Molecular Genetic Analysis of Drought Stress Response Traits in Brachypodium spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Seed Treatment and Seedling Development
2.3. Culture Solutions
2.4. Root and Shoot Analysis
2.5. SSR Analysis
2.6. Statistical Analysis
3. Results
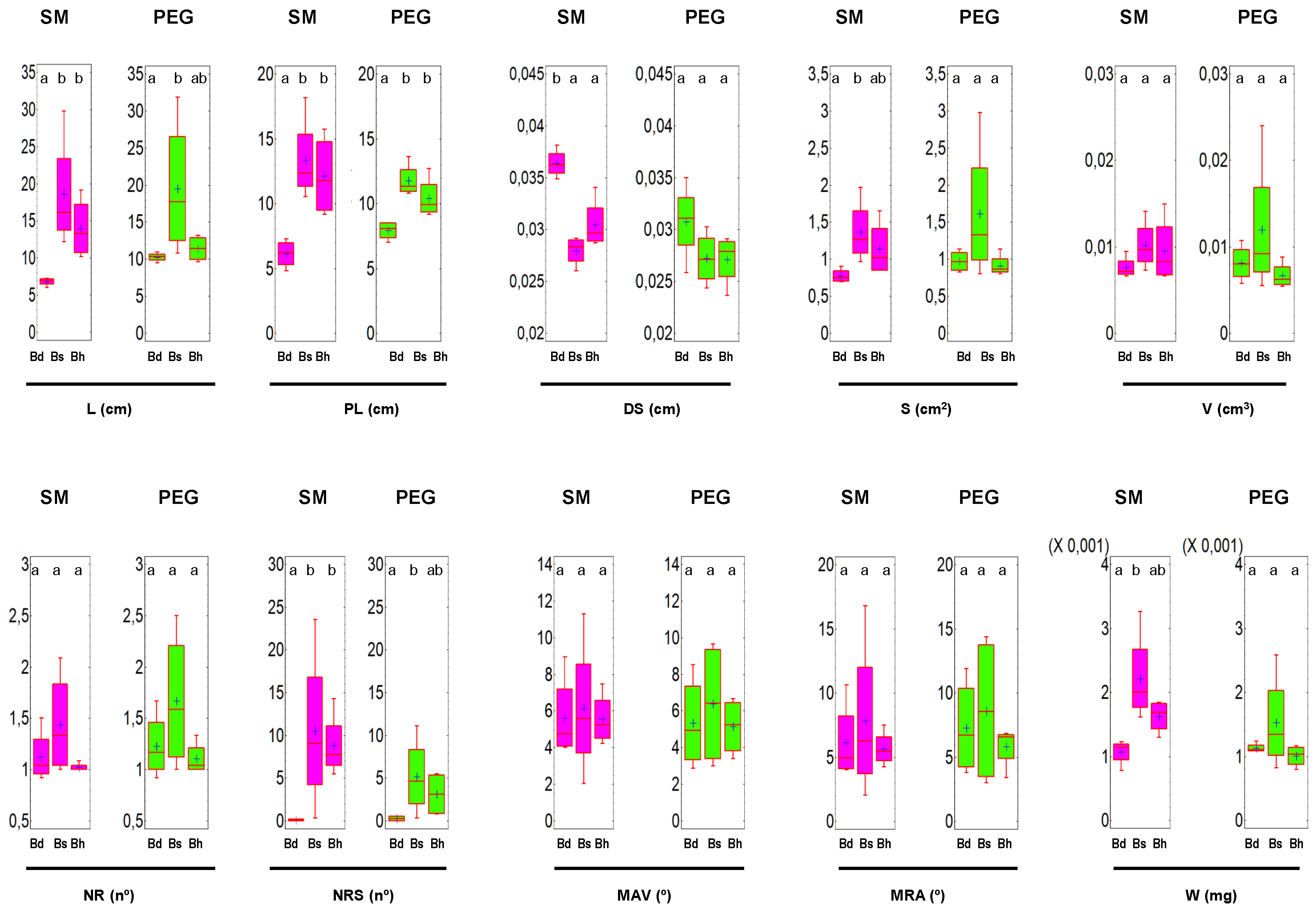
3.1. Analysis of the RSA and W in Brachypodium spp.
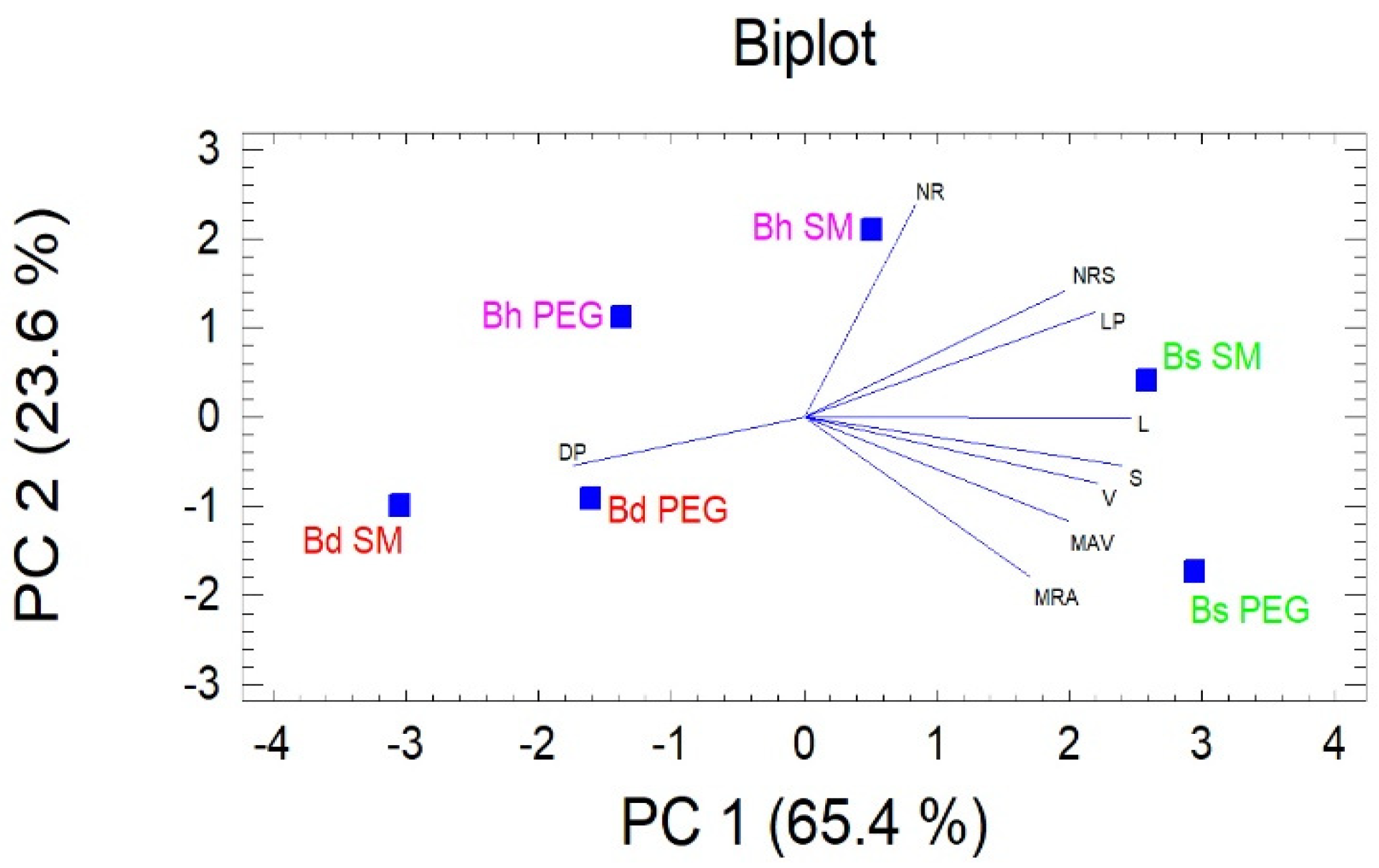
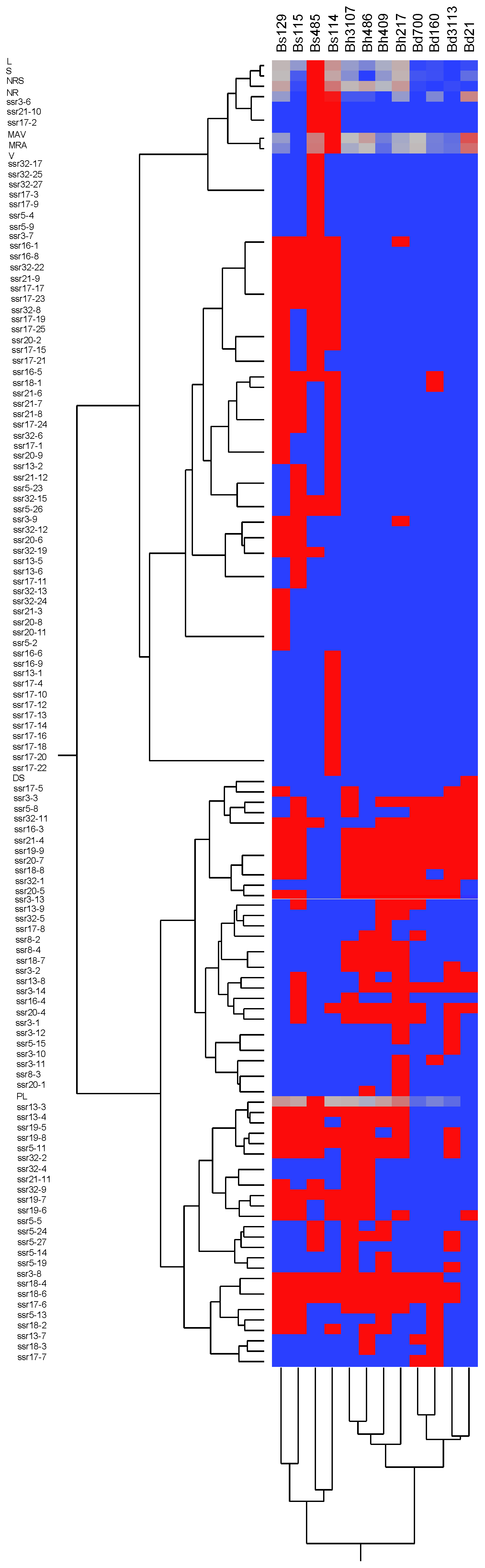
3.2. Analysis of SSR and RSA
4. Discussion
4.1. RSA Traits and Shoot Dry Weight Diversity in Brachypodium spp.
4.2. Drought Influence in RSA Traits and Shoot Dry Weight in Brachypodium spp.
4.3. Association between RSA Traits and SSRs.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Dorlodot, S.; Forster, B.; Pagês, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Brown, K.M. New roots for agriculture: Exploiting the root phenome. Philos. Trans. R. Soc. B 2012, 367, 1598–1604. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; George, T.S.; Dupuy, L.X.; Karley, A.J.; Valentine, T.A.; Wiesel, L.; Wishart, J. Root traits for infertile soils. Front. Plant Sci. 2013, 4, 193. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Root architecture and plant productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef]
- Manschadi, A.M.; Hammer, G.L.; Christopher, J.T.; deVoil, P. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 2008, 303, 115–129. [Google Scholar] [CrossRef]
- Nakhforoosh, A.; Grausgruber, H.; Kaul, H.P.; Bodner, G. Wheat root diversity and root functional characterizacion. Plant Soil 2014, 380, 211–229. [Google Scholar] [CrossRef]
- Narayanan, S.; Mohan, A.; Gill, K.S.; Prasad, P.V.V. Variability of Root Traits in Spring Wheat Germplasm. PLoS ONE 2014, 9, e100317. [Google Scholar] [CrossRef]
- González, J.M.; Friero, E.; Selfa, L.; Froilán, S.; Jouve, N. A comparative study of root system architecture in seedlings of Brachypodium spp. using three plant growth supports. Cereal Res. Commun. 2016, 44, 69–78. [Google Scholar] [CrossRef]
- Ruiz, M.; Giraldo, P.; González, J.M. Phenotypic variation in root architecture traits and their relationship with eco-geographical and agronomic features in a core collection of tetraploid wheat landraces (Triticum turgidum L.). Euphytica 2018, 214, 54. [Google Scholar] [CrossRef]
- Adeleke, E.; Millas, R.; McNeal, W.; Faris, J.; Taheri, A. Variation analysis of root system development in wheat seedlings using root phenotyping system. Agronomy 2020, 10, 206. [Google Scholar] [CrossRef]
- Rich, S.M.; Watt, M. Soil conditions and cereal root system architecture: Review and considerations for linking Darwin and Weaver. J. Exp. Bot. 2013, 64, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; George, T.S.; Gregory, P.J.; Bengough, A.G.; Hallett, P.D.; McKenzie, B.M. Matching roots to their environment. Ann. Bot. 2013, 112, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Wojciechowski, T. Opportunities and challenges in the subsoil: Pathways to deeper rooted crops. J. Exp. Bot. 2015, 66, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Ehdaie, B.; Andrew, P.; Waines, J.G. Root system plasticity to drouht influences grain yield in bread wheat. Euphytica 2012, 186, 219–232. [Google Scholar] [CrossRef]
- Uga, Y.; Kitomi, Y.; Ishikawa, S.; Yano, M. Genetic improvement for root growth angle to enhance crop production. Breed. Sci. 2015, 65, 111–119. [Google Scholar] [CrossRef]
- Tuberosa, R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012, 3, 347. [Google Scholar] [CrossRef]
- Wasson, A.P.; Richards, R.A.; Chatrath, R.; Misra, S.C.; Sai Prasad, S.V.; Rebetzke, G.J.; Kirkegaard, J.A.; Christopher, J.; Watt, M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012, 63, 3485–3498. [Google Scholar] [CrossRef]
- Ali, M.L.; Luetchens, J.; Singh, A.; Shaver, T.M.; Kruger, G.R.; Lorenz, A.J. Greenhouse screening of maize genotypes for deep root mass and related root traits and their association with grain yield under water-deficit conditions in the field. Euphytica 2016, 207, 79–94. [Google Scholar] [CrossRef]
- Tomar, R.S.S.; Tiwari, S.; Vinod Naik, B.K.; Chand, S.; Deshmukh, R.; Mallick, N.; Singh, S.; Singh, N.K.; Tomar, S.M.S. Molecular and Morpho-Agronomical Characterization of Root Architecture at Seedling and Reproductive Stages for Drought Tolerance in Wheat. PLoS ONE 2016, 11, e0156528. [Google Scholar] [CrossRef]
- Ayalew, H.; Liu, H.; Yan, G. Identification and validation of root length QTLs for water stress resistance in hexaploid wheat (Titicum aestivum L.). Euphytica 2017, 213, 126. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Petersony, D.G.; Estill, J.C.; Chapman, B.A. Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 2003, 13, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Draper, J.; Mur, L.A.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P.M. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Hill, T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 2008, 27, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Hus, K.; Rojek-Jelonek, M.; Kurczynska, E.; Nibau, C.; Doonan, J.H.; Hasterok, H. In Vitro Tissue Culture in Brachypodium: Applications and Challenges. Int. J. Mol. Sci. 2020, 21, 1037. [Google Scholar] [CrossRef]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Scholthof, K.B.; Irigoyen, S.; Catalán, P.; Mandadi, K.K. Brachypodium: A monocot grass model genus for plant biology. Plant Cell 2018, 30, 1673–1694. [Google Scholar] [CrossRef] [PubMed]
- Joint Genome Institute. Available online: https://jgi.doe.gov/our-science/science-programs/plant-genomics/brachypodium/ (accessed on 7 February 2019).
- Idiziak, D.; Betekhtin, A.; Wolny, E.; Lesniewska, K.; Wright, J.; Febrer, M.; Bevan, M.W.; Jenkins, G.; Hasterok, R. Painting the chromosomes of Brachypodium-current status and future prospects. Chromosoma 2011, 120, 469–479. [Google Scholar] [CrossRef]
- Filiz, E.; Ozdemir, B.S.; Budak, F.; Vogel, J.P.; Tuna, M.; Budak, H. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 2009, 52, 876–890. [Google Scholar] [CrossRef]
- Vogel, J.P.; Tuna, M.; Budak, H.; Huo, N.; Gu, Y.Q.; Steinwand, M.A. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol. 2009, 9, 88. [Google Scholar] [CrossRef]
- Hammami, R.; Jouve, N.; Cuadrado, A.; Soler, C.; González, J.M. Prolamin storage proteins and alloploidy in wild populations of the small grass Brachypodium distachyon (L.) P. Beauv. Plant Syst. Evol. 2011, 297, 99–111. [Google Scholar] [CrossRef]
- Jaroszewicz, A.M.; Kosina, R.; Stankiewicz, P.R. RAPD, karyology and selected morphological variation in a model grass, Brachypodium distachyon. Weed Res. 2012, 52, 204–216. [Google Scholar] [CrossRef]
- Hammami, R.; Jouve, N.; Soler, C.; Friero, E.; González, J.M. Genetic diversity of SSR and ISSR markers in wild populations of Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). Plant Syst. Evol. 2014, 300, 2029–2040. [Google Scholar] [CrossRef]
- Catalán, P.; Müller, J.; Hasterok, R.; Jenkins, G.; Mur, L.A.J.; Langdon, T.; Betekhtin, A.; Siwinska, D.; Pimentel, M.; López-Alvarez, D. Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 2012, 109, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Catalán, P.; López-Álvarez, D.; Bellosta, C.; Villar, L. Updated taxonomic descriptions, iconography, and habitat preferences of Brachypodium distachyon, B. stacei, and B. hybridum (Poaceae). Anales del Jardin Botánico de Madrid 2016, 73, e028. [Google Scholar] [CrossRef]
- Catalán, P.; Chalhoub, B.; Chochois, V.; Garvin, D.F.; Hasterok, R.; Manzaneda, A.J.; Mur, L.A.J.; Pecchioni, N.; Rasmussen, S.K.; Vogel, J.P.; et al. Update on the genomics and basic biology of Brachypodium. Trends Plant Sci. 2014, 19, 414–418. [Google Scholar] [CrossRef]
- Chochois, V.; Vogel, J.P.; Rebetzke, G.J.; Watt, M. Variation in Adult Plant Phenotypes and Partitioning among Seed and Stem-Borne Roots across Brachypodium distachyon Accessions to Exploit in Breeding Cereals for Well-Watered and Drought Environments. Plant Physiol. 2015, 168, 953–967. [Google Scholar] [CrossRef]
- Le Marié, C.; Kirchgessner, N.; Marschall, D.; Walter, A.; Hund, A. Rhizoslides: Paper-based growth system for non-destructive, high throughput phenotyping of root development by means of image analysis. Plant Methods 2014, 10, 13. [Google Scholar] [CrossRef]
- Aniol, A. Induction of aluminum tolerance in wheat seedlings by low doses of aluminum in the nutrient solution. Plant Physiol. 1984, 75, 551–555. [Google Scholar] [CrossRef]
- Lobet, G.; Pagès, L.; Draye, X. A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 2011, 157, 29–39. [Google Scholar] [CrossRef]
- SAS Institute Inc. SAS/STAT® 13.2 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2014. [Google Scholar]
- Sanguineti, M.; Li, S.; Maccaferri, M.; Corneti, S.; Rotondo, F.; Chiari, T.; Tuberosa, R. Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann. Appl. Biol. 2007, 151, 291–305. [Google Scholar] [CrossRef]
- Moghe, G.D.; Shiu, S.H. The causes and molecular consequences of polyploidy in flowering plants. Ann. N. Y. Acad. Sci. 2014, 1320, 16–34. [Google Scholar] [CrossRef] [PubMed]
- López-Álvarez, D.; Manzaneda, A.J.; Rey, P.J.; Giraldo, P.; Benavente, E.; Allainguillaume, J.; Mur, L.A.J.; Caicedo, A.L.; Hazen, S.P.; Breiman, A.; et al. Environmental niche variation and evolutionary diversification of the Brachypodium distachyon grass complex species in their native circum-Mediterranean range. Am. J. Bot. 2015, 102, 1–16. [Google Scholar] [CrossRef] [PubMed]
- González-Moreno, A.; Casanova, C.; Gascó, A.; Rodríguez, J.A. Brachypodium hybridum Plant Cover Improves Water Infiltration in Mediterranean Crop Soils. J. Plant Chem. Ecophysiol. 2016, 1, 1008. [Google Scholar]
- Ruiz-Colmenero, M.; Bienes, R.; Eldridge, D.J.; Marques, M.J. Vegetation cover reduces erosion and enhances soil organic carbon in a vineyard in the central Spain. CATENA 2012, 104, 153–160. [Google Scholar] [CrossRef]
- Ehdaie, B.; Mohammadi, S.A.; Nouraein, M. QTLs for root traits at mid-tillering and for root and shoot traits at maturity in a RIL population of spring bread wheat grown under well-watered conditions. Euphytica 2016, 211, 17–38. [Google Scholar] [CrossRef]
- Ma, J.; Luo, W.; Zhang, H.; Zhou, X.H.; Qin, N.N.; Wei, Y.M.; Liu, Y.X.; Jiang, Q.T.; Chen, G.Y.; Zheng, Y.L.; et al. Identification of quantitative trait loci for seedling root traits from Tibetan semi-wild wheat (Triticum aestivum subsp. tibetanum). Genome 2017, 60, 1068–1075. [Google Scholar] [CrossRef]
- Uga, Y.; Assaranurak, I.; Kitomi, Y.; Larson, B.G.; Craft, E.J.; Shaff, J.E.; McCouch, S.R.; Kochian, L.V. Genomic regions responsible for seminal and crown root lengths identified by 2D & 3D root system image analysis. BMC Genom. 2018, 19, 273. [Google Scholar]
| Spp. | Genotype | Geographic Origin: Longitude/Latitude | Locality (Spain Province) | Altitude (m.) |
|---|---|---|---|---|
| B. distachyon | Bd21 | Iraq | - | - |
| Bd160 | 38°57′00″/2°31′59″ | Bonillo (Albacete) | 1035 | |
| Bd700 | 40°55′34″/2°55′29″ | Jadraque (Guadalajara) | 701 | |
| Bd3113 | 35°56′00″/2°53′00″ | Segóbriga (Cuenca)lbaceteongitud y latitudic callialnenan im the immature embryos of Brachypodium. in vitro | 830 | |
| = 855.3 | ||||
| B. stacei | Bd114 | 37°59′00″/3°28′00″ | Baeza (Jaén) | 661 |
| Bd115 | 38°40′00″/2°29′00″ | Alcaráz (Albacete) | 949 | |
| Bd129 | 37°20′00″/3°47′00″ | Moclín (Granada) | 1084 | |
| Bd485 | 38°44′18″/0°13′56″ | Cabo de la Nao (Alicante) | 42 | |
| = 684 | ||||
| B. hybridum | Bd217 | 37°27′28″/2°14′57″ | Oria (Almería) | 829 |
| Bd409 | 39°14′56″/1°03′59″ | Cofrentes (Valecia) | 452 | |
| Bd486 | 41°10′40″/−1°28′28″ | Roda de Bará (Tarragona) | 23 | |
| Bd3107 | 37°35′00″/4°35′00″ | Córdoba (Córdoba) | 445 | |
| = 437.2 | ||||
| spp. | Cult. sol. | L | S | V × 10−3 | DS × 10−2 | PL | NR | NRS | MAV | MRA | W × 10−3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bd | SM | 6.81 a | 0.77 a | 7.5 a | 3.6 b | 6.1 a | 1.06 a | 0.16 a | 5.62 a | 6.12 a | 1.1 a |
| PEG | 10.27 b | 0.97 b | 8.1 a | 3.1 a | 7.92 b | 1.10 a | 0.25 a | 5.32 a | 7.28 a | 1.1 a | |
| Bs | SM | 18.54 a | 1.36 a | 10.1 a | 2.8 a | 13.33 a | 1.43 a | 10.5 a | 6.12 a | 7.83 a | 2.2 a |
| PEG | 19.52 a | 1.61 a | 11.99 a | 2.7 a | 11.76 a | 1.66 a | 5.17 a | 6.37 a | 8.61 a | 1.5 a | |
| Bh | SM | 13.95 a | 1.13 a | 9.5 b | 3.04 b | 12.11 a | 1.02 a | 8.78 b | 5.54 a | 5.64 a | 1.6 b |
| PEG | 11.41 a | 0.91 a | 6.7 a | 2.71 a | 10.41 a | 1.10 a | 3.13 a | 5.13 a | 5.83 a | 1.00 a |
| Cult. Sol. | Genot. | L | S | V × 10−3 | DS × 10−2 | PL | NR | NRS | MAV | MRA | W × 10−3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bd | SM | 21 | 6.92 a | 0.71 a | 6.96 a | 3.60 a | 4.79 a | 1.5 b | 0.17 a | 8.96 b | 10.64 b | 1.2 bc |
| 160 | 7.27 a | 0.90 a | 9.45 a | 3.81 a | 7.27 b | 1.00 ab | 0.00 a | 4.01 a | 4.01 a | 1.2 c | ||
| 700 | 6.02 a | 0.69 a | 6.63 a | 3.64 a | 5.76 ab | 1.08 ab | 0 a | 5.42 a | 5.71 a | 0.78 a | ||
| 3113 | 7.02 a | 0.78 a | 7.26 a | 3.48 a | 6.66 ab | 1.08 a | 0.17 a | 4.10 a | 4.14 a | 1.1 b | ||
| PEG | 21 | 10.38 a | 1.13 a | 10.6 b | 3.49 b | 7.02 a | 1.6 b | 0.00 a | 8.53 c | 11.89 b | 1.1 a | |
| 160 | 10.25 a | 0.82 a | 5.72 a | 2.58 a | 8.50 a | 1.25 ab | 0.00 a | 3.75 ab | 4.65 a | 1.1 a | ||
| 700 | 10.96 a | 1.05 a | 8.65 ab | 3.11 ab | 8.48 a | 1.42 a | 0.50 a | 6.12 bc | 8.78 ab | 1.1 a | ||
| 3113 | 9.51 a | 0.88 a | 7.39 ab | 3.11 ab | 7.66 a | 1.58 ab | 0.50 a | 2.88 a | 3.79 a | 1.2 a | ||
| Bs | SM | 114 | 17.00 a | 1.33 a | 10.1 ab | 2.78 a | 10.53 a | 2.08 b | 8.08 b | 11.29 b | 16.78 b | 2.1 ab |
| 115 | 12.17 a | 0.96 a | 7.27 a | 2.91 a | 12.15 a | 1.33 ab | 0.33 a | 2.04 a | 2.04 a | 1.6 a | ||
| 129 | 15.25 a | 1.20 a | 9.26 ab | 2.87 a | 12.49 a | 1.08 a | 10.08 b | 5.35 ab | 5.35 ab | 1.9 ab | ||
| 485 | 29.76 b | 1.97 b | 14.1 b | 2.60 a | 18.16 a | 1.58 ab | 23.50 c | 5.81 ab | 7.16 ab | 3.3 b | ||
| PEG | 114 | 21.22 b | 1.48 c | 9.69 b | 2.43 a | 11.06 a | 1.92 bc | 11.08 b | 9.00 b | 12.16 b | 1.5 b | |
| 115 | 10.80 a | 0.80 a | 5.47 a | 2.61 ab | 10.77 a | 1.33 ab | 0.33 a | 3.00 a | 3.00 a | 0.82 a | ||
| 129 | 14.22 a | 1.17 b | 8.66 b | 2.81 bc | 11.60 a | 1.25 a | 3.67 b | 3.81 a | 3.93 a | 1.2 b | ||
| 485 | 31.82 c | 2.98 d | 24.1 c | 3.02 c | 13.63 b | 2.5 c | 5.58 b | 9.66 b | 14.36 b | 2.6 c | ||
| Bh | SM | 217 | 19.13 c | 1.65 c | 14.9 c | 3.40 b | 15.74 c | 1 a | 14.25 b | 5.66 a | 5.66 a | 1.8 bc |
| 409 | 15.22 b | 1.18 b | 9.65 b | 3.01 a | 13.74 b | 1 a | 7.50 a | 4.22 a | 4.22 a | 1.8 c | ||
| 486 | 11.26 a | 0.85 a | 6.60 a | 2.87 a | 9.81 a | 1 a | 7.92 ab | 7.47 a | 7.47 a | 1.3 a | ||
| 3107 | 10.21 a | 0.85 a | 6.85 a | 2.91 a | 9.15 a | 1.08 a | 5.50 a | 4.81 a | 5.23 a | 1.6 b | ||
| PEG | 217 | 12.63 bc | 0.87 a | 5.40 a | 2.36 a | 10.24 a | 1.33 a | 0.78 a | 4.27 ab | 6.42 a | 0.79 a | |
| 409 | 9.67 a | 0.85 a | 6.53 a | 2.91 b | 9.54 a | 1 a | 1.00 a | 3.38 a | 3.38 a | 0.96 ab | ||
| 486 | 10.16 ab | 0.80 a | 5.89 a | 2.72 b | 9.18 a | 1.08 a | 5.50 b | 6.22 bc | 6.84 a | 1.1 bc | ||
| 3107 | 13.17 c | 1.13 b | 8.80 b | 2.85 b | 12.68 b | 1 a | 5.25 b | 6.67 c | 6.67 a | 1.2 c |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, J.M.; Redondo-Pedraza, J.; Loarce, Y.; Hammami, R.; Friero, E.; Jouve, N. Molecular Genetic Analysis of Drought Stress Response Traits in Brachypodium spp. Agronomy 2020, 10, 518. https://doi.org/10.3390/agronomy10040518
González JM, Redondo-Pedraza J, Loarce Y, Hammami R, Friero E, Jouve N. Molecular Genetic Analysis of Drought Stress Response Traits in Brachypodium spp. Agronomy. 2020; 10(4):518. https://doi.org/10.3390/agronomy10040518
Chicago/Turabian StyleGonzález, Juan M., Jaime Redondo-Pedraza, Yolanda Loarce, Rifka Hammami, Eva Friero, and Nicolás Jouve. 2020. "Molecular Genetic Analysis of Drought Stress Response Traits in Brachypodium spp." Agronomy 10, no. 4: 518. https://doi.org/10.3390/agronomy10040518
APA StyleGonzález, J. M., Redondo-Pedraza, J., Loarce, Y., Hammami, R., Friero, E., & Jouve, N. (2020). Molecular Genetic Analysis of Drought Stress Response Traits in Brachypodium spp. Agronomy, 10(4), 518. https://doi.org/10.3390/agronomy10040518





