Abstract
Radish flower color, bolting time, and flowering time are important traits for attracting certain pollinators and affect fleshy root quality. In this study, an analysis of the anthocyanidins in radish flowers by high-performance liquid chromatography revealed that differences in the cyanidin content are likely to be associated with the variability in radish flower colors (i.e., purple and white petals). A quantitative trait loci (QTL) analysis identified nine QTLs on three Raphanus sativus linkage groups. Three QTLs—qRFC1, qRBT1, and qRFT1—which were consistently detected and explained a high proportion of the observed variation (10.30% to 34.57%), were considered as the major QTLs responsible for flower color, bolting time, and flowering time, respectively. A total of 16 and 11 candidate genes within the major QTL regions for flower color and bolting/flowering times, respectively, were preliminarily annotated. Six genes (Rs018140, Rs018950, Rs019220, Rs020080, Rs020590, and Rs021450) related to flower color were differentially expressed in the parental lines. On the basis of nucleotide and amino acid sequence diversity between the parental lines, Rs314940, Rs315000, Rs315310, and Rs315960 were identified as candidate genes mediating the radish bolting and flowering times. This study revealed the genetic complexity of the radish flower color, bolting time, and flowering time traits. The identified candidate genes in the QTL regions may be useful for radish breeding programs and also for functional characterization in radish.
1. Introduction
Radish (Raphanus sativus, 2n = 18), which belongs to the family Brassicaceae, is one of the most popular vegetable crops worldwide, but especially in eastern Asia. Flower-related traits—such as color, bolting time, and flowering time—are important radish characteristics. More specifically, certain flower colors can attract suitable pollinators, while also repelling herbivorous insects and animals [1,2,3,4]. Bolting and flowering occur in a critical period in which plants transition from the vegetative phase to the reproductive phase of the life cycle [5]. There are relatively few reports describing research on the mechanism underlying the inheritance of flower-related traits, especially flower color, in radish. Thus, clarifying the genetic basis of flower coloration, bolting time, and flowering time will provide plant breeding programs with useful information.
Flower color is an important trait for attracting pollinators. In Brassica, flower color is influenced by two independently inherited pigment systems. Carotenoid pigments produce yellow petals, whereas the absence of carotenoids results in white petals. Anthocyanin pigments produce pink and purple petals, with the intensity of the colors and the distribution of the pigments varying substantially in the petals [6,7,8]. To date, there are many reports describing the mechanism mediating the inheritance of flower coloration in Brassica species. In Brassica rapa, a single dominant gene controls the development of white flowers, with no apparent cytoplasmic effects [9,10]. Interestingly, a genetic analysis conducted by Zhang et al. (2020) revealed two recessive genes that control the white flower trait [11]. In Brassica carinata, one gene mediating the yellow flower trait exhibits incomplete dominance [12]. The development of white flowers is controlled by a single nuclear gene in Brassica napus [13,14,15], but it is the result of two independent recessive genes in Brassica juncea [16,17]. Regarding Brassica oleracea, a single recessive gene (cpc-1) is associated with yellow flower coloration and BoCCD4 is responsible for the production of white or yellow petals according to map-based clones [18,19]. Similarly, Xu et al. (2019) and Yan et al. (2020) identified the recessive genes responsible for yellow flowers in Chinese kale and cauliflower [20,21]. However, the molecular mechanism and genetic characteristics regulating flower coloration in radish remain uncharacterized.
Bolting and flowering are important changes in the life cycle of Brassica species. The associated transition from vegetative growth to reproductive growth involves a series of complex processes. In the model plant species, Arabidopsis thaliana, six major pathways (i.e., photoperiod, vernalization, ambient temperature, aging, autonomous, and gibberellin pathways) regulate the bolting and flowering times [22,23]. Genes controlling flowering traits have been extensively explored in A. thaliana, resulting in the identification of almost 300 genes that modulate the flowering process either directly or indirectly (http://www.phytosystems.ulg.ac.be/florid/, accessed on 10 February 2020). The inheritance of the bolting and flowering traits in some Brassica species has been investigated. The cloning and subsequent analysis of Brassica FLC genes resulted in the identification of four, nine, and three FLC homologs in B. rapa [24], B. napus [25,26], and B. oleracea [27], and R. sativus [28], respectively. Some quantitative trait loci (QTLs) and genetic variations related to bolting and flowering times have been reported for Brassica species. Many QTLs for these two traits were identified in B. rapa, with several QTLs co-localized with BrFLC1, BrFLC2, and BrFLC5. In B. oleracea, six [27] and five [29] QTLs related to bolting and flowering were detected by different researchers. In B. napus, more than 30 relevant QTLs were identified, and the key flowering time-related genes PSEUDO-RESPONSE REGULATOR 7 (PRR7) and FY are reportedly located in a major QTL region on C02 [30,31,32]. Regarding R. sativus, several analyses of QTLs and genetic variations for bolting and flowering times have been reported. For example, Kitashiba (2017) and Wang (2018) detected RsFLC2 in a QTL region [33,34]. Moreover, by combining RNA sequencing and quantitative real-time polymerase chain reaction (qRT-PCR) analyses, many research groups have isolated genes related to bolting and flowering times and the associated regulatory network (e.g., FLC, FT, SOC1, CO, and LFC) [35,36,37,38].
In this study, using the F2:3 population derived from ‘YR4’ (white flowers, late bolting and flowering) and ‘YR18’ (purple flowers, early bolting and flowering) inbred lines, we investigated the inheritance of flower coloration, bolting time, and flowering time in radish. To detect flowering regulators related to vernalization and ambient conditions, phenotypic data were collected and a QTL analysis was performed to identify genomic regions associated with R. sativus floral traits. The results of this study will help breeders develop new germplasm with desirable flower colors and bolting and flowering times on the basis of marker-assisted selection and the fine-mapping of flowering-related loci in the radish genome.
2. Materials and Methods
2.1. Plant Materials
Radish inbred line ‘YR4’ (white flowers, late bolting and flowering) was crossed with ‘YR18’ (purple flowers, early bolting and flowering). The F1 generation was self-pollinated to produce 180 F2 progeny, which was then self-pollinated to generate the 180 F2:3 lines used for investigating traits.
2.2. Anthocyanidin Extraction and Analysis
As described in the color grade, we select the flower samples from the lines which were recorded as “0” in flower index following the description of Figure 1 legends, and those were regarded as ‘white type’. Likewise, the flowers of lines which has flower index “4” were selected and those were regarded as ‘purple type’. In each type, 15 plant lines were selected based on the flower index, respectively. Five individual plants per each line were selected as a replication for the anthocyanidin compositions and contents analysis. Anthocyanidins were extracted from freeze-dried petals, which were collected at the flowering stage, 14 DAF (days after flowering), and detected as previously described [39]. Briefly, anthocyanidins were analyzed using the Agilent 1200 series high-performance liquid chromatography (HPLC) system. Anthocyanidins were identified on the basis of the typical retention times for the standard compounds, including cyanidin, delphinidin, malvidin, pelargonidin, and peonidin. Samples were prepared and the HPLC system was operated according to established methods [40]. For each sample, the mean and standard deviation were calculated for three biological replicates.

Figure 1.
Flower color phenotypes of R. sativus. Flowers are arranged from left to right according to their petal color, which was characterized using the following 0–4 scale: 0, white petals; 1, little purple color from the edge covering <50% of total petals; 2, pink color on edge covering >50% of total petals; 3, pale purple color covering all of the petals; 4, dark purple petals.
2.3. Evaluation of Flower-Related Traits
The two parental lines (‘YR4’ and ‘YR18’), their F1 lines, 180 F2 lines, and 180 F2:3 lines were evaluated in 2017 and 2018. Ten seeds of ‘YR4’, ‘YR18’, and F1 plants and five individuals from each F2:3 line were sown in a greenhouse on 19 February 2018 (average 0 to 11 °C, 11.1 to 12.4 h light within 6 weeks after sawing) and 21 June 2018 (average 24 to 28 °C, 14.3 to 13.5 h light within 6 weeks after sawing). The plants in the spring underwent a natural vernalization and the plants in the summer were subjected to a six-week artificial vernalization treatment (20 July to 31 August) at 4–5 °C.
We examined each plant in all generations regarding flower color (first mature flower), bolting time (number of days from the sowing date to when the main stalk grew to 2 cm in height), and flowering time (number of days from the sowing date to when the first flower bloomed). The flower index was calculated as described below (Figure 1). The data for the distribution of phenotypic traits were analyzed and the correlations among the flower color, bolting time, and flowering time were determined using the SPSS 24.0 program.
2.4. Genetic Map and QTL Analysis
To identify the QTLs associated with phenotypic traits data above-mentioned, we used the R. sativus genetic map which was constructed by Ma et al., previously (2021, under revision). This genetic map was analyzed by JoinMap software version 4.0 using 180 F2 population. Finally, the genetic map consist of 403 markers, and total distance is 1074.23 cM with an average distance of 2.67 cM. It covered about 83% of the radish genome. The inclusive composite interval mapping (ICIM) method of IciMapping 4.1 (http://www.isbreeding.net/software/, accessed on 4 February 2019) was used for mapping the QTLs of flower-related traits. Genome-wide threshold values (α = 0.05) were used to detect putative QTLs on the basis of 1000 permutations [41]. To eliminate false-positive QTLs, we also used Windows QTL Cartographer 2.5 [42] to detect putative QTLs.
2.5. Candidate Gene Prediction and Gene Sequence Variation Analysis
High-throughput paired-end whole-genome sequencing of two parents (‘YR4’ and ‘YR18’) were performed using an Illumina Hiseq2500 sequencer with 350 bp insertions. After bioinformatics analysis descripted as Ma et al. (2021), the resequencing genome coverage parental lines (YR4 and YR18) was 25X and 28X, then the whole-genome scaffolds were assembled base on the radish genome database [43]. The alignment to the reference genome indicated the read mapping rate for ‘YR4’ and ‘YR18’ was respectively 77.74% and 77.30%. Furthermore, for the identified candidate genes, we identified the orthologous gene pairs between R. sativus and A. thaliana using the MCScanX program. Specifically, the following parameters were used: e = 1e − 20, u = 1, and s = 5 [44]. The TAIR database was used to predict gene annotations (http://www.arabidopsis.org/, accessed on 10 February 2020). For the identified potential candidate genes, CLC genomic workbench 12.0 was used to investigate gene sequence and amino acid variation between parental lines.
2.6. Quantitative Real-Time PCR Analysis
To compare the expression level of candidate genes between two genotypes, quantitative real-time (qRT)-PCR analyses were performed. We collected the leaf and petal tissues from ‘YR4’ and ‘YR18’ plants at 14 DAF (days after flowering). Total RNA was isolated from ‘YR4’ and ‘YR18’ leaves and petals using the RNeasy® Mini kit (QIAGEN, Hilden, Germany). The RNA (2 µg) extracted from individual plants was used as the template in a 20-µL reverse transcription reaction, which was completed using the TOPscript™ RT DryMix kit (Enzynomic Co., Daejeon, Korea). The resulting cDNA was diluted 10-fold and then a 2-µL aliquot was used for a qRT-PCR analysis (20-µL reaction volume). The qRT-PCR primers were designed according to the available radish sequence information. Details regarding the primers are provided in Table S2. The qRT-PCR analysis was performed using the CFV96™ Real-Time System (Bio-Rad, Berkeley, CA, USA) and the SYBR Green Supermix (PhilKorea, Seoul, Korea). The PCR program was as follows: 95 °C for 3 min; 39 cycles of 95 °C for 15 s and 58 °C for 20 s. Data were acquired during the annealing/extension step and were analyzed using the CFX Manager software (version 2.1) (Bio-Rad). Three replicates of each sample were analyzed, and mean gene expression levels were normalized against 18S rRNA levels.
3. Results
3.1. Investigation of Flower Color and Anthocyanidin Accumulation in Purple and White Petals
There were considerable differences in flower color as shown in Figure 1. Based on the flower color index, the ‘YR4’ flowers having white petals was recorded as a ‘0’, and whereas the ‘YR18’ flowers representing purple petals was regarded as a ‘4’. The flowers of the F1 plants had pink petals, implying the gene(s) responsible for purple coloration exhibit incomplete dominance. Additionally, the color index of the F2 and F2:3 lines segregated continuously and exhibited a normal distribution in all environments, suggesting that both major and minor QTLs are involved in R. sativus flower coloration (Figure 2).
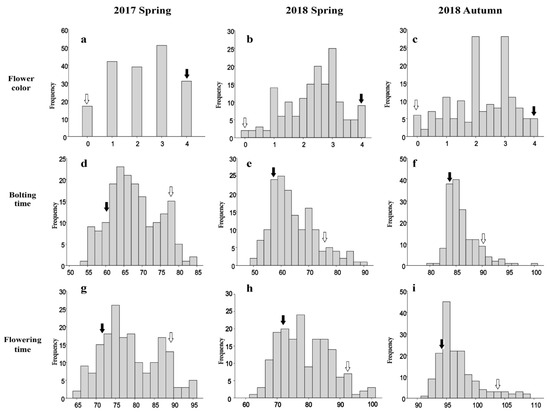
Figure 2.
Frequency distribution of the flower color, bolting time, and flowering time traits in radish populations over two years. (a,d,g): frequency distribution of the flower color, bolting time, and flowering time for the F2 population in spring 2017; (b,e,h): frequency distribution of the flower color, bolting time, and flowering time for the F2:3 population in spring 2018; (c,f,i): frequency distribution of the flower color, bolting time, and flowering time for the F2:3 population in autumn 2018. The trait frequencies were normally distributed. White and black arrows indicate P1 (‘YR4’) and P2 (‘YR18’), respectively.
The anthocyanidin compositions and contents in the purple and white petals at the flowering stage were analyzed using an HPLC system. The results indicated that the purple petals (‘YR18’) contained more anthocyanidins than the white petals (‘YR4’). Additionally, the anthocyanidins in the purple petals (‘YR18’) included cyanidin, peonidin, malvidin, and pelargonidin, whereas only cyanidin was detected in the white petals (‘YR4’). We made two types of samples by pooling, which were used flowers recorded as “0” and “4” in the F2:3 population as described in the color grade of Figure 1, and those were regarded as ‘white type’ and purple type’. Moreover, the pink petals of the flowers in the F1 plants, and the purple types of the F2:3 population also contained cyanidin, peonidin, malvidin, and pelargonidin. In particular, the anthocyanidin contents of purple petals (‘YR18’) and white petals (‘YR4’) showed the greatest difference with cyanidin contents of 7558.92 ± 212.44 and 24.52 ± 4.43 μg/g, respectively (Figure 3). This may suggest that the difference in cyanidin content could associated the difference in flower color between ‘YR18’ and ‘YR4’.
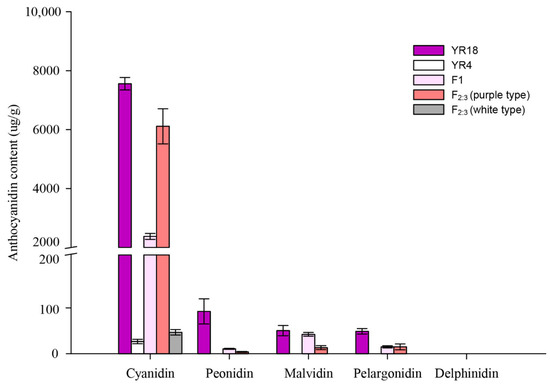
Figure 3.
Anthocyanidin compositions and contents of the flower petals in diverse genotypes. Inbred lines ‘YR18’ and ‘YR4’ were the parental lines used for constructing the mapping population. The F1 lines were derived from a cross between the parental lines. Both F2:3 (purple type) and F2:3 (white type) refer to bulk pools, which were collected according to the flower colors in the F2:3 population. The horizontal and vertical axes respectively present the main anthocyanidin compounds (e.g., cyanidin, peonidin, malvidin, pelargonidin, and delphinidin) and the anthocyanidin contents (µg/g). Three biological replicates were analyzed.
3.2. Differences in the Bolting and Flowering Times between the Parental Lines
The two parental lines (‘YR4’ and ‘YR18’) showed significant differences for bolting and flowering times. The results of these two traits were similar in the two times spring cultivations over two years. However, there was a tendency for the timing to be slightly delayed in the autumn (Table 1). In the two spring cultivations, the time was similar, but there was a tendency for the timing to be slightly delayed in the autumn. The range of bolting time and flowering time in the population represented a broader range than that of the parental lines, indicating transgressive segregation (Figure 2). The F2:3 plants varied considerably regarding bolting time (46–104 days) and flowering time (58–111 days) in spring 2018. In autumn 2018, the bolting time was 79–101 days and the flowering time was 91–116 days. These results suggest that bolting and flowering times are quantitative traits controlled by multiple genes. A Pearson correlation analysis of the three replicates for the analyzed populations revealed that the bolting and flowering times were highly positively correlated (r = 0.935, 0.910, and 0.870, p < 0.01) (Table S1), implying that different tightly linked QTLs govern the bolting and flowering time traits.

Table 1.
Summary of bolting and flowering time traits in the parental lines, F1 and F2:3 population.
3.3. Genetic Linkage Map and QTL Analysis
On the basis of the genetic linkage map [43] and the flower traits, we detected nine QTLs related to flower traits on three linkage groups of the R. sativus genome. These QTLs had a logarithm of odds (LOD) value of 4.86–17.72 and explained 7.32%–34.57% of the phenotypic variation (i.e., R2). The QTLs for each trait and their predicted locations are presented in Table 2 and Figure 4.

Table 2.
Details regarding the QTLs identified for radish flower color, bolting, and flowering time traits.
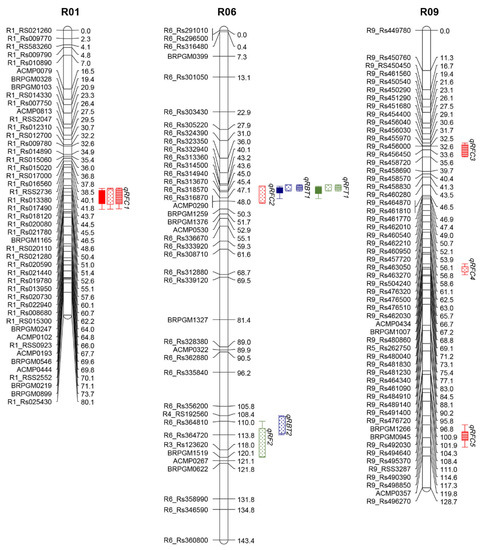
Figure 4.
Genetic linkage map and distribution of QTLs for flower color, bolting time, and flowering time in the F2 and F2:3 populations. Markers are presented on the left side of the linkage group. Genetic distances are provided as centimorgans (cM) on the right side of the linkage group. The identified QTLs are indicated by abbreviated trait names on the right side of the linkage group. Red, blue, and green boxes represent QTLs for radish flower color, bolting time, and flowering time, respectively. Filled boxes represent the F2 population in spring 2017, whereas dotted and lined boxes represent the F2:3 population in spring and autumn 2018, respectively.
3.4. Flower Color Trait
We detected five QTLs related to flower color on three chromosomes (R01, R06, and R09) (Figure S1). The LOD values for these QTLs ranged from 4.78 to 17.72 and the R2 was 6.78–34.57%. Additionally, qRFC1 located between markers R1_Rs020080 and R1_Rs021280 on chromosome R01 had a relatively high LOD value (8.83–17.72) and R2 (18.85–34.57%), suggesting that this locus is important for flower coloration. The QTLs detected in only one phenotypic test were considered to be minor QTLs related to the radish flower color.
3.5. Bolting and Flowering Time Traits
We detected two QTLs each for bolting and flowering times on chromosome R06 (Figure S2). The major QTLs (qRBT1 and qRFT1) were mapped together between markers R6_Rs332940 and R6_Rs318570 on chromosome R06. These QTLs had high LOD and R2 values. The remaining QTLs (qRBT1 for bolting time and qRFT1 for flowering time) were detected only in spring 2018. Interestingly, all QTLs for bolting and flowering times were localized at the same position on chromosome R06.
3.6. Identification of Potential Candidate Genes in the Major QTL Regions
We detected one QTL, qRFC1, in all three experiments, with an R2 of 18.85%, 32.45%, and 34.57%. Therefore, this QTL might be responsible for radish flower coloration. Additionally, we revealed 269 genes in the QTL region (1.83 Mb) of chromosome R01 using the R. sativus database (http://radish-genome.org/About_us/, accessed on 12 March 2020). By annotating these genes according to the radish and A. thaliana (TAIR) databases, we determined that 16 of the genes are involved in anthocyanin synthesis (Table S3). Furthermore, we aligned these gene sequences with the sequences of the parental lines and identified nine genes with sequence variations, and amino acid variation. A qRT-PCR analysis was performed to compare the expression patterns of these nine candidate genes between the parental lines. Although we observed sequence variation but the differential expression of three genes (Rs019190, Rs019780, and Rs020110) between parental lines were not detected. Finally, six genes represented the significantly different gene expression between the parental lines (Figure 5). The data indicated Rs018140 (sucrose transport protein; SUC2), Rs018950 (glutathione s-transferase 12; AT1G27130), Rs019220 (Acyl-CoA N-acyltransferases, NAT, superfamily protein; AT1G24040), Rs020080 (uncharacterized PKHD-type hydroxylase; At1g22950-like), Rs020590 (ethylene-responsive transcription factor; ERF118), and Rs021450 (WD-40 repeat family protein; AT1G27470) were relatively highly expressed in ‘YR18’ flower petals. Our results suggest that these potentially testable six genes are likely to be associated with radish flower coloration.
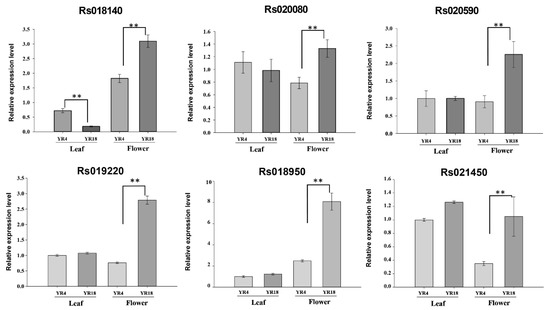
Figure 5.
Expression levels of six candidate genes in ‘YR4’ and ‘YR18’ leaves and flowers. Error bars indicate the standard error of the mean of three biological replicates (n = 5), and asterisks indicate a significant difference between the ‘YR4’ and ‘YR18’ lines based on a t-test (independent). Double asterisks (**) indicates p < 0.01.
The QTLs qRBT1 (related to bolting time) and qRFT1 (related to flowering time) were mapped to the same region of chromosome R06 and had relatively high LOD and R2 values. Moreover, this locus included 330 genes, of which 11 genes are related to bolting and flowering times according to the Flowering Interactive Database (www.phytosystems.ulg.ac.be/florid/, accessed on 10 February 2020) (Table S4). Among 11 genes, 4 genes represented sequence variation between the parental lines (Figure S3); Rs314940 (C2H2-like zinc finger protein), Rs315000 (flowering promoting factor 1), Rs315310 (AGAMOUS-like 79), and Rs315960 (brassinosteroid-6-oxidase 2). Consistent with this finding, the predicted amino acid sequences of these genes also varied between the parents. Accordingly, we speculate that Rs314940, Rs315000, Rs315310, and Rs315960 are potentially important for the radish bolting and flowering times.
4. Discussion
Flower color, bolting time, and flowering time are important agronomic traits for radish because they are related to seed yield and quality as well as antioxidant activities and herbivore resistance [3,4,45,46]. In the present study, an HPLC analysis revealed cyanidin as the main pigment may be associated with the production of purple radish flowers. A genetic linkage map comprising 403 markers (SSRs and SNPs) was used to analyze the differences between parental traits [43]. Genetic and QTL analyses of the F2 and F2:3 populations derived from a cross between two inbred lines (‘YR4’ and ‘YR18’) detected nine QTLs for three flower-related traits. The region corresponding to the QTL position was found in the Reference Genome, and candidate genes related to flower-related characteristics could be easily selected using the reference genome information.
4.1. Trait Variations in the Parental Lines and Generated Populations
To improve the accuracy of QTL analyses, biparental populations consisting of more than 100 individuals should be used [47,48]. Therefore, we determined the segregation ratios for flower-related traits using a large biparental population (180 individuals from each segregating population) with two replications. The two parental lines—as well as the F1, F2,, and F2:3 lines—were grown in different seasons for phenotypic examinations. The flower color index segregated continuously in the F2 and F2:3 populations, with a similar distribution in different environment plots. This implies that both major and minor QTLs influence R. sativus flower coloration. In this study we have conducted QTL analysis with three seasonal replications data using F2 and F3 populations. In the results, a single robust QTL region was identified in R1 that has shown higher LOD value in all of the replications. However, we also identified a minor QTL in R6. Even though the minor QTL on R6 was detected without replication, we have scavenged this region and identified 316 genes out of which few genes CHS, MYB78, MYB110, 2 F3′5′H5 genes were related to anthocyanin biosynthesis [49,50,51,52]. However, the distribution of the color index varied among seasons, possibly because of the effects of temperature and light on the anthocyanin content, which resulted in small changes to flower color [49]. Additionally, the flowers on F1 plants had purple petals that differed from the parental petals in terms of color, which may reflect the incomplete or partial dominance of the gene responsible for flower coloration. A lack of clear segregation ratios for the flower color phenotypes of the F2 and F2:3 lines suggests many genes might influence the radish flower color trait. The mechanism mediating the inheritance of the flower color trait differs between radish and other Brassicaceae crops. Flower color is a qualitative trait in B. rapa, B. carinata, B. napus, and B. juncea [13,15,16,21,50,51]. This implies that the molecular mechanism underlying trait inheritance is more complex in radish than in other Brassicaceae species.
Transgressive segregation (i.e., the phenotypes of individuals in a segregating hybrid population differ from the phenotypes of the parental lines) was observed for the bolting and flowering time traits in spring and autumn 2018. The trait frequency was normally distributed in the F2 and F2:3 populations. Our results suggest that bolting and flowering time traits are generally influenced by multiple genes, leading to a continuous variation in the phenotypes. Most radishes required 20 days at 5 °C for complete vernalization of seeds at the seeding stage [52,53]. The artificial vernalization at 4 °C from 20 July to 31 August in 2018 resulted in bolting and flowering times that differed from the corresponding times in the spring. Additionally, we observed that bolting time is positively correlated with flowering time in all three environments. Similarly, our genetic analysis indicated the QTLs related to bolting and flowering times are co-localized, which is consistent with the findings of an earlier study by Wang et al. (2018) [34]. Moreover, bolting and flowering times are controlled by QTLs on the same linkage group (R06 and R07). The co-localization of QTLs that govern different traits within the same genetic interval may be due to polygenic inheritance or a pleiotropic effect. A single gene or genetic loci that are closely linked might control multiple traits. They may also be inherited similarly, as suggested by a high positive correlation between the traits. Some of the QTLs (qRFC2, qRFC3, qRFC4, qRFC5, qRBT2, and qRFT2) were inconsistently detected in three experiments (Table 2), likely because of environmental effects. For example, the plants grown in autumn 2018 were vernalized in the cold room (4 °C). The genotype × environment interaction is expected to lead to inconsistencies if the environmental conditions change. However, we were able to identify the genomic regions responsible for specific traits under different environmental conditions because we conducted experiments in different years and seasons.
4.2. Functional Genes Were Detected in the Major QTL Block of the Radish Genome
The qRFC1 was identified as a main locus associated with flower color. Although the current study was unable to identify the structural candidate genes of anthocyanin biosynthesis pathway responsible for flower coloration in the QTL region. However, several regulatory genes of the biosynthetic pathway were detected, on the basis of primer information and the reference genome sequence. In total we detected 269 genes in the major QTLs, 16 of which are related to anthocyanin biosynthesis. Furthermore, the sequences of 9 of these 16 genes varied between the parental lines (Table S3). Six of the genes (Rs018140, Rs018950, Rs019220, Rs020080, Rs020590, and Rs021450) were differentially expressed in the petals of the parental lines. The Rs018950 gene is homologous to the A. thaliana gene AT1G27130, which was annotated with glutathione s-transferase 12. A glutathione S-transferase gene, GhGSTF12, has been reported to be involved for anthocyanin accumulation in cotton, and the expression of this gene represented complement of the defective phenotype of Arabidopsis tt19 mutant [54]. The Rs021450 gene is a homolog of the A. thaliana gene AT1G27470 which is annotated as WD-40 repeat family protein. In Camellia sinensis, a WD40 repeat protein functions as part of the MYB-bHLH-WD40 ternary complexes to control anthocyanin and proanthocyanidin accumulation [55]. The Rs019220 gene is a homolog of the A. thaliana gene, Acyl-CoA N-acyltransferases (NAT) superfamily protein (AT1G24040), which has a role in anthocyanin modifications [56].
The major QTLs for bolting and flowering times were co-localized on chromosome R06 of the radish genome (Table 2 and Figure 4). Our data analysis revealed a positive correlation between bolting and flowering times. Using the radish genome database, we narrowed down the candidate gene region in the QTLs according to gene functions. On the basis of the R. sativus reference genome, we detected 11 genes possibly related to bolting and flowering times (Table S4) [57,58,59,60,61,62,63,64,65,66,67]. An alignment of the genes with the sequences in the parental lines revealed variations in Rs314940, Rs315000, Rs315310, and Rs315960 as well as in the encoded amino acid sequences. The Rs314940 gene is a homolog of the A. thaliana gene AT5G48890 (C2H2-like zinc finger protein), which is involved in molecular mechanisms regulating leaf vascular bundles and the shoot apical meristem under long-day conditions (i.e., it functions as a floral repressor) [57]. The Rs315000 gene is a homolog of the A. thaliana gene AT5G24860 (flowering promoting factor 1), which regulates flowering and modulates gibberellin signal transduction [58]. The Rs315310 gene is homologous to AT3G30260, which encodes a protein (AGAMOUS-like 79) that delays flowering in A. thaliana [59]. The Rs315960 gene is homologous to AT3G30180, which encodes a cytochrome P450 involved in the production of a brassinolide that inhibits flowering by activating the floral repressor FLOWERING LOCUS C [60]. Accordingly, we propose that these four radish genes likely affect bolting and flowering times (Figure S1). Combining the QTL analysis in this study with transcriptome sequencing and fine-mapping approaches will be useful for precisely identifying the radish genes associated with flower color, bolting time, and flowering time as well as for developing tightly linked molecular markers.
5. Conclusions
In this study, we determined that the diversity in radish flower colors might be mainly the result of differences in cyanidin contents. Additionally, a QTL analysis detected nine QTLs associated with flower-related traits, including three important QTLs, qRFC1, qRBT2, and qRFT2, on R. sativus chromosomes 01 and 06 that respectively explained 34.42%, 18.54%, and 18.16% of the phenotypic variation. Thus, qRFC1, qRBT2, and qRFT2 are likely the major QTLs responsible for radish flower color, bolting time, and flowering time, respectively. A qRT-PCR analysis indicated Rs018140, Rs018950, Rs019220, Rs020080, Rs020590, and Rs021450 expression levels are closely related to radish flower coloration. Furthermore, on the basis of a sequence variation analysis, we speculate that Rs314940, Rs315000, Rs315310, and Rs315960 may have important functions related to radish bolting and flowering times. This study revealed the genomic regions and genetic complexity associated with the radish flower color, bolting time, and flowering time traits. The data presented herein provide the foundation for future molecular and functional analyses, as well as for the development of molecular markers for the marker-assisted selection of cultivars exhibiting bolting resistance in the spring.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agronomy11081623/s1, Figure S1: QTLs identified for radish flower color trait. Flower color trait was evaluated in 2017 and 2018 at Chungnam National University with the parental lines and 180 F2 and F2:3 population. The red and green color lines represented the QTL results analyzed using data evaluated in spring and autumn 2018. The blue color line means the QTL results based on the data investigated in spring 2017. Figure S2: QTLs identified for radish flowering and bolting times traits. Flowering and bolting time traits were evaluated in 2017 and 2018 at Chungnam National University with the parental lines and 180 F2 and F2:3 population. The red and blue color lines represented the bolting time related QTL results analyzed using data evaluated in spring and autumn of 2018 using F2:3 population. The light green color line represented the bolting time related QTL results analyzed using data evaluated in spring 2017 using F2 population. The grey and yellow color lines represented the flowering time related QTL results analyzed using data evaluated in spring and autumn of 2018 using F2:3 population. The dark blue color line represented the bolting time related QTL results analyzed using data evaluated in spring 2017 using F2 population. Figure S3: Sequence variations in four candidate genes between ‘YR4’ and ‘YR18’. Table S1: Pearson correlation coefficients for the bolting and flowering times in two populations. Table S2: qRT-PCR primer sequence information for the validation of potential candidate genes expression level for flower color trait. Table S3: List of potential candidate genes in major QTL region related to radish flower color. Table S4: List of potential candidate genes in the major QTL related to radish flowering and bolting times [57,58,59,60,61,62,63,64,65,66,67].
Author Contributions
S.R.C. and Y.P.L. designed the experiment. Y.M. carried out the experiment, analyzed all data, and drafted the manuscript. S.S.C. participated in data analysis and modification of the manuscript. J.J.R. participated in the candidate gene identification. S.K. generated and provided plant materials, and line characteristic information. Y.M., S.R.C., and T.H.G. prepared the samples for anthocyanin analysis, and phenotypic investigation. S.R.C. and Y.P.L. conceived and designed the study, participated as a director, and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries through the Golden Seed Project, which is funded by the Ministry of Agriculture, Food and Rural Affairs (Grant numbers, 213006-05-5-SBO20 and 213006-05-5-SB110).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated in this study are included in this published article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| QTL | Quantitative trait locus |
| LG | Linkage group |
| SNP | Single nucleotide polymorphism |
| InDel | Insertion and deletion |
| HPLC | High performance liquid chromatography |
| MYB | Myeloblastosis |
| bHLH | Basic helix-loop-helix |
| LOD | Logarithm of odds |
| CIM | Composite interval mapping |
| ICIM | Inclusive composite interval mapping |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
References
- Proctor, M.; Yeo, P. The Pollination of Flowers; HarperCollins: Britain, UK, 1973. [Google Scholar]
- Conner, J.K. The Natural History of Pollination. Ecology 1997, 78, 327–329. [Google Scholar] [CrossRef]
- Irwin, R.E.; Strauss, S.Y.; Storz, S.; Emerson, A.; Guibert, G. The role of herbivores in the maintenance of a flower color polymorphism in wild radish. Ecology 2003, 84, 1733–1743. [Google Scholar] [CrossRef]
- Caruso, C.M.; Scott, S.L.; Wray, J.C.; Walsh, C.A. Pollinators, herbivores, and the maintenance of flower color variation: A case study with Lobelia siphilitica. Int. J. Plant Sci. 2010, 171, 1020–1028. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Kay, Q. Preferential pollination of yellow-flowered morphs of Raphanus raphanistrum by Pieris and Eristalis spp. Nature 1976, 261, 230. [Google Scholar] [CrossRef]
- Stanton, M.L. Reproductive biology of petal color variants in wild populations of Raphanus sativus: I. Pollinator response to color morphs. Am. J. Bot. 1987, 74, 178–187. [Google Scholar] [CrossRef]
- Irwin, R.E.; Strauss, S.Y. Flower color microevolution in wild radish: Evolutionary response to pollinator-mediated selection. Am. Nat. 2004, 165, 225–237. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.-C.; Byun, D.H.; Lee, D.Y.; Park, J.Y.; Lee, J.H.; Lee, H.O.; Sung, S.H.; Yang, T.-J. Association of molecular markers derived from the BrCRISTO1 gene with prolycopene-enriched orange-colored leaves in Brassica rapa. Theor. Appl. Genet. 2014, 127, 179–191. [Google Scholar] [CrossRef]
- Rahman, M. Inheritance of petal colour and its independent segregation from seed colour in Brassica rapa. Plant Breed. 2001, 120, 197–200. [Google Scholar] [CrossRef]
- Zhang, N.; Chen, L.; Ma, S.; Wang, R.; He, Q.; Tian, M.; Zhang, L. Fine mapping and candidate gene analysis of the white flower gene Brwf in Chinese cabbage (Brassica rapa L.). Sci. Rep. 2020, 10, 6080. [Google Scholar] [CrossRef]
- Jambhulkar, S.; Raut, R. Inheritance of flower colour and leaf waxiness in Brassica carinata A. Br. Crucif. Newsl 1995, 17, 66–67. [Google Scholar]
- Liu, X.-P.; Tu, J.-X.; Chen, B.-Y.; Fu, T.-D. Identification of the linkage relationship between the flower colour and the content of erucic acid in the resynthesized Brassica napus L. Acta Genet. Sin. 2004, 31, 357–362. [Google Scholar]
- Huang, T.; Böhlenius, H.; Eriksson, S.; Parcy, F.; Nilsson, O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 2005, 309, 1694–1696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, C.; Wang, Y.; Yao, X.; Wang, F.; Wu, J.; King, G.J.; Liu, K. Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 2015, 206, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, R.; Chen, L.; Niu, S.; Li, Q.; Xu, K.; Wen, J.; Yi, B.; Ma, C.; Tu, J. Inheritance and gene mapping of the white flower trait in Brassica juncea. Mol. Breed. 2018, 38, 20. [Google Scholar] [CrossRef]
- Singh, K.; Chauhan, J. Genetics of flower colour in Indian mustard (Brassica juncea L. Czern & Coss). Indian J. Genet. Plant Breed. 2011, 71, 377–378. [Google Scholar]
- Han, F.-q.; Yang, C.; Fang, Z.-y.; Yang, L.-m.; Zhuang, M.; Lv, H.-h.; Liu, Y.-m.; Li, Z.-s.; Liu, B.; Yu, H.-l. Inheritance and InDel markers closely linked to petal color gene (cpc-1) in Brassica oleracea. Mol. Breed. 2015, 35, 1–8. [Google Scholar] [CrossRef]
- Han, F.; Cui, H.; Zhang, B.; Liu, X.; Yang, L.; Zhuang, M.; Lv, H.; Li, Z.; Wang, Y.; Fang, Z. Map-based cloning and characterization of BoCCD4, a gene responsible for white/yellow petal color in B. oleracea. BMC Genom. 2019, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Luo, W.; Guo, J.; Chen, H.; Akram, W.; Xie, D.; Li, G. Fine mapping and candidate gene analysis of the yellow petal gene c kpc in Chinese kale (Brassica oleracea L. var. alboglabra Bailey) by whole-genome resequencing. Mol. Breed. 2019, 39, 1–11. [Google Scholar] [CrossRef]
- Yan, C.; Huang, Y.; Liu, Z.; Guo, F.; Jiao, Z.; Yang, W.; Zhu, F.; Qiu, Z. Rapid identification of yellow-flowered gene Bofc in cauliflower (Brassica oleracea var. botrytis) by bulked segregant analysis and whole-genome resequencing. Euphytica 2020, 216. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550–550. e522. [Google Scholar] [CrossRef] [PubMed]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arab. Thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Quijada, P.; Sung, S.-B.; Lukens, L.; Amasino, R.; Osborn, T.C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 2002, 162, 1457–1468. [Google Scholar] [CrossRef]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Stoutjesdijk, P.; Dennis, E.S.; Peacock, W.J. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 2001, 28, 545–553. [Google Scholar] [CrossRef]
- Zou, X.; Suppanz, I.; Raman, H.; Hou, J.; Wang, J.; Long, Y.; Jung, C.; Meng, J. Comparative analysis of FLC homologues in Brassicaceae provides insight into their role in the evolution of oilseed rape. PLoS ONE 2012, 7, e45751. [Google Scholar] [CrossRef]
- Okazaki, K.; Sakamoto, K.; Kikuchi, R.; Saito, A.; Togashi, E.; Kuginuki, Y.; Matsumoto, S.; Hirai, M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 2007, 114, 595–608. [Google Scholar] [CrossRef]
- Yi, G.; Park, H.; Kim, J.-S.; Chae, W.B.; Park, S.; Huh, J.H. Identification of three FLOWERING LOCUS C genes responsible for vernalization response in radish (Raphanus sativus L.). Hortic. Environ. Biotechnol. 2014, 55, 548–556. [Google Scholar] [CrossRef]
- Li, G.; Zhang, G.; Zhang, Y.; Liu, K.; Li, T.; Chen, H. Identification of quantitative trait loci for bolting and flowering times in Chinese kale (Brassica oleracea var. alboglabra) based on SSR and SRAP markers. J. Hortic. Sci. Biotechnol. 2015, 90, 728–737. [Google Scholar] [CrossRef]
- Li, B.; Zhao, W.; Li, D.; Chao, H.; Zhao, X.; Ta, N.; Li, Y.; Guan, Z.; Guo, L.; Zhang, L. Genetic dissection of the mechanism of flowering time based on an environmentally stable and specific QTL in Brassica napus. Plant Sci. 2018, 277, 296–310. [Google Scholar] [CrossRef]
- Jian, H.; Zhang, A.; Ma, J.; Wang, T.; Yang, B.; Shuang, L.S.; Liu, M.; Li, J.; Xu, X.; Paterson, A.H. Joint QTL mapping and transcriptome sequencing analysis reveal candidate flowering time genes in Brassica napus L. Bmc Genom. 2019, 20, 21. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, B.; Ma, N.; Liu, X.; Qin, M.; Zhang, Y.; Wang, K.; Guo, N.; Zuo, K.; Liu, X.; et al. Quantitative Trait Locus Mapping and Identification of Candidate Genes Controlling Flowering Time in Brassica napus L. Front Plant Sci 2020, 11, 626205. [Google Scholar] [CrossRef] [PubMed]
- Nishio, T.; Kitashiba, H. The Radish Genome; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Wang, Q.; Zhang, Y.; Zhang, L. A naturally occurring insertion in the RsFLC2 gene associated with late-bolting trait in radish (Raphanus sativus L.). Mol. Breed. 2018, 38, 137. [Google Scholar] [CrossRef]
- Nie, S.; Xu, L.; Wang, Y.; Huang, D.; Muleke, E.M.; Sun, X.; Wang, R.; Xie, Y.; Gong, Y.; Liu, L. Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.). Sci. Rep. 2015, 5, 14034. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Xu, L.; Nie, S.; Chen, Y.; Liang, D.; Sun, X.; Karanja, B.K.; Luo, X.; Liu, L. Genome-wide characterization of the MADS-box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Li, C.; Xu, L.; Wang, Y.; Huang, D.; Muleke, E.M.; Sun, X.; Xie, Y.; Liu, L. De novo transcriptome analysis in radish (Raphanus sativus L.) and identification of critical genes involved in bolting and flowering. BMC Genom. 2016, 17, 389. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, S.; Xu, W.; Liu, X. Genome-wide transcriptome profiling of radish (Raphanus sativus L.) in response to vernalization. PLoS ONE 2017, 12, e0177594. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Yeo, H.J.; Kim, N.S.; Eun, P.Y.; Kim, S.-J.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.-Y.; Kim, J.K.; Park, S.U. Metabolic profiling of pale green and purple kohlrabi (Brassica oleracea var. gongylodes). Appl. Biol. Chem. 2017, 60, 249–257. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Choi, S.R.; Chhapekar, S.S.; Kim, M.-S.; Singh, S.; Yi, S.Y.; Oh, S.H.; Kim, H.; Lee, C.Y.; Oh, M.-H. Red Chinese cabbage transcriptome analysis reveals structural genes and multiple transcription factors regulating reddish purple color. Int. J. Mol. Sci. 2020, 21, 2901. [Google Scholar] [CrossRef]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Basten, C.; Zeng, Z. Windows QTL Cartographer 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2012. [Google Scholar]
- Ma, Y.; Chhapekar, S.S.; Lu, L.; Yu, X.; Kim, S.; Choi, G.J.; Lim, Y.P.; Choi, S.R. QTL mapping for Fusarium wilt resistance based on the whole-genome resequencing and their association with functional genes in Raphanus sativus. Theor. Appl. Genet. 2021, 1–16. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, M.; Wang, X.; Tong, C.; Huang, S.; Tehrim, S.; Liu, Y.; Hua, W.; Liu, S. Bolbase: A comprehensive genomics database for Brassica oleracea. BMC Genom. 2013, 14, 664. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Clegg, M.T. Influence of flower color polymorphism on genetic transmission in a natural population of the common morning glory, Ipomoea purpurea. Evolution 1984, 38, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.; Aida, R.; Kishimoto, S.; Ishiguro, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Ohmiya, A. Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol. 2013, 54, 1684–1695. [Google Scholar] [CrossRef]
- Hill, W.G. Understanding and using quantitative genetic variation. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Bunger, L. Inferences on the genetics of quantitative traits from long-term selection in laboratory and domestic animals. Plant Breed. Rev. 2004, 24, 169–210. [Google Scholar]
- Nozaki, K.; Takamura, T.; Fukai, S. Effects of high temperature on flower colour and anthocyanin content in pink flower genotypes of greenhouse chrysanthemum (Chrysanthemum morifolium Ramat.). J. Hortic. Sci. Biotechnol. 2006, 81, 728–734. [Google Scholar] [CrossRef]
- Huang, Z.; Ban, Y.; Bao, R.; Zhang, X.; Xu, A.; Ding, J. Inheritance and gene mapping of the white flower in Brassica napus L. New Zealand J. Crop Hortic. Sci. 2014, 42, 111–117. [Google Scholar] [CrossRef]
- Jia, L.; Wang, J.; Wang, R.; Duan, M.; Qiao, C.; Chen, X.; Ma, G.; Zhou, X.; Zhu, M.; Jing, F.; et al. Comparative transcriptomic and metabolomic analyses of carotenoid biosynthesis reveal the basis of white petal color in Brassica napus. Planta 2021, 253, 8. [Google Scholar] [CrossRef]
- SAGWANSUPYAKORN, C.; SHINOHARA, Y.; SuzuKI, Y. Effects of light intensity and temperature on devernalization of Japanese radish. J. Jpn. Soc. Hortic. Sci. 1986, 55, 56–61. [Google Scholar] [CrossRef][Green Version]
- KAYMAK, H.Ç.; GÜVENÇ, İ. The influence of vernalization time and day length on flower induction of radish (Raphanus sativus L.) under controlled and field conditions. Turk. J. Agric. For. 2010, 34, 401–413. [Google Scholar]
- Shao, D.; Li, Y.; Zhu, Q.; Zhang, X.; Liu, F.; Xue, F.; Sun, J. GhGSTF12, a glutathione S-transferase gene, is essential for anthocyanin accumulation in cotton (Gossypium hirsutum L.). Plant Sci. 2021, 305, 110827. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hou, H.; Jiang, X.; Wang, P.; Dai, X.; Chen, W.; Gao, L.; Xia, T. A WD40 repeat protein from Camellia sinensis regulates anthocyanin and Proanthocyanidin accumulation through the formation of MYB–bHLH–WD40 ternary complexes. Int. J. Mol. Sci. 2018, 19, 1686. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Nishizaki, Y.; Ozeki, Y.; Miyahara, T. The role of acyl-glucose in anthocyanin modifications. Molecules 2014, 19, 18747–18766. [Google Scholar] [CrossRef] [PubMed]
- Weingartner, M.; Subert, C.; Sauer, N. LATE, a C2H2 zinc-finger protein that acts as floral repressor. Plant J. 2011, 68, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Melzer, S. An α-crystallin gene, ACD31. 2 from Arabidopsis is negatively regulated by FPF1 overexpression, floral induction, gibberellins, and long days. J. Exp. Bot. 2004, 55, 1433–1435. [Google Scholar] [CrossRef]
- Gao, R.; Wang, Y.; Gruber, M.Y.; Hannoufa, A. miR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front. Plant Sci. 2018, 8, 2226. [Google Scholar] [CrossRef]
- Li, Z.; Ou, Y.; Zhang, Z.; Li, J.; He, Y. Brassinosteroid signaling recruits histone 3 lysine-27 demethylation activity to FLOWERING LOCUS C chromatin to inhibit the floral transition in Arabidopsis. Mol. Plant 2018, 11, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Staiger, D. SRR1 is essential to repress flowering in non-inductive conditions in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5811–5822. [Google Scholar] [CrossRef][Green Version]
- Niu, L.; Zhang, Y.; Pei, Y.; Liu, C.; Cao, X. Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 2008, 148, 490–503. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Hu, Y.; Yang, Y.; Cai, J.; Liu, H.; Zhang, C.; Liu, X.; Hou, X. Arabidopsis NF-YCs play dual roles in repressing brassinosteroid biosynthesis and signaling during light-regulated hypocotyl elongation. Plant Cell 2021, koab112. [Google Scholar]
- Liu, L.; Zhang, J.; Adrian, J.; Gissot, L.; Coupland, G.; Yu, D.; Turck, F. Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PLoS ONE 2014, 9, e89799. [Google Scholar] [CrossRef] [PubMed]
- Lolas, I.B.; Himanen, K.; Grønlund, J.T.; Lynggaard, C.; Houben, A.; Melzer, M.; Van Lijsebettens, M.; Grasser, K.D. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010, 61, 686–697. [Google Scholar] [CrossRef]
- Lee, J.H.; Jung, J.H.; Park, C.M. INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS C-mediated flowering pathways in Arabidopsis. Plant J. 2015, 84, 29–40. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).