) as detailed in the manufacturer’s protocol. Beta-actin was used as the gene of the internal control in T. erecta. The qRT-PCR experiments were performed with three replicates for each sample. The relative gene expression level was quantified via the 2–ΔΔCT method. The primers were listed in Supplementary Table S2.
2.6. Scanning Electron Microscopy
Because mutations in stamens and pistils render transgenic T. erecta lines sterile, all phenotypic analyses were confined to the T0-generation transgenic T. erecta lines. The four-wheel floral organs of RNAi-TeAG1/TeAG2 transgenic T. erecta lines and wild-type plants were harvested and immersed in 2.5% (v/v) glutaraldehyde at 4 °C overnight, then subjected to a graded ethanol dehydration series (30–100%), with 15 min changes at each concentration. After critical-point drying (CPD 020; Balzers Union, Balzers, Liechtenstein), the specimens were gold-sputter-coated (Nanotech SEMPrep II, Nanotech Ltd., Prestwick, UK) and mounted on stubs with double-sided conductive tape. Imaging was performed with a LEO 435VP scanning electron microscope (LEO Electron Microscopy Ltd., Oberkochen, Germany) [5].
To ensure the heritability of phenotypic changes in transgenic N. tabacum plants, the documentation and statistical analysis of these phenotypic changes were performed using trait statistics from T1-generation transgenic N. tabacum lines. For each transgenic N. tabacum line, 12 or more plants were selected for phenotypic observation and data analysis. The phenotypic characterizations of transgenic N. tabacum lines primarily focused on flowering time and morphological alterations in floral organs. To conduct a detailed phenotypic analysis of transgenic plants, the four-whorl floral organs at full bloom from both wild-type and transgenic N. tabacum plants were collected and examined using SEM.
2.7. Statistical Analysis
All statistical analyses were conducted with SPSS 26. Statistical significance was evaluated by a one-way ANOVA. Statistically significant differences at p < 0.05 and p < 0.01 were indicated by lowercase and uppercase letters, respectively.
3. Results
3.1. Genome-Wide Identification of AG Genes in Asteraceae
A total of 52 AG-like genes were identified from 22 plant species across three Asteraceae subfamilies (Asteroideae, Carduoideae, and Cichorioideae) (Table S3). The copy number of the AG-like genes was unevenly distributed among different species in these three Asteraceae subfamilies, with most species possessing two copies (Table S3). Bidens alba contained the maximum number of AG-like genes (n = 4), whereas Smallanthus sonchifolius, Mikania micrantha, and Taraxacum mongolicum each possessed only a single-copy AG-like gene. In species such as T. erecta and Chrysanthemum lavandulifolium, two copies of AG were identified. ExPASy analysis revealed that all these AG-like proteins were hydrophilic unstable proteins with isoelectric points (pI) ranging from 6.3 to 9.4 (Table S3). These AG-like proteins exhibited considerable variation in amino acid length across different plant species. The longest isoform (Lactuca sativa var. angustana, LSA22458) comprised 449 residues (51.26 kDa), whereas the shortest (C. lavandulifolium, EVM0043976) contained 241 residues (27.93 kDa). Subcellular localization analysis revealed that all these AG-like proteins were exclusively localized within the nucleus (Table S3).
3.2. Phylogenetic and Gene Structure Analysis of AG Gene in Asteraceae
Phylogenetic analysis indicated that these AG-like proteins were primarily clustered into two major clades (clade 1 and clade 2), with the proteins in each clade originating from three subfamilies within the Asteraceae family (Figure 1). Notably, in some species, two or more AG homologs were distributed across both clades. For example, the AG proteins (KAK1422760.1 and KAK1424857.1) from T. erecta were clustered within clade 1 and clade 2, respectively (Figure 1). Furthermore, within each major clade, AG-like proteins from the same subfamily typically formed a monophyletic subclade (Figure 1). For instance, all proteins in subclade 1 were exclusively derived from species of the Carduoideae subfamily (Figure 1).
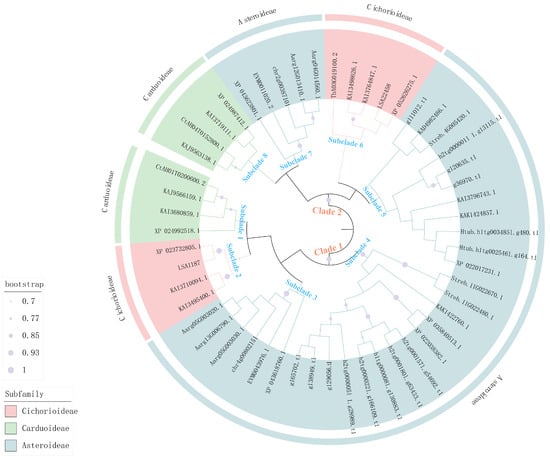
Figure 1.
Phylogenetic analysis of AG-like proteins from 22 plant species across Asteroideae, Carduoideae, and Cichorioideae subfamilies.
Motif prediction demonstrated a high conservation of motifs 1–9 across Asteraceae AG-like proteins, with primary divergence occurring at motif 10 (Figure 2a). For example, both AG proteins in T. erecta possessed motifs 1–9, but they differed in the presence of motif 10, which was found in KAK1422760.1 but was absent in KAK1424857.1 (Figure 2a). Interestingly, proteins containing motif 10 co-clustered within the same phylogenetic branch (Figure 1). Gene structure analysis showed that most AG-like proteins contained 7–9 exons and 6–8 introns (Figure 2b). However, the AG-like protein (LSA22458) in L. sativa var. angustana represented an exception, possessing 14 exons and 13 introns (Figure 2b).
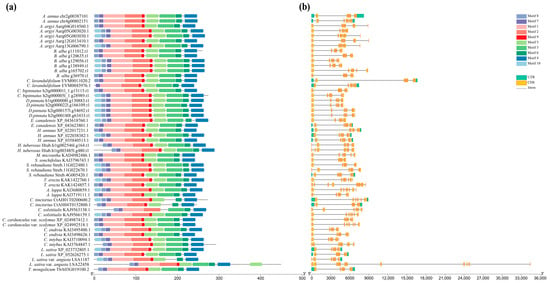
Figure 2.
Motif and structure analysis of AG-like genes from 22 plant species across Asteroideae, Carduoideae, and Cichorioideae subfamilies. (a) Motif analysis of AG-like proteins from 22 plant species across Asteroideae, Carduoideae, and Cichorioideae subfamilies; (b) structure analysis of AG-like genes from 22 plant species across Asteroideae, Carduoideae, and Cichorioideae subfamilies.
3.3. TeAG1 and TeAG2 Redundantly Affect Stamen and Pistil Development
In this study, we generated transgenic T. erecta containing the RNAi-TeAG1, RNAi-TeAG2, and TeAG1/TeAG2 double RNAi constructs. Nine RNAi-TeAG1 transgenic lines, eleven RNAi-TeAG2 transgenic lines, and four RNAi-TeAG1/TeAG2 double RNAi transgenic lines were obtained. QRT-PCR analysis showed that the expressions of TeAG1 and TeAG2 had varying degrees of decline in transgenic plants containing RNAi-TeAG1 and RNAi-TeAG2 constructs (Figure S1a,b). However, compared with the wild-type T. erecta, the transgenic lines containing a single RNAi construct, either RNAi-TeAG1 or RNAi-TeAG2, did not exhibit any visible morphological changes.
Compared to wild-type plants, two of the RNAi-TeAG1/TeAG2 transgenic lines (RNAi-TeAG1/TeAG2#1 and RNAi-TeAG1/TeAG2#2) displayed severe phenotypes (named RNAi-TeAG1/TeAG2#S), and two transgenic lines (RNAi-TeAG1/TeAG2#3 and RNAi-TeAG1/TeAG2#4) showed weak phenotypes (named RNAi-TeAG1/TeAG2#W) (Figure 3). In RNAi-TeAG1/TeAG2#S transgenic T. erecta lines, the pistils of ray florets and disk florets and the stamen of disk florets were converted into corolla-like organs (Figure 3e–h). However, in RNAi-TeAG1/TeAG2#W transgenic lines, only the stamens and pistils of the inner disk florets turned into corolla-like organs (Figure 3i,k,l), whereas the pistils of peripheral ray florets displayed no visible morphological alterations (Figure 3j).

Figure 3.
Phenotype analysis of RNAi-TeAG1/TeAG2 transgenic T. erecta plants. (a–d) Phenotypes of inflorescences and individual flowers of wild-type plants. (e–h) Phenotypes of inflorescences and individual flowers of RNAi-TeAG1/TeAG2#S (RNAi-TeAG1/TeAG2#2) lines. (i–l) Phenotypes of inflorescences and individual flowers in RNAi-TeAG1/TeAG2#W (RNAi-TeAG1/TeAG2#4) transgenic T. erecta lines. Bar = 0.5 cm. The red arrows point to the stamens and the white arrows point to the pistils.
To confirm that stamens and pistils in the RNAi-TeAG1/TeAG2#S transgenic lines undergo floral organ homeotic transformation, the floral organ epidermal cell structures of RNAi-TeAG1/TeAG2#2 (RNAi-TeAG1/TeAG2#S) transgenic lines and wild-type T. erecta were observed by SEM (Figure 4). SEM observations revealed that, in wild-type plants, the anthers on the upper part of the stamens carried conspicuous pollen grains (Figure 4d), and the epidermal cells of the lower filament appeared as wrinkled, ribbon-like structures (Figure 4e). In contrast, in RNAi-TeAG1/TeAG2#2 transgenic lines, the upper epidermis of the stamens consisted of tightly arranged papillate cells (Figure 4j), while the epidermal cells of the lower filament were composed of irregular, rough-surfaced clump-like structures (Figure 4k), thus exhibiting a morphology closely resembling the corolla structure in wild-type T. erecta (Figure 4b,c,h,i). Moreover, the pistils in RNAi-TeAG1/TeAG2#2 transgenic lines had undergone a homeotic conversion in which the original columnar papillate cells of the style were replaced by domed papillae characteristic of the petal abaxial epidermis (Figure 4b,f,h,l).
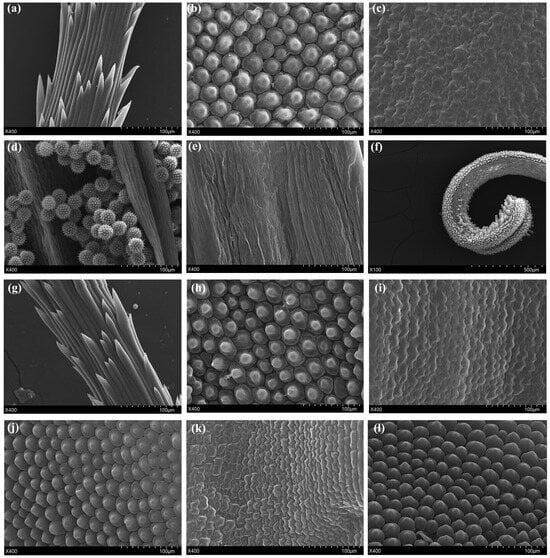
Figure 4.
Scanning electron micrograph of floral organs in T. erecta transgenic lines. (a–f) Scanning electron micrograph of floral organs in wild-type plants. (a) Epidermis cells of pappus in wild-type plants. (b,c) Adaxial (b) and abaxial (c) epidermis cells of corolla of wild-type plants. (d,e) Epidermis cells of anthers (d) and filaments (e) of wild-type plants. (f) Epidermis cells of pistils of wild-type plants. (g–l) Scanning electron micrograph of floral organs in RNAi-TeAG1/TeAG2#2 transgenic T. erecta lines. (g) Epidermis cells of pappus in wild-type plants. (h,i) Adaxial (h) and abaxial (i) epidermis cells of corolla of RNAi-TeAG1/TeAG2#2 transgenic T. erecta lines. (j,k) Epidermis cells of anthers and filaments of RNAi-TeAG1/TeAG2#2 transgenic T. erecta lines. (l) Epidermis cells of pistils of RNAi-TeAG1/TeAG2#2 transgenic T. erecta lines. Bar = 100 μm.
3.4. Dramatic Effect of Overexpression of TeAG1 and TeAG2 in N. tabacum on Sepal and Petal Identities
To further explore the conserved function of the AG genes in T. erecta in regulating the development of the stamen and pistil, we ectopically expressed TeAG1 and TeAG2 in N. tabacum under the control of the CaMV 35S promoter. A total of twenty-seven 35S:TeAG1 and thirty-three 35S:TeAG2 transgenic N. tabacum lines were obtained. Phenotypic observed showed that five of the 35S:TeAG1 transgenic lines displayed severe phenotypes (named OE-TeAG1#S), thirteen showed weak phenotypes (named OE-TeAG1#W), and nine had no remarkable phenotypic changes. Among the thirty-three 35S:TeAG2 transgenic N. tabacum lines, four exhibited a strong phenotype (named OE-TeAG2#S), fourteen displayed a moderate phenotype (named OE-TeAG2#M), eight showed a weak phenotype (named OE-TeAG2#W), and seven were phenotypically indistinguishable from the wild type. Additionally, compared with wild-type N. tabacum, both 35S:TeAG1 and 35S:TeAG2 transgenic plants exhibited an early-flowering phenotype (Figure S2a,b,d–f, Table S4). The transcripts of these two genes in N. tabacum were tested by semi-qRT-PCR (Figure S2c).
The overexpression of TeAG1 in N. tabacum affected the development of corollas. Compared to wild-type plants (Figure 5a,d), the corolla in 35S:TeAG1 transgenic lines underwent a morphological shift from a funnel-shaped structure to a trumpet-shaped form (Figure 5b,e). Additionally, the apical corollas developed wrinkled, ligulate structures (Figure 5b,e), and an anther-like structure emerged at the corolla–corolla junctions (Figure 5e). Consistent with the phenotype observed in OE-TeAG1#S transgenic N. tabacum lines, corollas in the OE-TeAG1#W transgenic N. tabacum lines also adopted a tongue-like morphology (Figure 5c,f). Notably, in the OE-TeAG1#W transgenic lines, the corolla color shifted from purplish-red to pinkish-white (Figure 5f). However, in contrast to strong-phenotype lines, no conspicuous anther-like structures were detectable at the corolla–corolla junctions (Figure 5f).

Figure 5.
A phenotype analysis of transgenic N. tabacum plants overexpressing T. erecta class C genes. (a–c) The inflorescence of wild-type plants (a), OE-TeAG1#S transgenic lines (b), and OE-TeAG1#W transgenic lines (c). (d–f) The corollas of wild-type plants (d), OE-TeAG1#S transgenic lines (e), and OE-TeAG1#W transgenic lines (f). (g–i) The inflorescence of OE-TeAG2#S (g), OE-TeAG2#M transgenic lines (h), and OE-TeAG2#W transgenic lines (i). (j–l) The corollas of OE-TeAG2#S (j), OE-TeAG2#M transgenic lines (k), and OE-TeAG2#W transgenic lines (l). Bar = 1 cm.
The overexpression of TeAG2 in N. tabacum affected the development of sepals and corollas. In OE-TeAG2#S and OE-TeAG2#M transgenic N. tabacum lines, the five apical lobes of the sepals were transformed into ten lobes (Figure 5g,h). The apices of these ten lobes exhibited stigma-like structures (Figure 5g,h,j). Furthermore, the five apical lobes of the corolla were conspicuously converted into anther-like structures (Figure 5j,k). Compared with the observations in OE-TeAG2#S and OE-TeAG2#M transgenic N. tabacum lines, OE-TeAG2#W transgenic N. tabacum lines exhibited no alteration in the number of apical sepal lobes (Figure 5i,l). Additionally, their apical sepal lobes lacked conspicuous stigma-like structures at the tips (Figure 5i,l). Interestingly, the corolla in OE-TeAG2#W transgenic N. tabacum lines closely resembled that observed in OE-TeAG1#W transgenic N. tabacum lines. Specifically, the apical region of the corolla was divided into five ligulate petal lobes (Figure 5i,l), and their color was altered to white (Figure 5l).
To further confirm the homeotic transformation of sepals and petals in OE-TeAG1#S and OE-TeAG2#S transgenic N. tabacum lines, we performed scanning electron microscopy (SEM) to analyze the cellular architecture of all four floral whorls in both strong-phenotype transgenic plants and wild-type N. tabacum. In wild-type N. tabacum, epidermal cells at both the adaxial (inner) and abaxial (outer) surfaces of the corolla apex junction exhibited rough conical protrusions (Figure 6a2,a3). Additionally, the abaxial surface displayed conspicuous trichomes and stomata (Figure 6a3). In wild-type N. tabacum, the sepal epidermis exhibited smooth rectangular cells (Figure 6a1). In contrast, OE-TeAG1#S transgenic N. tabacum lines developed sepals with rough-surfaced rectangular epidermal cells (Figure 6b1). Additionally, in the OE-TeAG1#S transgenic N. tabacum lines, the adaxial cells at the junction of the corolla apex were composed of numerous columnar cells, and the dorsal epidermis contained some spherical protruding cells and trichome-like structures (Figure 6b2,b3), resembling the epidermal cell structure of the wild-type anther (Figure 6a6).
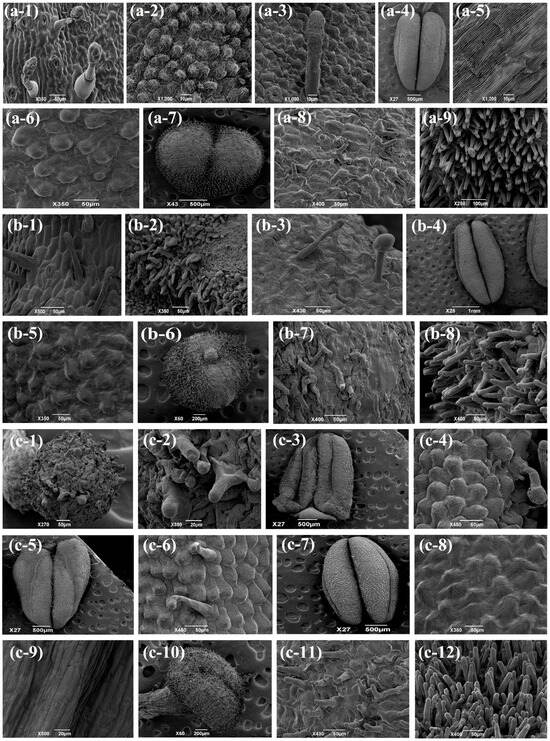
Figure 6.
A scanning electron micrograph of floral organs in transgenic N. tabacum and wild-type N. tabacum. (a1–a9) A scanning electron micrograph of the floral organs of wild-type N. tabacum. (a1) The epidermis cells of sepals. (a2,a3) The adaxial (a2) and abaxial (a3) epidermis cells of the corolla. (a4) A scanning electron micrograph of the anther. (a5) The epidermis cells of the anther. (a6) The epidermis cells of anther filaments. (a7) A scanning electron micrograph of the stigma. (a8) The epidermal cells at the top of the stigma. (a9) The epidermal cells at the edge of the stigma. (b1–b8) A scanning electron micrograph. (b1) The epidermis cells of the sepals of OE-TeAG1#S transgenic gene N. tabacum. (b2,b3) The adaxial (b2) and abaxial (b3) epidermis cells of the corolla. (b4) A scanning electron micrograph of the anther. (b5) The epidermis cells of the corolla. (b6) A scanning electron micrograph of the stigma. (b7,b8) The epidermal cells at the top (b7) and edge (b8) of the stigma. (c1–c12) A scanning electron micrograph of the floral organs of OE-TeAG2#S transgenic gene N. tabacum. (c1) A scanning electron micrograph of the sepal. (c2) The epidermis cells of sepals. (c3,c4) A scanning electron micrograph of the adaxial (c3) and abaxial (c4) corolla. (c5,c6) A scanning electron micrograph of the abaxial (c5) and abaxial (c6) corolla. (c7) A scanning electron micrograph of the anther. (c8,c9) The epidermis cells of the anther (c8) and anther filament (c9). (c10) A scanning electron micrograph of the stigma. (c11,c12) The epidermal cells at the top (c11) and edge (c12) of the stigma. Bar = 50 μm.
In contrast to OE-TeAG1#S transgenic lines, both sepals and corollas in the OE-TeAG2#S transgenic N. tabacum lines underwent conspicuous homeotic transformations (Figure 6c1–c6), and stamens and pistils developed normally without morphological alterations (Figure 6c7–c12). In OE-TeAG2#S transgenic lines, the sepal epidermis contained papillate cells that were unique to stigmas (Figure 6a7–a9,c1,c2), and the ventral and dorsal epidermal cells of the corolla were both transformed into structures resembling the epidermal cells of the wild-type anther (Figure 6a4,a5,c3–c4).
3.5. Analysis of Endogenous Gene Expression in Transgenic Plants
According to the ABC model of flower development, the identity of floral organs is determined by the combinations of class A, B, and C genes. To elucidate the molecular basis of floral organ transformations in transgenic T. erecta lines, the expression levels of class A, B, and C floral organ identity genes were analyzed in both transgenic and wild-type T. erecta plants. Compared with the wild-type T. erecta plants, the expression levels of class C genes TeAG1 and TeAG2 were significantly reduced in both RNAi-TeAG1/TeAG2#S and RNAi-TeAG1/TeAG2#W transgenic T. erecta lines (Figure 7g,h). In addition, the transcript of TeAG2 in the RNAi-TeAG1/TeAG2#S transgenic T. erecta lines was significantly lower than that in the RNAi-TeAG1/TeAG2#W transgenic T. erecta lines (Figure 7h). Conversely, the expression levels of the class A genes TeAP1-1 and TeAP1-2 were significantly upregulated in the RNAi-TeAG1/TeAG2 transgenic T. erecta lines, with the expression levels of TeAP1-1 and TeAP1-2 in the RNAi-TeAG1/TeAG2#S transgenic T. erecta lines being significantly higher than those in the weak-phenotype lines (Figure 7a,b). The expression level of the class B gene TeTM6 was significantly higher in RNAi-TeAG1/TeAG2 transgenic T. erecta lines than in the wild-type T. erecta plants (Figure 7e), while the expression levels of TePI and TeAP3 did not show significant changes compared with the wild-type T. erecta plants (Figure 7c,d,f). Notably, the transcript of TeTM6 in the RNAi-TeAG1/TeAG2#S transgenic T. erecta lines was significantly higher than that in the RNAi-TeAG1/TeAG2#W transgenic T. erecta lines (Figure 7e).
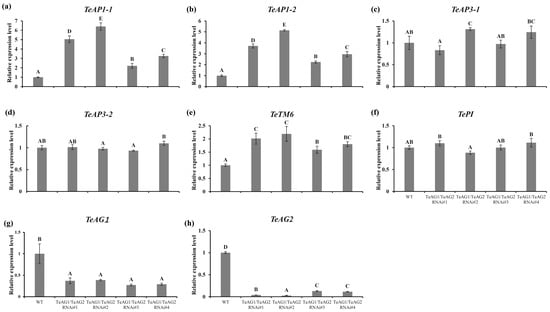
Figure 7.
A qRT-PCR analysis of the expression level of endogenous floral development-related genes in T. erecta transgenic lines. The transgenic T. erecta lines RNAi-TeAG1/TeAG2#1 and RNAi-TeAG1/TeAG2#2 showed severe phenotypes. The transgenic T. erecta lines RNAi-TeAG1/TeAG2#3 and RNAi-TeAG1/TeAG2#4 showed weak phenotypes. (a,b) The expression level of the class A genes TeAP1-1 (a) and TeAP1-2 (b) in transgenic T. erecta lines. (c–f) The expression level of the class B genes TeAP3-1 (c), TeAP3-2 (d), TeTM6 (e), and TePI (f) in transgenic T. erecta lines. (g,h) The expression level of the class C genes TeAG1 (g) and TeAG2 (h) in transgenic T. erecta lines. Based on the ABC model of floral development, A genes alone determine sepal identity; A + B genes specify petal identity; B + C genes specify stamen identity; the C gene alone determines pistil identity. Values are the means ± SE. Different letters indicate significant differences. Capital letters indicate p < 0.01.
The overexpression of both the TeAG1 and TeAG2 genes in N. tabacum led to a significant downregulation of NtAP1 transcripts in the sepals and corollas of transgenic N. tabacum lines (Figure 8a), with the expression levels of NtAP1 in 35S:TeAG1 transgenic N. tabacum lines being higher than those in the 35S:TeAG2 transgenic N. tabacum lines (Figure 8a). The expression levels of the class B genes NtAP3 and NtPI were also altered in the transgenic N. tabacum lines (Figure 8b,c). Specifically, NtAP3 expression in the corollas of 35S:TeAG1 and 35S:TeAG2 transgenic N. tabacum lines was significantly lower than that in wild-type plants (Figure 8b). Compared with NtAP3, the transcript of NtPI was only significantly upregulated in the sepals of 35S:TeAG2 transgenic plants, while it was downregulated in the corolla (Figure 8c). Specifically, in 35S:TeAG2 transgenic N. tabacum lines, NtPI expression in the sepals was positively correlated with the expression level of the introduced TeAG2 gene (Figure 8c and Figure S2c). The expression level of NtAG in the sepals and corollas of 35S:TeAG1 and 35S:TeAG2 transgenic N. tabacum lines was significantly upregulated (Figure 8d). Moreover, the transcript of NtAG in these floral organs was positively correlated with the expression levels of TeAG genes (Figure 8d and Figure S2c).
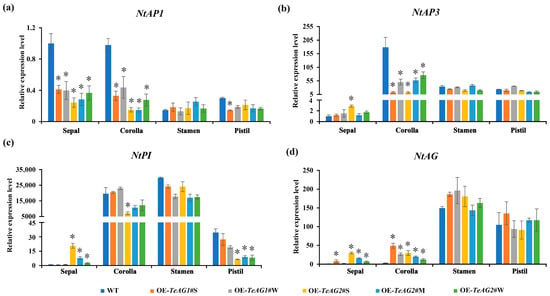
Figure 8.
A qRT-PCR analysis of the expression level of endogenous floral development-related genes in transgenic tobacco lines. (a) The expression level of the class A gene NtAP1 in transgenic tobacco lines. (b,c) The expression level of the class B genes NtAP3 (b) and NtPI (c) in transgenic tobacco lines. (d) The expression level of the class C gene NtAG in transgenic tobacco lines. Bars represent the means ± SD of three biological replicates. Asterisks indicate statistically significant differences, defined as an expression level of endogenous genes in transgenic plants that are 2 times higher or 1/2 lower than that in wild-type plants.
4. Discussion
4.1. Conservation and Divergence in the AGAMOUS Subfamily of MADS-Box Genes in Asteraceae
The MADS-box gene AG plays a key role in determining floral meristem and organ identities [10,12]. In this study, a total of 52 AG-like genes were identified from three Asteraceae subfamilies, which were present in unequal portions in these plants (Table S3). For example, six AG-like genes were identified in B. alba from the Asteroideae subfamily, whereas only one AG-like gene was selected in M. micrantha and S. sonchifolius (Tables S2 and S3). Many studies have revealed that Asteraceae species have undergone multiple genome-wide replication events [33,34]. Therefore, we speculated that this variation in gene copy number found in AG-like genes likely arose from multiple independent duplication events of AG-like genes during the evolution of the Asteraceae family. Motif analysis revealed a high conservation of motifs within AG proteins across these 22 species from Asteraceae (Figure 2a). Protein structure determines its function, and proteins with similar structures often have similar functions [35]. This indicated that the motif structures of AG proteins are highly conserved during the evolution of the Asteraceae family, implying that they might possess similar functions. Furthermore, existing studies in Asteraceae have reported that AG-like proteins are involved in regulating petal and stamen development, which is further support for our findings [16,36]. Notably, the differences in motifs among these Asteraceae plants mainly manifested the presence or absence of certain motif 10 (Figure 2a). Moreover, proteins containing motif 10 clustered together within a single clade on the phylogenetic tree (Figure 1). This high degree of structural conservation and the association of a specific motif (motif 10) with a distinct phylogenetic lineage collectively imply that AG genes might perform conserved and divergent biological functions in Asteraceae plants. Phylogenetic analysis revealed that the two AG proteins in T. erecta fell into clade 1 and clade 2, respectively. Therefore, investigating the functions of these two AG proteins in T. erecta will enhance our understanding of AG protein functions across different clades.
4.2. TeAGs Play a Conserved and Redundancy Function in Determining Stamen and Pistil Identities
Class C genes play a conserved role in regulating stamen and carpel identity. In this study, silencing class C genes TeAG1 and TeAG2 simultaneously in T. erecta resulted in the homeotic transformation of stamens and carpels into corolla structures (Figure 1). These phenotype changes were consistent with those observed upon the downregulation of C-class genes in other plant species. For instance, the suppression of the class C gene CAG in C. morifolium leads to the complete conversion of stamens and carpels into corolla-like organs [16]. In Medicago truncatula, the mtagamtagb class C double mutant exhibits a complete homeotic conversion of stamens and carpels into petaloid structures [37]. Moreover, the double-flower phenotype in the woody species Xanthoceras sorbifolium is caused by mutations in the class C gene XsAG1, resulting in the petaloid conversion of stamens and carpels [38]. Collectively, these findings demonstrated that class C genes in T. erecta function conservatively in regulating stamen and carpel development. Notably, mutations in class C genes in A. thaliana and other species lead to a petal-like stamen and sepal-like carpel [13,39], whereas silencing class C genes in T. erecta only caused the petaloid transformation of stamens and pistils (Figure 3 and Figure 4), implying the functional divergence of AG genes in governing stamen and carpel development across plant lineages.
Many AG genes exhibit conserved functions with partial genetic redundancy [36,40,41]. For example, the overexpression of either the GAG1 or GAG2 gene in G. hybrida leads to the transformation of the pappus into a pistil and the corolla into a stamen [27]. In this study, the ectopic expression of both the TeAG1 and TeAG2 genes in N. tabacum led to homeotic transformations in the sepals and corollas of the transgenic N. tabacum lines (Figure 4 and Figure 5), indicating that TeAG1 and TeAG2 shared similar functions in regulating the development of stamens and pistils. Furthermore, the individual silencing of TeAG1 or TeAG2 in T. erecta did not affect any floral organ development; only the simultaneous silencing of both TeAG1 and TeAG2 led to mutations in stamens and pistils (Figure 3 and Figure 4). Previous studies have shown that TeAG1 and TeAG2 shared similar expression patterns which are specifically and highly expressed in stamens and pistils [5], further indicating that TeAG1 and TeAG2 had redundant roles in regulating the development of these reproductive organs. Similar results have also been reported in Papaver somniferum [42] and Medicago sativa [37]. For instance, stamens and pistils undergo complete homeotic conversion into petaloid organs in the M. sativa mtagamtagb double mutant, whereas the floral organs in the inner two whorls fail to alter their identity in single mutants of either C-class gene [37]. In the ABC model of flower development, class A and class C genes exhibit antagonistic interactions. Specifically, the overexpression of class C genes in sepals and petals can suppress the expression of class A genes in the outer two whorls of floral organs, thereby leading to the transformation of sepals into pistils and petals into stamens [13,43,44]. In this study, the suppression of the transcripts of TeAG genes in T. erecta led to the upregulation of the expression of class A genes TeAP1-1 and TeAP1-2 (Figure 7). The ectopic expressions of TeAG1 and TeAG2 in N. tabacum caused a significant downregulation of the endogenous class A gene NtAP1 in sepals and corollas (Figure 8). This demonstrated that the TeAG1 and TeAG2 genes possessed conserved repressive activity against class A genes, collectively indicating functional redundancy between TeAG1 and TeAG2 in regulating the stamen and pistil identity of T. erecta. Additionally, double flowers in plants typically refer to blossoms with multiple whorls of petals, a trait often caused by the mutation of stamens into petal-like structures. However, double flowers in Asteraceae plants refer to the presence of multiple whorls of ray florets or the mutation of internal disk florets into ray florets [45]. In this study, the simultaneous silencing of both TeAG1 and TeAG2 induced the stamens and pistils to be converted into corolla-like structures, resulting in true double-petaled flowers (Figure 9).
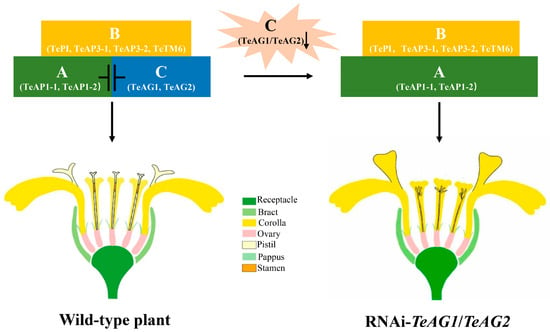
Figure 9.
Schematic presentation of C functions in T. erecta at level of flowers. The capital letters A, B and C represent the class A, B, and C genes in the ABC model of flower development. A down-regulated black arrow at TeAG1/TeAG2, indicating the co-silencing of TeAG1 and TeAG2 genes.
4.3. TeAG Genes Play Divergent Functions in Determining Floral Organs
Research has demonstrated functional divergence among different class C genes within the same plant [15,17,46]. In Oryza sativa, the mads3 single mutant exhibits the homeotic conversion of stamens into lodicule-like organs, whereas the mads58 mutant displays impaired carpel development with indeterminate floral meristems [17]. Similarly, in Cyclamen persicum, CpAG1 silencing induces the homeotic conversion of stamens into petaloid organs, whereas CpAG2 knockdown causes the developmental arrest of stamens and carpels [46]. In this study, TeAG2 expression in RNAi-TeAG1/TeAG2#S transgenic T. erecta was significantly lower than that in RNAi-TeAG1/TeAG2#W transgenic T. erecta (Figure 7h). In contrast, class A genes TeAP1-1 and TeAP1-2 exhibited an opposing expression trend (Figure 7a,b). Notably, no significant difference in the expression level of TeAG1 was observed between these transgenic T. erecta lines (Figure 7g), suggesting that the development of pistils in ray florets was primarily regulated by TeAG2. Furthermore, phenotypic alterations in sepals and corollas were more pronounced in TeAG2-overexpressing transgenic N. tabacum than in TeAG1-overexpressing lines (Figure 5 and Figure 6). Additionally, the ectopic expression of TeAG2 in N. tabacum significantly altered NtPI transcript accumulation in the sepals and pistils of OE-TeAG2#S transgenic lines (Figure 8c). Conversely, the overexpression of TeAG1 in N. tabacum had no significant effect on NtPI expression in the corresponding tissues of OE-TeAG1#S transgenic lines (Figure 8c). These findings demonstrated that there was functional divergence between the class C paralogs TeAG1 and TeAG2 in regulating stamen and pistil identity specification during T. erecta floral development.
5. Conclusions
In this study, we identified and systematically analyzed AG-like genes from 22 species across three subfamilies of the Asteraceae family. Our results indicated that AG-like genes in these species may have conserved and specific biological functions in Asteraceae species. Furthermore, a functional analysis of AG-like genes in T. erecta revealed that TeAG1 and TeAG2 played conserved, redundant, and divergent functions in the regulation of stamen and pistil development. This finding expands our understanding of the functions of class C genes in Asteraceae plants and provides theoretical support for the artificial breeding of cultivars with double flowers.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/agronomy15102379/s1, Figure S1. The expression level of TeAG1 and TeAG2 in transgenic T. erecta lines and wild-type plants by qRT-PCR. Figure S2. The early flowering phenotype of overexpressing marigold class C genes. Table S1. Basic information of Asteraceae class C genes. Table S2. Sequence of primers. Table S3. Features of class C genes identified in Asteraceae. Table S4. Flowering phenotypes of T2 transgenic tobacco lines.
Author Contributions
Conceptualization, Y.H. and C.Z.; methodology, C.Z., C.H., K.Z. and H.L.; software, C.Z., C.H., H.L. and S.X.; validation, C.Z., C.H. and K.Z.; formal analysis, C.Z., C.H., Z.T., K.Z. and Y.H.; investigation, Z.T., C.Z., C.H., K.Z. and S.X.; resources, Z.T., Y.H. and C.Z.; data curation, C.Z., C.H., K.Z., S.X. and H.L.; writing—original draft preparation, C.Z.; writing—review and editing, C.Z., Y.H. and C.H.; visualization, C.Z., C.H. and S.X.; supervision, Y.H. and C.Z.; project administration, C.Z.; funding acquisition, C.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (Program No. 32202534) and National Natural Science Foundation of Hunan province (Program No. 2023JJ41042).
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
Author Zhengguo Tao was employed by the company Guangzhou Leader Bio-Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Endress, P.K. Origins of flower morphology. J. Exp. Zool. 2001, 291, 105–115. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31. [Google Scholar] [CrossRef]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Bowman, J.L.; Moyroud, E. Reflections on the ABC model of flower development. Plant Cell 2024, 36, 1334–1357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wei, L.; Wang, W.; Qi, W.; Cao, Z.; Li, H.; Bao, M.; He, Y. Identification, characterization and functional analysis of AGAMOUS subfamily genes associated with floral organs and seed development in Marigold (Tagetes erecta). BMC Plant Biol. 2020, 20, 439. [Google Scholar] [CrossRef] [PubMed]
- Cong, D.; Zhao, X.; Ni, C.; Li, M.; Han, L.; Cheng, J.; Liu, H.; Liu, H.; Yao, D.; Liu, S. The SEPALLATA-like gene HrSEP1 in Hippophae rhamnoides regulates flower development by interacting with other MADS-box subfamily genes. Front. Plant Sci. 2025, 15, 1503346. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Y.; Zhang, Y.; Liu, G.; Yu, S.; Zheng, Z. The overall regulatory network and contributions of ABC (D) E model genes in yellowhorn flower development. BMC Plant Biol. 2024, 24, 1081. [Google Scholar] [CrossRef]
- Ali, Z.; Raza, Q.; Atif, R.M.; Aslam, U.; Ajmal, M.; Chung, G. Genetic and molecular control of floral organ identity in cereals. Int. J. Mol. Sci. 2019, 20, 2743. [Google Scholar] [CrossRef]
- Irish, V. The ABC model of floral development. Curr. Biol. 2017, 27, R887–R890. [Google Scholar] [CrossRef]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, P. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, R.; Xu, Y.; Wang, M.; Zhang, J.; Bai, M.; Han, C.; Xiang, F.; Wang, Z.-Y.; Mysore, K.S.; et al. AGLF provides C-function in floral organ identity through transcriptional regulation of AGAMOUS in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2019, 116, 5176–5181. [Google Scholar] [CrossRef] [PubMed]
- Pelayo, M.A.; Yamaguchi, N.; Ito, T. One factor, many systems: The floral homeotic protein AGAMOUS and its epigenetic regulatory mechanisms. Curr. Opin. Plant Biol. 2021, 61, 102009. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar][Green Version]
- Mizukami, Y.; Huang, H.; Tudor, M.; Hu, Y.; Ma, H. Functional domains of the floral regulator AGAMOUS: Characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell 1996, 8, 831–845. [Google Scholar] [PubMed][Green Version]
- Mena, M.; Ambrose, B.A.; Meeley, R.B.; Briggs, S.P.; Yanofsky, M.F.; Schmidt, R.J. Diversification of C-function activity in maize flower development. Science 1996, 274, 1537–1540. [Google Scholar] [CrossRef]
- Aida, R.; Komano, M.; Saito, M.; Nakase, K.; Murai, K. Chrysanthemum flower shape modification by suppression of chrysanthemum-AGAMOUS gene. Plant Biotechnol. J. 2008, 25, 55–59. [Google Scholar] [CrossRef]
- Dreni, L.; Pilatone, A.; Yun, D.; Erreni, S.; Pajoro, A.; Caporali, E.; Zhang, D.; Kater, M.M. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 2011, 23, 2850–2863. [Google Scholar] [CrossRef]
- Yellina, A.L.; Orashakova, S.; Lange, S.; Erdmann, R.; Leebens-Mack, J.; Becker, A. Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica). EvoDevo 2010, 1, 13. [Google Scholar] [CrossRef]
- Mandel, J.R.; Dikow, R.B.; Siniscalchi, C.M.; Thapa, R.; Watson, L.E.; Funk, V.A. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc. Natl. Acad. Sci. USA 2019, 116, 14083–14088. [Google Scholar] [CrossRef]
- Funk, V.A.; Susanna, A.; Steussy, T.F.; Robinson, H.E. Classification of compositae. In Systematics, Evolution, and Biogeography of Compositae; International Association for Plant Taxonomy: Vienna, Austria, 2009; pp. 171–192. [Google Scholar]
- Kazeminia, M.; Mehrabi, A.; Mahmoudi, R. Chemical composition, biological activities, and nutritional application of Asteraceae family herbs: A systematic review. Trends Phytochem. Res. 2022, 6, 187–213. [Google Scholar]
- Zhang, T.; Elomaa, P. Development and evolution of the Asteraceae capitulum. New Phytol. 2024, 242, 33–48. [Google Scholar] [CrossRef]
- Zhang, T.; Elomaa, P. Don’t be fooled: False flowers in Asteraceae. Curr. Opin. Plant Biol. 2021, 59, 101972. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, O.; Garcia-Jacas, N.; Garnatje, T.; Susanna, A.; Siljak-Yakovlev, S. Karyological evolution in Rhaponticum Vaill. (Asteraceae, Cardueae) and related genera. Bot. J. Linn. Soc. 2007, 153, 193–201. [Google Scholar] [CrossRef][Green Version]
- Liu, C.; Wang, F.; Li, R.; Zhu, Y.; Zhang, C.; He, Y. Marigold (Tagetes erecta) MADS-Box Genes: A Systematic Analysis and Their Implications for Floral Organ Development. Agronomy 2024, 14, 1889. [Google Scholar] [CrossRef]
- Shchennikova, A.V.; Shulga, O.A.; Skryabin, K.G. Diversification of the Homeotic AP3 Clade MADS-Box Genes in Asteraceae Species Chrysanthemum morifolium L. and Helianthus annuus L. Dokl. Biochem. Biophys. 2018, 483, 348–354. [Google Scholar]
- Yu, D.; Kotilainen, M.; Pöllänen, E.; Mehto, M.; Elomaa, P.; Helariutta, Y.; Albert, V.A.; Teeri, T.H. Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J. 1999, 17, 51–62. [Google Scholar] [CrossRef]
- Shchennikova, A.; Shul’ga, O.; Sizeneva, E.; Perkovskaya, N.; Skryabin, K. Diversification of functional activity of the chrysanthemum homeotic MADS-box gene CDM37. Doklady. Biochem. Biophys. 2011, 436, 29–31. [Google Scholar] [CrossRef]
- Shulga, O.; Mitiouchkina, T.; Shchennikova, A.; Skryabin, K.; Dolgov, S. Chrysanthemum modification via ectopic expression of sunflower MADS-box gene HAM59. In Proceedings of the XXV International EUCARPIA Symposium Section Ornamentals: Crossing Borders, Melle, Belgium, 28 June–2 July 2015; Volume 1087, pp. 105–111. [Google Scholar]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Y.; Liu, Y.; Yi, Q.; Chen, W.; Zhu, Y.; Duan, F.; Zhang, L.; He, Y. Establishment of Agrobacterium tumefaciens-mediated Genetic Transformation System of Marigold (Tagetes erecta). J. Integr. Plant Biol. 2023, 58, 760–769. [Google Scholar] [CrossRef]
- Gallois, P.; Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mol. Biol. 1995, 49, 39–48. [Google Scholar] [PubMed]
- Kong, X.; Zhang, Y.; Wang, Z.; Bao, S.; Feng, Y.; Wang, J.; Yu, Z.; Long, F.; Xiao, Z.; Hao, Y.; et al. Two-step model of paleohexaploidy, ancestral genome reshuffling and plasticity of heat shock response in Asteraceae. Hortic. Res. 2023, 10, uhad073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, T.; Luebert, F.; Xiang, Y.; Huang, C.-H.; Hu, Y.; Rees, M.; Frohlich, M.W.; Qi, J.; Weigend, M. Asterid phylogenomics/phylotranscriptomics uncover morphological evolutionary histories and support phylogenetic placement for numerous whole-genome duplications. Mol. Biol. Evol. 2020, 37, 3188–3210. [Google Scholar] [CrossRef] [PubMed]
- Whisstock, J.C.; Lesk, A.M. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003, 36, 307–340. [Google Scholar] [CrossRef]
- Sizeneva, E.S.; Shulga, O.A.; Shchennikova, A.V.; Skryabin, K.G. Functional diversification of two MADS-Box genes, HAM45 and HAM59, in sunflower. Dokl. Biol. Sci. 2013, 451, 221–224. [Google Scholar] [CrossRef]
- Zhu, B.; Li, H.; Wen, J.; Mysore, K.S.; Wang, X.; Pei, Y.; Niu, L.; Lin, H. Functional specialization of duplicated AGAMOUS homologs in regulating floral organ development of Medicago truncatula. Front. Plant Sci. 2018, 9, 854. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Y.; Zhang, T.; Liu, Z.; Cao, L.; Chang, Q.; Liu, Y.; Lu, X.; Yu, S.; Li, H.; et al. The double flower variant of yellowhorn is due to a LINE1 transposon-mediated insertion. Plant Physiol. 2023, 191, 1122–1137. [Google Scholar] [CrossRef]
- Pan, I.L.; McQuinn, R.; Giovannoni, J.J.; Irish, V.F. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 2010, 61, 1795–1806. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Z.-x.; Ma, J.; Song, Y.; Chen, F. Alternative splicing of the AGAMOUS orthologous gene in double flower of Magnolia stellata (Magnoliaceae). Plant Sci. 2015, 241, 277–285. [Google Scholar] [CrossRef]
- Wang, Q.; Dan, N.; Zhang, X.; Lin, S.; Bao, M.; Fu, X. Identification, characterization and functional analysis of C-class genes associated with double flower trait in carnation (Dianthus caryphyllus L.). Plants 2020, 9, 87. [Google Scholar] [CrossRef]
- Hands, P.; Vosnakis, N.; Betts, D.; Irish, V.F.; Drea, S. Alternate transcripts of a floral developmental regulator have both distinct and redundant functions in opium poppy. Ann. Bot. 2011, 107, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Drews, G.N.; Meyerowitz, E.M. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 1991, 3, 749–758. [Google Scholar] [PubMed]
- Mizukami, Y.; Ma, H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol. Biol. 1995, 28, 767–784. [Google Scholar] [CrossRef] [PubMed]
- Elomaa, P. My favourite flowering image: A capitulum of Asteraceae. J. Exp. Bot. 2019, 70, e6496–e6498. [Google Scholar] [CrossRef]
- Tanaka, Y.; Oshima, Y.; Yamamura, T.; Sugiyama, M.; Mitsuda, N.; Ohtsubo, N.; Ohme-Takagi, M.; Terakawa, T. Multi-petal cyclamen flowers produced by AGAMOUS chimeric repressor expression. Sci. Rep. 2013, 3, 2641. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).