Abstract
This study proposes the machine learning based classification of medical plant leaves. The total six varieties of medicinal plant leaves-based dataset are collected from the Department of Agriculture, The Islamia University of Bahawalpur, Pakistan. These plants are commonly named in English as (herbal) Tulsi, Peppermint, Bael, Lemon balm, Catnip, and Stevia and scientifically named in Latin as Ocimum sanctum, Mentha balsamea, Aegle marmelos, Melissa officinalis, Nepeta cataria, and Stevia rebaudiana, respectively. The multispectral and digital image dataset are collected via a computer vision laboratory setup. For the preprocessing step, we crop the region of the leaf and transform it into a gray level format. Secondly, we perform a seed intensity-based edge/line detection utilizing Sobel filter and draw five regions of observations. A total of 65 fused features dataset is extracted, being a combination of texture, run-length matrix, and multi-spectral features. For the feature optimization process, we employ a chi-square feature selection approach and select 14 optimized features. Finally, five machine learning classifiers named as a multi-layer perceptron, logit-boost, bagging, random forest, and simple logistic are deployed on an optimized medicinal plant leaves dataset, and it is observed that the multi-layer perceptron classifier shows a relatively promising accuracy of 99.01% as compared to the competition. The distinct classification accuracy by the multi-layer perceptron classifier on six medicinal plant leaves are 99.10% for Tulsi, 99.80% for Peppermint, 98.40% for Bael, 99.90% for Lemon balm, 98.40% for Catnip, and 99.20% for Stevia.
1. Introduction
Living things on earth depend on the oxygen produced by plants. There are many different types of plants, all of them playing an important role in maintaining the earth’s biodiversity by providing air and water to living humans [1]. Medicinal plants are plants used in the treatment and prevention of certain diseases and conditions that affect humans [2]. There are many different types of herbal remedies and they can vary from place to place, resulting in a similar pattern of “size” and “shapes” [3]. These plants have excellent medicinal properties from roots to leaves. The leaves of some herbs such as Karpooravalli (Coleus ambonicus), Podina (Mentha arvensis), Neem (Adidirachta indica), Thudhuvalai (Solanum trilobatum), Basil (Ocimum sanctum), etc. [4], are used in our life today. Some leaves have their own medicinal properties such as skin diseases, colds, blood purifier, indigestion [2].
Thus, medical plants are plants used for their particular properties beneficial to human health, even animal health. First called “simple” from the Middle Ages in medieval medicine, today they correspond to products from traditional or modern herbal medicine. The plant is rarely used whole; at least one of its parts (leaf, stem, root, etc.) can be used to heal itself. Different parts of the same plant can have different uses [5]. Plants with medicinal properties can also have food or condiment uses or even be used in the preparation of sanitary drinks. Since Antiquity, the theory of signatures, systematized in the 16th century, has played a major role in the distinction by analogy of plants necessary for human healing, before being extended, contested from the 17th century and completely abandoned by the elite community in the 18th century [6].
According to dissemination data, around the world, 14–28% of plants are listed as having medicinal use. Surveys carried out at the beginning of the 21st century revealed that 3–5% of patients in Western countries, 80% of rural populations in developing countries, and 85% of populations in southern Sahara use medicinal plants as their main treatment [7,8].
Nowadays, the herbal medicine market is filled with counterfeit or low-quality products, affecting human health and sustainable development from around the world. Therefore, it becomes a hot area of research to develop tools aiming to classify herbal medicines. It is now admitted that the leaf of the plant has characteristics that are easy to extract and analyze. Therefore, it is naturally used as the main basis for the identification of all medicinal plants. With the increasing development of image processing, automatic computer image recognition is now widely used in this regard [9,10].
Image processing algorithms are used to identify the leaf images [11,12,13,14,15,16,17,18,19,20]. The background behind this claim is developed below. As a first fact, medicinal plants are difficult to identify because most of them are found in deep forests, and the leaves look the same. If one chooses the wrong herb by mistake, one may have a serious health problem, which can lead to loss of human life. There are many ways to identify a plant. Plants are currently manually identified and subject to human error [11]. To avoid this, several researchers have developed an automated system identification system [12]. Many researchers work on plant leaf disease classification, segmentation, and quality assessment, for instance, Reference [13] proposed a medicinal plant classification framework using the shape and color feature of the leaf. They deployed a Support Vector Machine (SVM) classifier on the optimized features dataset and obtained 96.66% accuracy. Reference [14] proposed a plant recognition system using leaves. They used 50 medicinal images collected from google images and employed edge detection algorithms. The texture patch using Convolution Neural Networks (CNN) approach was deployed for classification and observed a 97.80% accuracy result. Reference [15] proposed a system for the classification of sugarcane leaf disease based on fungi. They used the triangle threshold approach for the segmentation of sugarcane leaves and obtained 98.60% accuracy. Reference [16] proposed a leaf image classification process using CNN. They collect 12,673 samples of the leaf of soybean and design LeNet architecture and obtain a 98.32% classification result. Reference [17] presented a Romanian medicinal plant recognition framework. They used color and gray level (GL) images for his experimentation. The fused approach of Linear Discriminant Analysis (LDA) and Principal Component Analysis (PCA) are deployed and 92.9% accuracy is observed. Reference [18] suggested a novel plant disease approach utilizing a fuzzy logic-based segmentation approach. They extracted fused features that are a combination of color texture and histogram features and deployed various Machine Learning (ML) classifiers. The random forest classifier gives a very promising accuracy that is 98.4%. Reference [19] proposed the Local Binary Patterns (LBP) approach for plant classification using leaves images. They dealt with texture feature extraction, salt and pepper noise removal using ML techniques. Finally, they achieved a 93.5% cumulative classification result. Reference [20] proposed a leaf-based classification of citrus plants. They used digital images and extracted 57 multi-features datasets, then optimized these features into 15 features via the ML approach. For the classification purpose, they employed various classifiers and it has been observed that MLP gives a higher performance which is 98.14% on a region of interest size of (256 × 256).
Contribution
The main aim of this study is to propose a framework for the classification of medicinal plant leaves based on multispectral and texture features using a ML approach. This study contains six steps which are given below:
- Collect multi-spectral and digital image dataset via computer vision laboratory setup.
- Crop exactly leaf region, and transform into the gray level format with (800 × 800) resolution.
- Employ seeds intensity-based edge/line detection utilizing Sobel filter.
- Draw 5 regions of observation on each image and extract fused features from the dataset.
- Optimize fused features dataset using chi-square feature selection approach.
- Apply machine learning based classifiers for observing medicinal plant leaves classification.
2. Materials and Methods
We recall that the medicinal plant leaves were collected from the Department of Agriculture, The Islamia University of Bahawalpur, Pakistan located at 29°23′44″ N and 71°41′1″ E [21]. The foundation of the dataset holds six types of medicinal plants leaves commonly English named as (herbal) Tulsi, Peppermint, Bael, Lemon balm, Catnip, and Stevia and scientifically named as (in Latin) Ocimum sanctum, Mentha balsamea, Aegle marmelos, Melissa officinalis, Nepeta cataria, and Stevia rebaudiana, respectively. These plants are described in Figure 1.

Figure 1.
Sample of the medicinal plants leaves belonging to the dataset.
In the experimentation, we used 50 fresh leaves of medicinal plants for each variety. Firstly, 100 digital images (50 front and 50 back) are taken for each variety. So, the size of digital image dataset of 100 × 6 = 600 colored images of pixel-dimension 1280 × 1024 where size of individual pixel is 0.26 mm was developed to perform further experiments. The multi spectral dataset was collected via a multi-spectral radiometer (MSR5) with 4 feet height. They extract five bands with range of 460 nm to 1560 nm known as “Red” (R), “Blue” (B), “Green” (G), “Near Infrared” (NIR) and “Spectral Bands Shortwave Infrared” (SWIR). In this regard, a total of 1200 samples were acquired, 600 samples of digital image, and 600 samples of multispectral dataset per each type of medicinal plant leaves utilizing computer vision laboratory setup as explained in Figure 2.
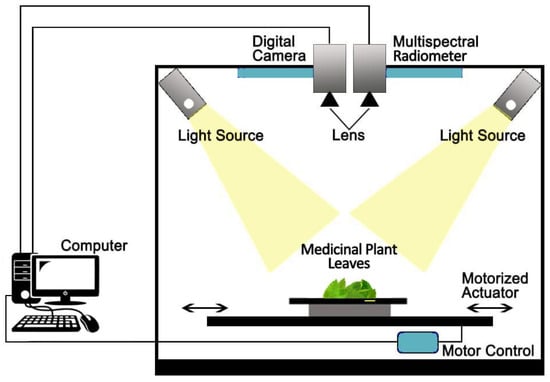
Figure 2.
Computer vision laboratory setup for medicinal plant leaves image acquisition.
The collected dataset is noise free due to the computer vision laboratory setup. For image pre-processing, the medicinal leaves digital image dataset is examined in OpenCV, computer vision library [22]. Also, the Sobel filter was employed for edge/line detection as shown in Figure 3.
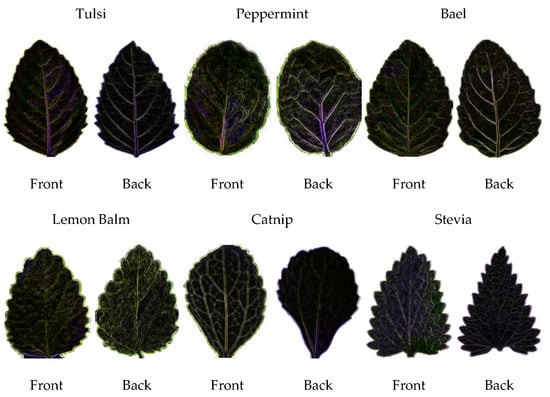
Figure 3.
Sample of Transformation RGB to edge/line detection process.
After that, we crop exactly the leaf region with (800 × 800) resolution, all the digital color images being transformed into the 8-bit gray-level format, and draw five regions of observation (ROO’s) on each sample image. The procedure of taking ROOs is divided into two steps; the first step, we take the size of ROOs is (220 × 220) and, in the second step, we take the size of ROOs is (280 × 280) and finally get the different datasets for experimentations. A total of 1000 (5 × 200) ROO’s have been generated for each medicinal plant leaf as represented in Figure 4. In this manner, a total of 6000 (6 × 1000) ROO’s has been generated on 6 varieties of medical plant leaves.
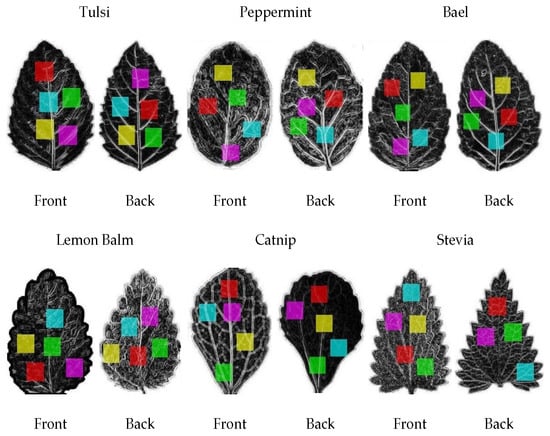
Figure 4.
Sample of transformation into gray level and draw 5 colorful regions of observation on medicinal plant leaves dataset.
2.1. Proposed Methodology
The proposed methodology for the medicinal plant leaves classification is described below. In the first step, all the images acquired in the dataset were examined in OpenCV, computer vision software library [22]. Then, the Sobel filter was employed for edge/line detection. This process is based on the seed intensity (pixel threshold value) of connected pixels of an image; if the threshold value is greater than six then mark the region called a region of observation (ROO’s). The graphical representation of the proposed methodology for the classification of medicinal plant leaves based on fused features using machine learning techniques is described in Figure 5.
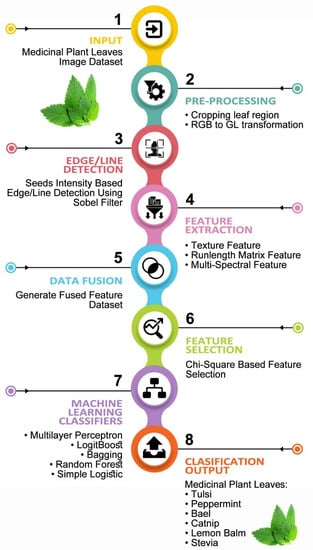
Figure 5.
Proposed framework for the classification of medicinal plant leaves based on multispectral and texture feature using machine learning approach.
2.2. Fused Features Extraction
The OpenCV, computer vision software library [22] was used for the fused feature extraction process that holds texture, spectral, and gray level run length matrix features. A total of 65 fused features were extracted from each ROO’s, which is grouped as 40 texture features, 5 multi-spectral features, and 20 run-length matrix features as described below. The extracted dataset has a large features vector space (FVS) size of 390,000 (6000 × 65) for medicinal plant leaves varieties classification.
2.2.1. Texture Feature
The texture features are based on GL co-occurrence matrix [23,24,25], which is calculated via 4 dimensions (0, 45, 90, 135) degrees and distance between seeds. In this study, we used 5 average features known as energy (), inertia (), entropy (), inverse difference (IDE), and correlation (). First, energy is defined by
where u and v are the spatial coordinates and is gray level values. The correlation is specified by
Also, the formula of the entropy is the following:
The IDE can be defined as
Finally, the inertia is obtained as
2.2.2. Spectral Features
The frequency domain features, known as spectral features, are used in texture analysis. These features are calculated as power of different areas (A) also known as rings [26]. The numerical explanation is given as
where is the frequency domain.
2.2.3. Gray Level Run-Length Matrix (GLRLM)
Galloway [27,28] introduced the Gray Level Run Length Matrix (GLRM), a section of gray also known as run length. It can be described as a linear multitude of continuous pixels with the same gray level in a particular direction. The basics on this approach is recalled below. Let be the number of seeds in the image, be the number of discrete run lengths in the image, be the run length matrix for an arbitrary direction , be the number of runs in the image along angle and be the number of discrete intensity values in the image. Then, the short run emphasis is described as
Long run emphasis is given by
Gray level non-uniformity corresponds to
Run percentage can be defined as
Low gray level run emphasis is described as follows:
High gray level run emphasis is obtained as
Short run low gray level emphasis is defined by
Long run low gray level emphasis is determined as
Finally, run length variance is presented below:
2.3. Feature Selection
The feature selection (FS) process is the most important part of the ML based classification. This process aims to select the most valid and remove the extra features with no importance in the classification process [29]. In this study, we observe that a total of 65 fused features dataset has been extracted from each ROO’s with a large FVS size of 390,000 (6000 × 65) for medicinal plant leaves varieties classification that takes too much time in classification. The feature selection should be to identify the minimum number of columns/features from the data source that are significant in building a model [30]. Our goal in this research is to achieve better accuracy in less time. It is observed that without feature selection, the Multi-Layer Perceptron (MLP) classifier gives 98.81% accuracy results where the size of ROO is 280 × 280, but it takes a lot of time (4.83 Seconds) due to large number features. But when we go with selected features, we obtain higher accuracy in less time. There many ML based features selection approaches such as PCA technique provided excellent results on linearly separated dataset, also used in the selection of features [31]. The PCA method is an unsupervised approach [32], but the medicinal plant leaves varieties dataset is labeled, and the PCA results were not as promising on the labeled data. To solve this problem, ML based supervised feature selection techniques, namely, chi-square attribute evaluator with ranker search method [33] were used to select optimize features from the large FVS. This approach was better compared to PCA and was able to obtain the sub-dataset with the optimal characteristics for this large dataset. The chi-square attribute evaluator with ranker search method is used in ML to rank the independence of two discrete properties [34]. In FS, we specifically check whether the presence of a particular term and the presence of a particular class is independent. Formally, when a document is given to , we estimate the following amounts for each term and rank them according to their scores through the following formula:
where N is the observed frequency and E is the expected frequency, if the document contains the terms i and zero, then the value of is 1 and if the document is in class j and zero, the value of is 1. The chi-square feature selection technique deployed on the medicinal plant leaves dataset reduces the FVS, and gives 14 optimized features with FVS size of 84,000 (6000 × 14) for medicinal plant leaves classification. Figure 6 shows the three-dimensional (3D) representation of the optimized features dataset within six classes using PCA. The MDF1, MDF2, and MDF3 are three different dimensions (like x, y, z) of most discriminant features.
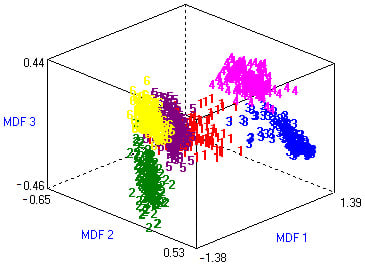
Figure 6.
The 3D visualization of medicinal plant leaves dataset.
The fused optimized features for the classification of medicinal plant leaves dataset are shown in Table 1.

Table 1.
Chi-square based fused optimized feature for the classification medicinal plant leaves.
2.4. Classification
Five machine learning classifiers named as multi-layer perceptron (MLP), LogitBoost (LB), Bagging (B), Random Forest (RF), and Simple Logistic (SL) are deployed on the medicinal plant leaves dataset. It is observed the MLP performed well as compared to the other implemented classifiers [35]. The mathematical foundations of the MLP are given below. The production of input weight and bias are summed using the summation function () defined by
where is the input variable I, k is the number of inputs, is the weight, and is the bias term. The activation functions of MLP is chosen as
The output of neuron j can be obtained as
The medicinal plant leaves classification MLP framework with all regulation parameters are shown in Figure 7. The deployed MLP classifier with all parameters is defined in Table 2.
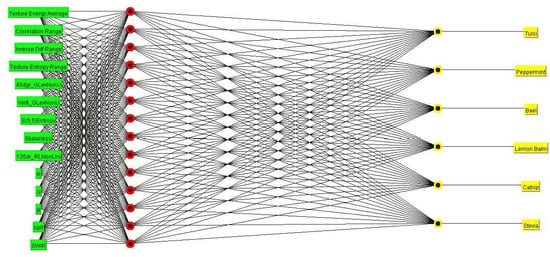
Figure 7.
MLP Based classification of medicinal plant leaves framework.

Table 2.
Deployed MLP classifier parameters.
The deployed MLP classifier with all parameters is defined in Table 2. It depends on threshold point values which are selected manually. In this study, the experiments were performed with different values but, after a large number of tests, these selected values bring complete satisfaction. If we increase or decrease these values, our accuracy will be disturbed.
3. Results and Discussion
Five ML classifiers namely multi-layer perceptron (MLP), LogitBoost (LB), Bagging (B) with REPTree, Random Forest (RF), and Simple Logistic (SL), deployed on fused optimized features dataset for the classification of medicinal plant leaves. The foundation of the dataset holds six types of medicinal plant leaves named as Tulsi, Peppermint, Bael, Lemon Balm, Catnip, and Stevia. The medicinal leaves classification based on fused features is performed using cross-validation (10-fold) data splitting approach. Different testing parameters such as “Receiver Operating Characteristic” (ROC), “Kappa Statistics”, “False Positive (FP), “Recall” (R), “True Positive” (TP), and “F-Measure” is observed [27]. Firstly, an experiment performed on ROO’s size (220 × 220) for the classification of medicinal plant leaves and observed a well-organized accuracy which is 95.87%, 95.04%, 94.21%, 93.38%, and 92.56% using MLP, LB, B, RF, and SL, respectively, as shown in Table 3.

Table 3.
Classification of medicinal plant leaves using five ML classifiers with ROO’s size of (220 × 220).
It is observed that the MLP performs efficiently insisted of other employed classifiers when the size of ROO’s 220 × 220, as shown in Figure 8.
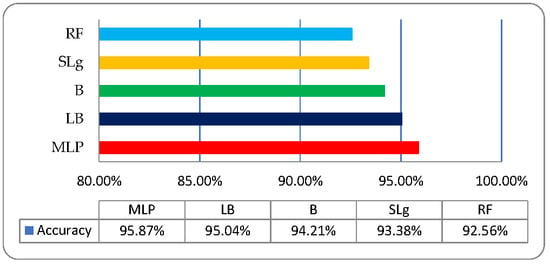
Figure 8.
Medicinal plant leaves classification graphic on ROI’s (220 × 200).
For improvement in classification result, the proposed approach employed on medicinal plant leaves dataset where the size of ROO’s is (280 × 280). We observe very promising results which are 99.01%, 98.01%, 97.02%, 96.03%, and 95.04% using MLP, LB, B, RF, and SL, respectively, as shown in Table 4.

Table 4.
Classification of medicinal plant leaves using five ML classifiers with ROO’s size of (280 × 280).
It is observed that MLP effectively emphasizes the other classifiers when the size of ROO is 280 × 280, as shown in Figure 9.
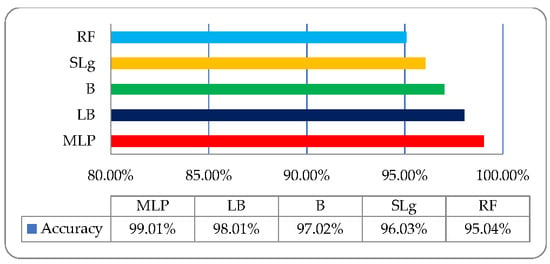
Figure 9.
Medicinal plant leaves classification graphic on ROI’s (280 × 280).
The Confusion Matrix (CM) of MLP classifier on fused medicinal plant leaves dataset using ROO’s size 280 × 280. The diagonal cells contain the number of correctly identified plants and the rest of the cells contain the number of misclassified plants. This CM is presented in Table 5.

Table 5.
CM showing medicinal plant leaves classification on ROO’s size (280 × 280) using MLP.
The distinct classification accuracy of six medicinal plant leaves, named as Tulsi, Peppermint, Bael, Lemon balm, Catnip, and Stevia, were 99.10%, 99.80%, 98.40%, 99.90%, 99.40%, and 99.20%, respectively, on ROO’s size 280 × 280 as shown in Figure 10.
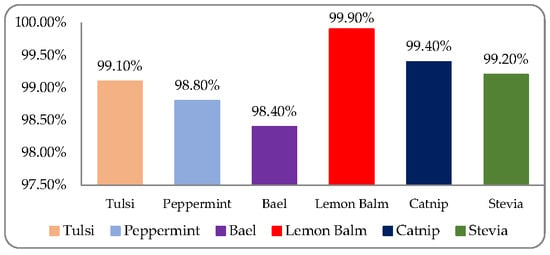
Figure 10.
MLP based classification of six medicinal plant leaf varieties on ROO’s size (250 × 250).
We started our experimentations with the size of ROOs 220 × 220. After that, we gradually increased the size of ROOs to achieve better accuracy. Finally, at the size of ROOs 280 × 280, we observe the promising accuracy because it covers maximum useful information. Further increase in the size of ROOs was causing a decrease in the accuracy due to speckle noise. Lastly, the comparative analysis performs for the classification of medicinal plant leaves with the sizes of ROO’s 220 × 220 and 280 × 280, respectively, as shown in Figure 11.
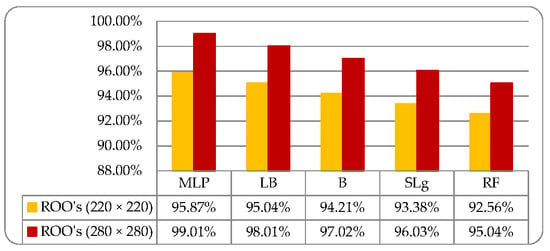
Figure 11.
Comparative analysis for the classification of medicinal plant leaves using ML based classifiers where ROO’s Size (220 × 220) and (280 × 280).
The methodology proposed is comparatively reliable and efficient from that described previously [13,14,16,17,18,19,20]. Furthermore, it is consistent, satisfactory, and better from the existing medicinal plant leaves classification. A comparative analysis of the proposed methodology with existing works is shown in Table 6.

Table 6.
Comparison of proposed methodology with existing methodologies.
4. Conclusions
In this study, we develop a machine learning (ML) based medical plants leaves classification utilizing multispectral and texture dataset. The main objective is to collect a refined and standardized dataset, edge/line detection, fused features extraction, optimized extracted features, and select the most valuable feature and select the efficient ML classifiers. The fused (multispectral + texture) feature dataset holds six types of medicinal leaves named Tulsi, Peppermint, Bael, Lemon Balm, Catnip, and Stevia collected via computer vision laboratory setup. Due to the complex laboratory setup, the collected dataset is very refined and standardized. The chi-square feature selection approach provides the 14 most worthful features that are useful to obtain better classification results. A total of five AI-based classifiers are considered, named as multi-layer perceptron (MLP), LogitBoost (LB), Bagging (B), Random Forest (RF), and Simple Logistic (SL). Firstly, an experiment is performed on ROO’s size (220 × 220) for the classification of medicinal plant leaves dataset. It is obtained a well-organized accuracy which are 95.87%, 95.04%, 94.21%, 93.38%, and 92.56% for MLP, LB, B, RF, and SL, respectively. Secondly, the same approach is employed on a medicinal plant leaves dataset where the size of ROO’s is (280 × 280). We obtain very promising results which are 99.01%, 98.01%, 97.02%, 96.03%, and 95.04% respectively. In addition, we observe that the MLP classifier performed well as compared to other implemented AI-based classifiers. This study opens a new horizon in the field of medicinal plant leaves classification. Also, it can be very helpful for pharmacists to recognize the correct medical plant and will help in the process of making medicine.
Limitation and Future Works
This study is limited to six medicinal plant leaves while there are millions of types of medicinal plant/herbs in the world. This is a pixel-based approach and in the future, we may wish to use an object-based approach. In future this proposed approach deployed on other medicinal plant leaves also proposed approach can be improved using hyper spectral and 3D digital image dataset.
Author Contributions
S.N., Data curation; A.A., Conceptualization, Methodology, Writing—original draft, Software; C.C., Writing—review and editing; M.H.T., Project administration; F.J., Validation, Formal analysis, Writing—review and editing; R.A.K.S., Supervision; M.U.H., Resources, Investigation; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data available on request due to restrictions eg privacy or ethical. The data presented in this study are available on request from the corresponding author. The data are not publicly available due to it is self-generated dataset.
Acknowledgments
The authors thank anonymous referees for careful reading of the manuscript and constructive comments, that significantly improved this paper. Samreen Naeem and Aqib Ali are thankful to their master supervisor, Salman Qadri, Department of Information Technology, The Islamia University of Bahawalpur, Pakistan for his motivational support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nawkar, M.G.; Maibam, P.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. UV-induced cell death in plants. Int. J. Mol. Sci. 2013, 14, 1608–1628. [Google Scholar] [CrossRef] [PubMed]
- Salehi, B.; Kumar, N.V.; Şener, B.; Sharifi-Rad, M.; Kılıç, M.; Mahady, G.B.; Vlaisavljevic, S.; Iriti, M.; Kobarfard, F.; Setzer, W.N.; et al. Medicinal plants used in the treatment of human immunodeficiency virus. Int. J. Mol. Sci. 2018, 19, 1459. [Google Scholar] [CrossRef] [PubMed]
- Nelly, A.; Annick, D.D.; Frederic, D. Plants used as remedies antirheumatic and antineuralgic in the traditional medicine of Lebanon. J. Ethnopharmacol. 2008, 120, 315–334. [Google Scholar]
- Raskin, I.; Ribnicky, D.M.; Komarnytsky, S.; Ilic, N.; Poulev, A.; Borisjuk, N.; Brinker, A.; Moreno, D.A.; Ripoll, C.; Yakoby, N.; et al. Plants and human health in the twenty-first century. Trends Biotechnol. 2002, 20, 522–531. [Google Scholar] [CrossRef]
- Leonti, M. The future is written: Impact of scripts on the cognition, selection, knowledge and transmission of medicinal plant use and its implications for ethnobotany and ethnopharmacology. J. Ethnopharmacol. 2011, 134, 542–555. [Google Scholar] [CrossRef]
- Mamedov, N. Medicinal plants studies: History, challenges and prospective. Med. Aromat. Plants 2012, 1, e133. [Google Scholar] [CrossRef]
- Amenu, E. Use and Management of Medicinal Plants by Indigenous People of Ejaji Area (Chelya Woreda) West Shoa, Ethiopia: An Ethnobotanical Approach. Master’s Thesis, University in Addis, Ababa, Ethiopia, 2007. [Google Scholar]
- Hu, R.; Lin, C.; Xu, W.; Liu, Y.; Long, C. Ethnobotanical study on medicinal plants used by Mulam people in Guangxi, China. J. Ethnobiol. Ethnomed. 2020, 16, 1–50. [Google Scholar] [CrossRef]
- Pferschy-Wenzig, E.M.; Bauer, R. The relevance of pharmacognosy in pharmacological research on herbal medicinal products. Epilepsy Behav. 2015, 52, 344–362. [Google Scholar] [CrossRef]
- Oppong, P.K. The Influence of Packaging and Brand Equity on Over-the-Counter Herbal Medicines in Kumasi, Ghana. Ph.D. Thesis, University of KwaZulu-Natal, Durban, South African, 2018. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, X. Classification and quality evaluation of tobacco leaves based on image processing and fuzzy comprehensive evaluation. Sensors 2011, 11, 2369–2384. [Google Scholar] [CrossRef]
- Rahmatullah, M.; Azam, M.N.; Rahman, M.M.; Seraj, S.; Mahal, M.J.; Mou, S.M.; Nasrin, D.; Khatun, Z.; Islam, F.; Chowdhury, M.H. Chowdhury. A survey of medicinal plants used by Garo and non-Garo traditional medicinal practitioners in two villages of Tangail district, Bangladesh. Am. Eurasian J. Sustain. Agric. 2011, 5, 350–357. [Google Scholar]
- Dahigaonkar, T.D.; Kalyane, R. Identification of ayurvedic medicinal plants by image processing of leaf samples. Int. Res. J. Eng. Technol. (Irjet) 2018, 5, 351–355. [Google Scholar]
- Sabarinathan, C.; Hota, A.; Raj, A.; Dubey, V.K.; Ethirajulu, V. Medicinal plant leaf recognition and show medicinal uses using convolutional neural network. Int. J. Glob. Eng. 2018, 1, 120–127. [Google Scholar]
- Khirade, S.D.; Patil, A.B. Plant disease detection using image processing. In Proceedings of the 2015 International Conference on Computing Communication Control and Automation, Pune, India, 26–27 February 2015; pp. 768–771. [Google Scholar]
- Wallelign, S.; Polceanu, M.; Buche, C. Soybean plant disease identification using convolutional neural network. In Proceedings of the Thirty-First International Flairs Conference, Melbourne, FL, USA, 10 May 2018. [Google Scholar]
- Simion, I.M.; Casoni, D.; Sârbu, C. Classification of Romanian medicinal plant extracts according to the therapeutic effects using thin layer chromatography and robust chemometrics. J. Pharm. Biomed. Anal. 2019, 163, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, G.; Kumar, V.; Joshi, H.D. A novel computer vision based neutrosophic approach for leaf disease identification and classification. Measurement 2019, 135, 782–794. [Google Scholar] [CrossRef]
- Turkoglu, M.; Hanbay, D. Leaf-based plant species recognition based on improved local binary pattern and extreme learning machine. Phys. A Stat. Mech. Appl. 2019, 527, 121297. [Google Scholar] [CrossRef]
- Qadri, S.; Furqan Qadri, S.; Husnain, M.; Saad Missen, M.M.; Khan, D.M.; Muzammil-Ul-Rehman Razzaq, A.; Ullah, S. Machine vision approach for classification of citrus leaves using fused features. Int. J. Food Prop. 2019, 22, 2072–2089. [Google Scholar] [CrossRef]
- The Islamia University of Bahawalpur, Pakistan. 2020. Available online: https://www.iub.edu.pk/faculty-of-agriculture-and-environmental-sciences?1=1forAgriculturalfarms (accessed on 1 October 2020).
- Bradski, G.; Kaehler, A. Learning OpenCV: Computer Vision with the OpenCV Library; O’Reilly Media, Inc.: Newton, MA, USA, 2008. [Google Scholar]
- Galloway, M. Texture analysis using gray level run lengths. Comput. Graph. Image Process 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Naeem, S.; Ali, A.; Qadri, S.; Mashwani, W.K.; Tairan, N.; Shah, H.; Fayaz, M.; Jamal, F.; Chesneau, C.; Anam, S. Machine-Learning Based Hybrid-Feature Analysis for Liver Cancer Classification Using Fused (MR and CT) Images. Appl. Sci. 2020, 10, 3134. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. ManCybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Bantan, R.A.; Ali, A.; Naeem, S.; Jamal, F.; Elgarhy, M.; Chesneau, C. Discrimination of sunflower seeds using multispectral and texture dataset in combination with region selection and supervised classification methods. Chaos Interdiscip. J. Nonlinear Sci. 2020, 30, 113142. [Google Scholar] [CrossRef]
- Ali, A.; Qadri, S.; Khan Mashwani, W.; Kumam, W.; Kumam, P.; Naeem, S.; Goktas, A.; Jamal, F.; Chesneau, C.; Anam, S.; et al. Machine Learning Based Automated Segmentation and Hybrid Feature Analysis for Diabetic Retinopathy Classification Using Fundus Image. Entropy 2020, 22, 567. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Z.; Rehman, M.; Najam, S.; Rizvi, S.M.D. An e_cient gray-level co-occurrence matrix (GLCM) based approach towards classification of skin lesion. In Proceedings of the 2019 Amity International Conference on Artificial Intelligence (AICAI), Dubai, UAE, 4–6 February 2019; pp. 317–320. [Google Scholar]
- Ali, A.; Mashwani, W.K.; Tahir, M.H.; Belhaouari, S.B.; Alrabaiah, H.; Naeem, S.; Nasir, J.A.; Jamal, F.; Chesneau, C. Statistical features analysis and discrimination of maize seeds utilizing machine vision approach. J. Intell. Fuzzy Syst. 2021, 40, 703–714. [Google Scholar] [CrossRef]
- Behura, A. The Cluster Analysis and Feature Selection: Perspective of Machine Learning and Image Processing. Data Anal. Bioinf. A Mach. Learn. Perspect. 2021, 249–280. [Google Scholar] [CrossRef]
- Cardinali, F.; Bracciale, M.P.; Santarelli, M.L.; Marrocchi, A. Principal Component Analysis (PCA) Combined with Naturally Occurring Crystallization Inhibitors: An Integrated Strategy for a more Sustainable Control of Salt Decay in Built Heritage. Heritage 2021, 4, 13. [Google Scholar] [CrossRef]
- Ali, A.; Qadri, S.; Mashwani, W.K.; Brahim Belhaouari, S.; Naeem, S.; Rafique, S.; Jamal, F.; Chesneau, C.; Anam, S. Machine learning approach for the classification of corn seed using hybrid features. Int. J. Food Prop. 2020, 23, 1110–1124. [Google Scholar] [CrossRef]
- Gnanambal, S.; Thangaraj, M.; Meenatchi, V.T.; Gayathri, V. Classification algorithms with attribute selection: An evaluation study using weka. Int. J. Adv. Netw. Appl. 2018, 9, 3640–3644. [Google Scholar]
- McHugh, M.L. The chi-square test of independence. Biochem. Med. 2013, 23, 143–149. [Google Scholar] [CrossRef]
- Ali, A.; Nasir, J.A.; Ahmed, M.M.; Naeem, S.; Anam, S.; Jamal, F.; Chesneau, C.; Zubair, M.; Anees, M.S. Machine Learning Based Statistical Analysis of Emotion Recognition using Facial Expression. RADS J. Biol. Res. Appl. Sci. 2020, 11, 39–46. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).