Structural Characterization and Molecular Docking Screening of Most Potent 1,2,4-Triazine Sulfonamide Derivatives as Anti-Cancer Agents
Abstract
1. Introduction
2. Materials and Methods
2.1. DFT Method
2.2. Molecular Docking through Maestro Schrödinger
3. Results
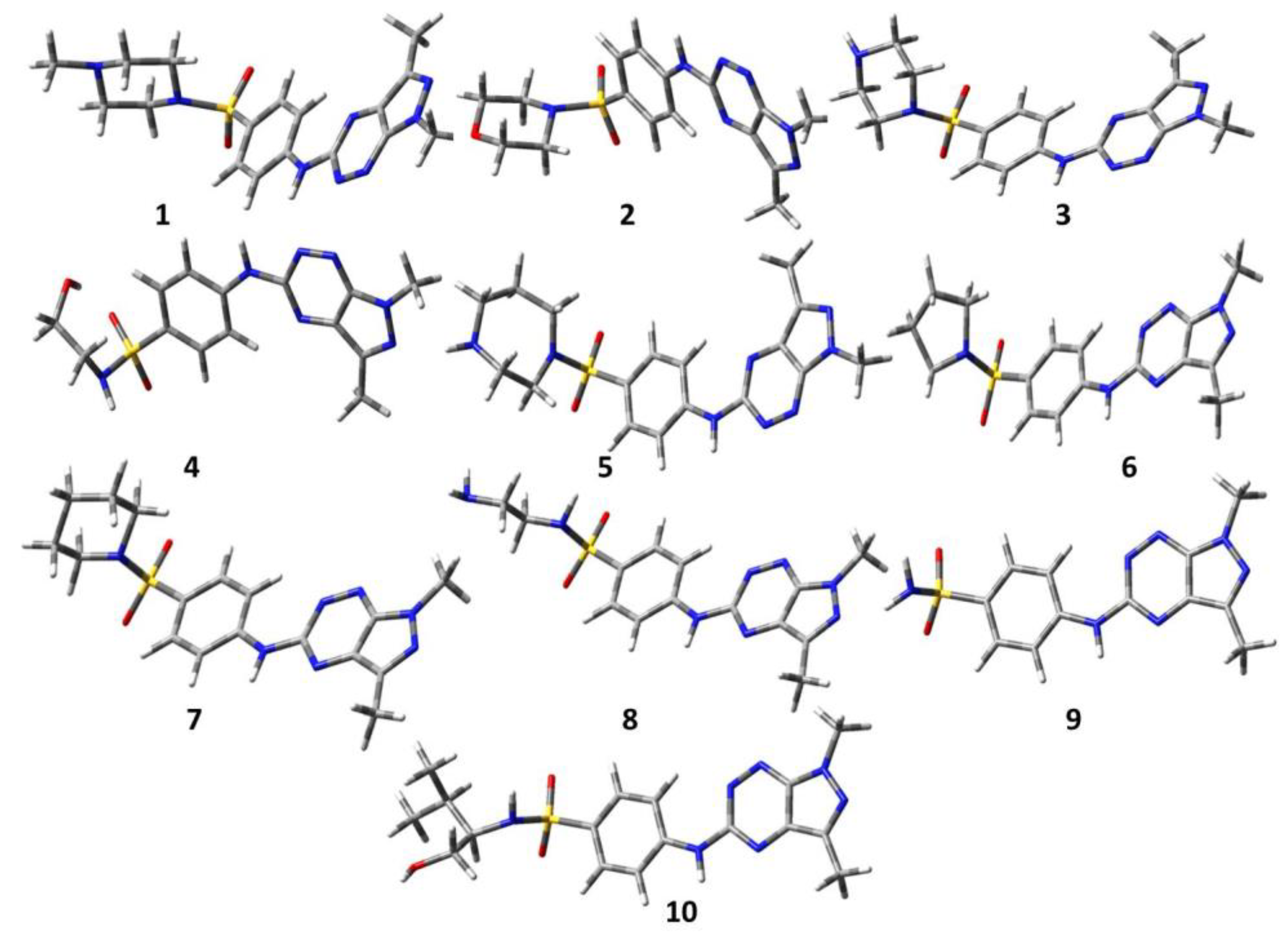
3.1. Optimization through Gaussian View
3.2. Descriptors of Global Reactivity
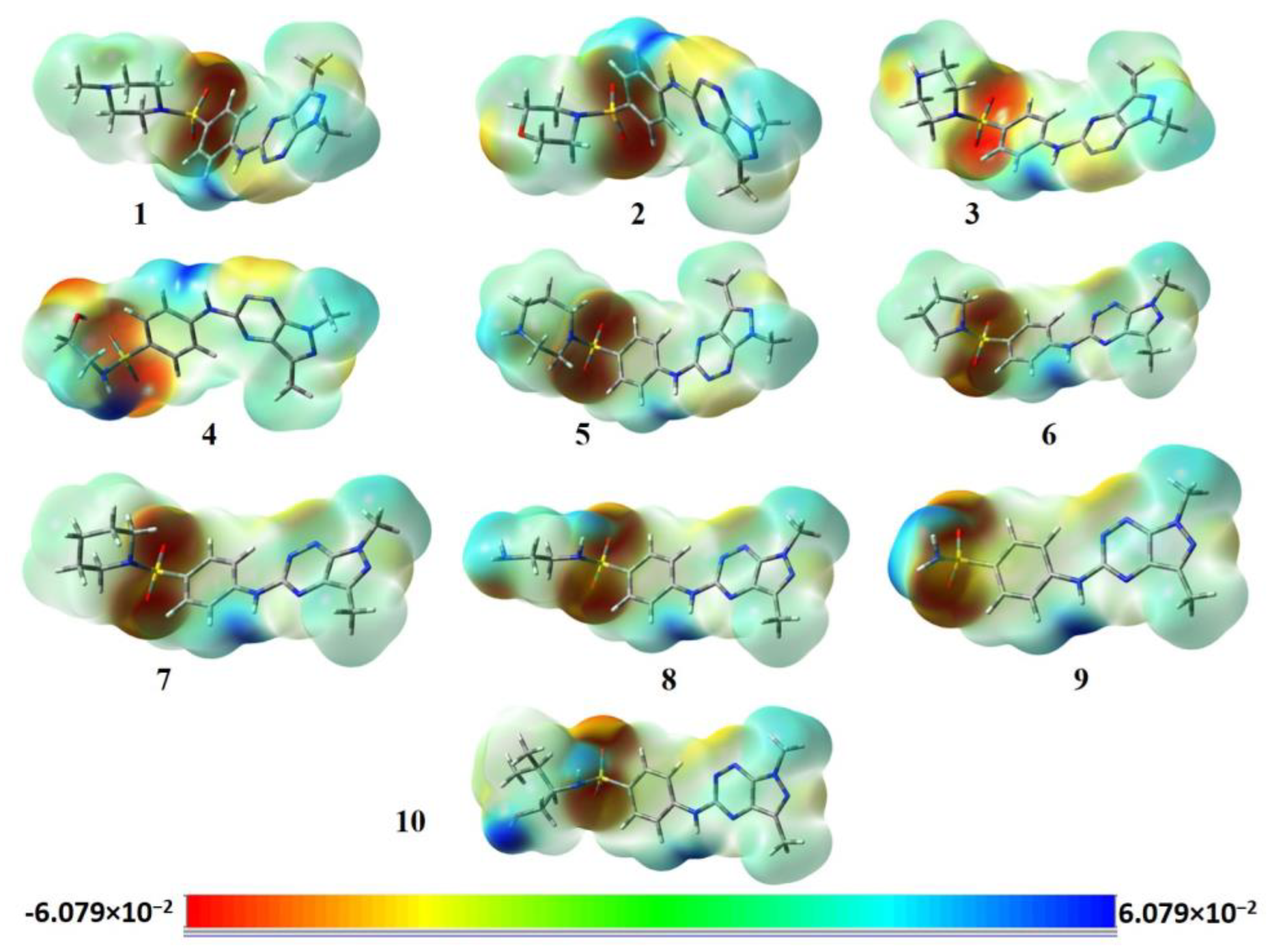
3.3. Molecular Electrostatic Potential (MEP) Analysis
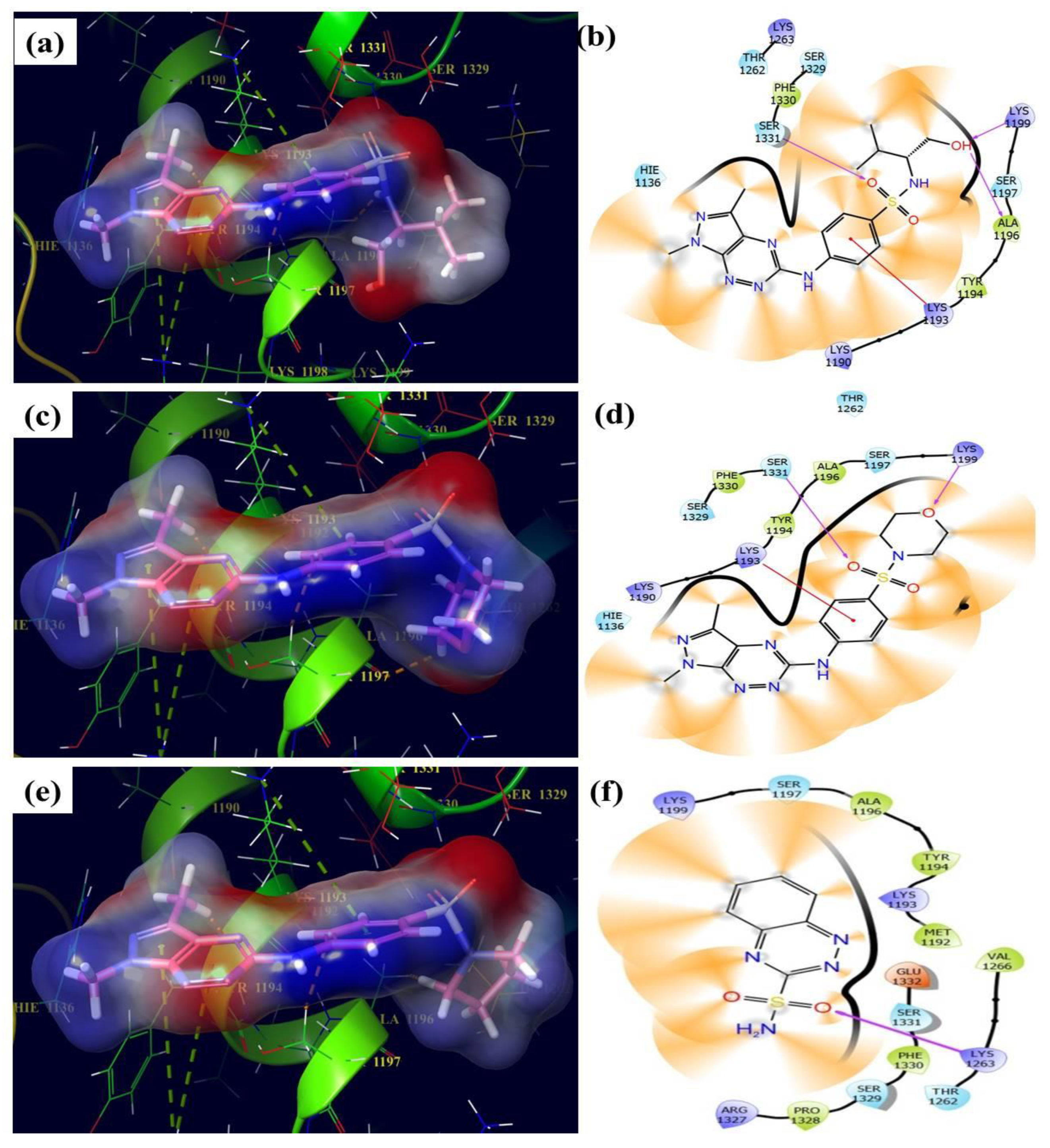
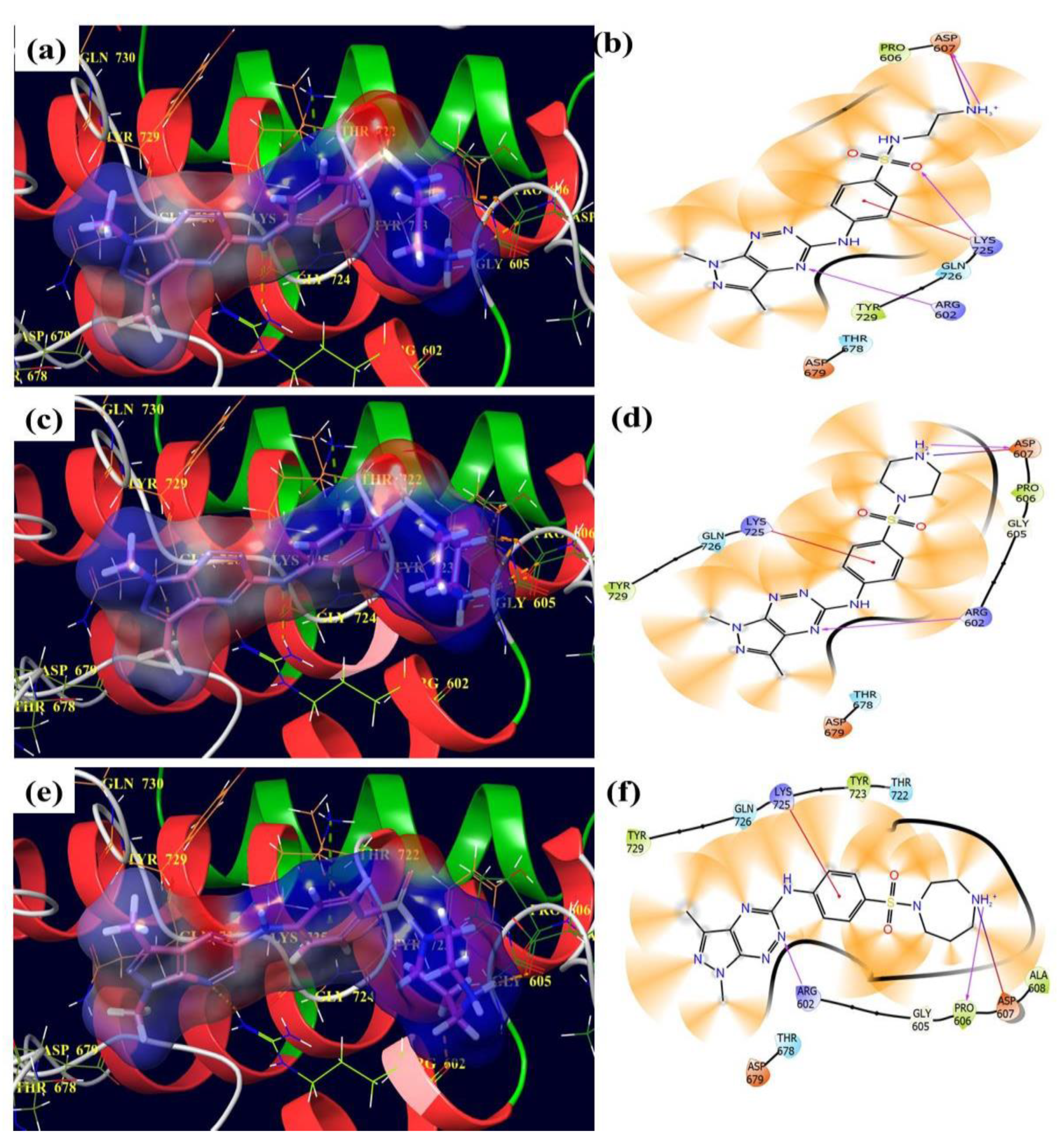
3.4. Molecular Docking Assay
3.5. Results of ADME
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kciuk, M.; Mujwar, S.; Szymanowska, A.; Marciniak, B.; Bukowski, K.; Mojzych, M.; Kontek, R. Preparation of Novel Pyrazolo [4, 3-e] tetrazolo [1, 5-b][1, 2, 4] triazine Sulfonamides and Their Experimental and Computational Biological Studies. Int. J. Mol. Sci. 2022, 23, 5892. [Google Scholar] [CrossRef]
- Villa, E.; Ali, E.S.; Sahu, U.; Ben-Sahra, I. Cancer cells tune the signaling pathways to empower de novo synthesis of nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Stewart, L.E.; Darley, B.A.; Pham, A.M.; Esteban, I.; Panda, S.S. Plant-Based Natural Products and Extracts: Potential Source to Develop New Antiviral Drug Candidates. Molecules 2021, 26, 6197. [Google Scholar] [CrossRef]
- Hong, P.; Liu, Q.-W.; Xie, Y.; Zhang, Q.-H.; Liao, L.; He, Q.-Y.; Li, B.; Xu, W.W. Echinatin suppresses esophageal cancer tumor growth and invasion through inducing AKT/mTOR-dependent autophagy and apoptosis. Cell Death Dis. 2020, 11, 524. [Google Scholar] [CrossRef]
- Kumar, R.; Saneja, A.; Panda, A.K. An Annexin V-FITC—Propidium Iodide-Based Method for Detecting Apoptosis in a Non-Small Cell Lung Cancer Cell Line. In Lung Cancer; Springer: Berlin/Heidelberg, Germany, 2021; pp. 213–223. [Google Scholar]
- Shamim, S.; Khan, K.M.; Ullah, N.; Chigurupati, S.; Wadood, A.; Rehman, A.U.; Ali, M.; Salar, U.; Alhowail, A.; Taha, M. Synthesis and screening of (E)-3-(2-benzylidenehydrazinyl)-5, 6-diphenyl-1, 2, 4-triazine analogs as novel dual inhibitors of α-amylase and α-glucosidase. Bioorganic Chem. 2020, 101, 103979. [Google Scholar] [CrossRef]
- Hermanowicz, J.M.; Szymanowska, A.; Sieklucka, B.; Czarnomysy, R.; Pawlak, K.; Bielawska, A.; Bielawski, K.; Kalafut, J.; Przybyszewska, A.; Surazynski, A. Exploration of novel heterofused 1, 2, 4-triazine derivative in colorectal cancer. J. Enzym. Inhib. Med. Chem. 2021, 36, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, C.; Layton, M. Science of Synthesis: Houben-Weyl Methods of Molecular Transformations; Weinreb, S.M., Ed.; Thieme: New York, NY, USA, 2004; Volume 17, p. 357. [Google Scholar]
- Khatir, Z.Z.; Irannejad, H. Pharmacologic Activities of 5, 6-Diaryl/heteroaryl-3-substituted-1, 2, 4-triazines as a Privileged Scaffold in Drug Development. Mini Rev. Med. Chem. 2021, 21, 2874–2928. [Google Scholar] [CrossRef] [PubMed]
- Okolotowicz, K.J.; Dwyer, M.; Ryan, D.; Cheng, J.; Cashman, E.A.; Moore, S.; Mercola, M.; Cashman, J.R. Novel tertiary sulfonamides as potent anti-cancer agents. Bioorganic Med. Chem. 2018, 26, 4441–4451. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Indisulam: An anticancer sulfonamide in clinical development. Expert Opin. Investig. Drugs 2003, 12, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Ghorab, M.M.; Ragab, F.A.; Heiba, H.I.; Elsayed, M.S.; Ghorab, W.M. Design, synthesis and molecular modeling study of certain 4-Methylbenzenesulfonamides with CDK2 inhibitory activity as anticancer and radio-sensitizing agents. Bioorganic Chem. 2018, 80, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Said, M.A.; Eldehna, W.M.; Nocentini, A.; Fahim, S.H.; Bonardi, A.; Elgazar, A.A.; Kryštof, V.; Soliman, D.H.; Abdel-Aziz, H.A.; Gratteri, P. Sulfonamide-based ring-fused analogues for CAN508 as novel carbonic anhydrase inhibitors endowed with antitumor activity: Design, synthesis, and in vitro biological evaluation. Eur. J. Med. Chem. 2020, 189, 112019. [Google Scholar] [CrossRef]
- Manasa, K.L. E7010: Investigational anticancer agents targeting the microtubules. Int. J. Pharm. Sci. Res. 2015, 6, 3713. [Google Scholar]
- Gornowicz, A.; Szymanowska, A.; Mojzych, M.; Czarnomysy, R.; Bielawski, K.; Bielawska, A. The Anticancer Action of a Novel 1, 2, 4-Triazine Sulfonamide Derivative in Colon Cancer Cells. Molecules 2021, 26, 2045. [Google Scholar] [CrossRef]
- Mojzych, M.; Šubertová, V.; Bielawska, A.; Bielawski, K.; Bazgier, V.; Berka, K.; Gucký, T.; Fornal, E.; Kryštof, V. Synthesis and kinase inhibitory activity of new sulfonamide derivatives of pyrazolo [4, 3-e][1, 2, 4] triazines. Eur. J. Med. Chem. 2014, 78, 217–224. [Google Scholar] [CrossRef]
- Evecen, M.; Çelik, F.; Bektaş, E.; Güler, H.İ.; Ünver, Y. Experimental and theoretical investigations, enzyme inhibition activity and docking study of 5-methyl-4-(2-(piperazin-1-yl) ethyl)-2, 4-dihydro-3H-1, 2, 4-triazol-3-one. J. Mol. Struct. 2023, 1275, 134679. [Google Scholar] [CrossRef]
- Wang, P.; Yan, F.; Dong, J.; Wang, S.; Shi, Y.; Zhu, M.; Ma, H.; Xue, R.; Zhai, D.; Song, X. A Multiple-step Screening Protocol to Identify Norepinephrine and Dopamine Reuptake Inhibitors for Depression. Phys. Chem. Chem. Phys. 2023, 25, 8341–8354. [Google Scholar] [CrossRef]
- Shaker, S.; Gardouh, A.R.; Ghorab, M.M. Factors affecting liposomes particle size prepared by ethanol injection method. Res. Pharm. Sci. 2017, 12, 346. [Google Scholar] [CrossRef]
- Mughal, E.U.; Ashraf, J.; Hussein, E.M.; Nazir, Y.; Alwuthaynani, A.S.; Naeem, N.; Sadiq, A.; Alsantali, R.I.; Ahmed, S.A. Design, Synthesis, and Structural Characterization of Thioflavones and Thioflavonols as Potential Tyrosinase Inhibitors: In Vitro and In Silico Studies. ACS Omega 2022, 7, 17444–17461. [Google Scholar] [CrossRef] [PubMed]
- Sikandar, S.; Zahoor, A.F.; Ghaffar, A.; Anjum, M.N.; Noreen, R.; Irfan, A.; Munir, B.; Kotwica-Mojzych, K.; Mojzych, M. Unveiling the Chemistry and Synthetic Potential of Catalytic Cycloaddition Reaction of Allenes: A Review. Molecules 2023, 28, 704. [Google Scholar] [CrossRef]
- Kohn, W.; Sham, L.J. Self-consistent equations including exchange and correlation effects. Phys. Rev. 1965, 140, A1133. [Google Scholar] [CrossRef]
- Haoyu, S.Y.; He, X.; Li, S.L.; Truhlar, D.G. MN15: A Kohn–Sham global-hybrid exchange–correlation density functional with broad accuracy for multi-reference and single-reference systems and noncovalent interactions. Chem. Sci. 2016, 7, 5032–5051. [Google Scholar]
- Eathiraj, S.; Palma, R.; Volckova, E.; Hirschi, M.; France, D.S.; Ashwell, M.A.; Chan, T.C. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J. Biol. Chem. 2011, 286, 20666–20676. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wang, A.; Wu, H.; Qi, Z.; Li, X.; Yan, X.-E.; Chen, C.; Yu, K.; Zou, F.; Wang, W. Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode. Oncotarget 2017, 8, 18359. [Google Scholar] [CrossRef]
- Son, J.; Jang, J.; Beyett, T.S.; Eum, Y.; Haikala, H.M.; Verano, A.; Lin, M.; Hatcher, J.M.; Kwiatkowski, N.P.; Eser, P.Ö. A Novel HER2-Selective Kinase Inhibitor Is Effective in HER2 Mutant and Amplified Non–Small Cell Lung Cancer. Cancer Res. 2022, 82, 1633–1645. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, G.; Bowen, S.J.; Zhu, Y.; Roush, N.; Zachary, T.; Javens, C.; Williams, T.; Janssen, A.; Gonzales, A. Type 2 inhibitor leads of human tropomyosin receptor kinase (hTrkA). Bioorg. Med. Chem. Lett. 2019, 29, 126624. [Google Scholar] [CrossRef]
- Roskoski Jr, R. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef]
- Sogabe, S.; Kawakita, Y.; Igaki, S.; Iwata, H.; Miki, H.; Cary, D.R.; Takagi, T.; Takagi, S.; Ohta, Y.; Ishikawa, T. Structure-based approach for the discovery of pyrrolo [3, 2-d] pyrimidine-based EGFR T790M/L858R mutant inhibitors. ACS Med. Chem. Lett. 2013, 4, 201–205. [Google Scholar] [CrossRef]
- Dorsch, D.; Schadt, O.; Stieber, F.; Meyring, M.; Grädler, U.; Bladt, F.; Friese-Hamim, M.; Knühl, C.; Pehl, U.; Blaukat, A. Identification and optimization of pyridazinones as potent and selective c-Met kinase inhibitors. Bioorganic Med. Chem. Lett. 2015, 25, 1597–1602. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.A.S.; Nawaz, M.; Izzeldin, I.; Abubshait, H.A.; Alsadig, A.; Gomaa, M.S.; Abubshait, S.A.; Alsewdan, D. Molecular docking and Anticancer Activity of Some Synthesized 1,4- naphthoquinone Derivatives against Human Cancer Cell Line. J. Mol. Struct. 2023, 1275, 134702. [Google Scholar] [CrossRef]
- Strzelecka, M.; Glomb, T.; Drąg-Zalesińska, M.; Kulbacka, J.; Szewczyk, A.; Saczko, J.; Kasperkiewicz-Wasilewska, P.; Rembiałkowska, N.; Wojtkowiak, K.; Jezierska, A. Synthesis, Anticancer Activity and Molecular Docking Studies of Novel N-Mannich Bases of 1, 3, 4-Oxadiazole Based on 4, 6-Dimethylpyridine Scaffold. Int. J. Mol. Sci. 2022, 23, 11173. [Google Scholar] [CrossRef]
- Tokalı, F.S.; Taslimi, P.; Demircioğlu, İ.H.; Şendil, K.; Tuzun, B.; Gülçin, İ. Novel phenolic Mannich base derivatives: Synthesis, bioactivity, molecular docking, and ADME-Tox Studies. J. Iran. Chem. Soc. 2022, 19, 563–577. [Google Scholar] [CrossRef]
- Demir, Y. Naphthoquinones, benzoquinones, and anthraquinones: Molecular docking, ADME and inhibition studies on human serum paraoxonase-1 associated with cardiovascular diseases. Drug Dev. Res. 2020, 81, 628–636. [Google Scholar] [CrossRef] [PubMed]

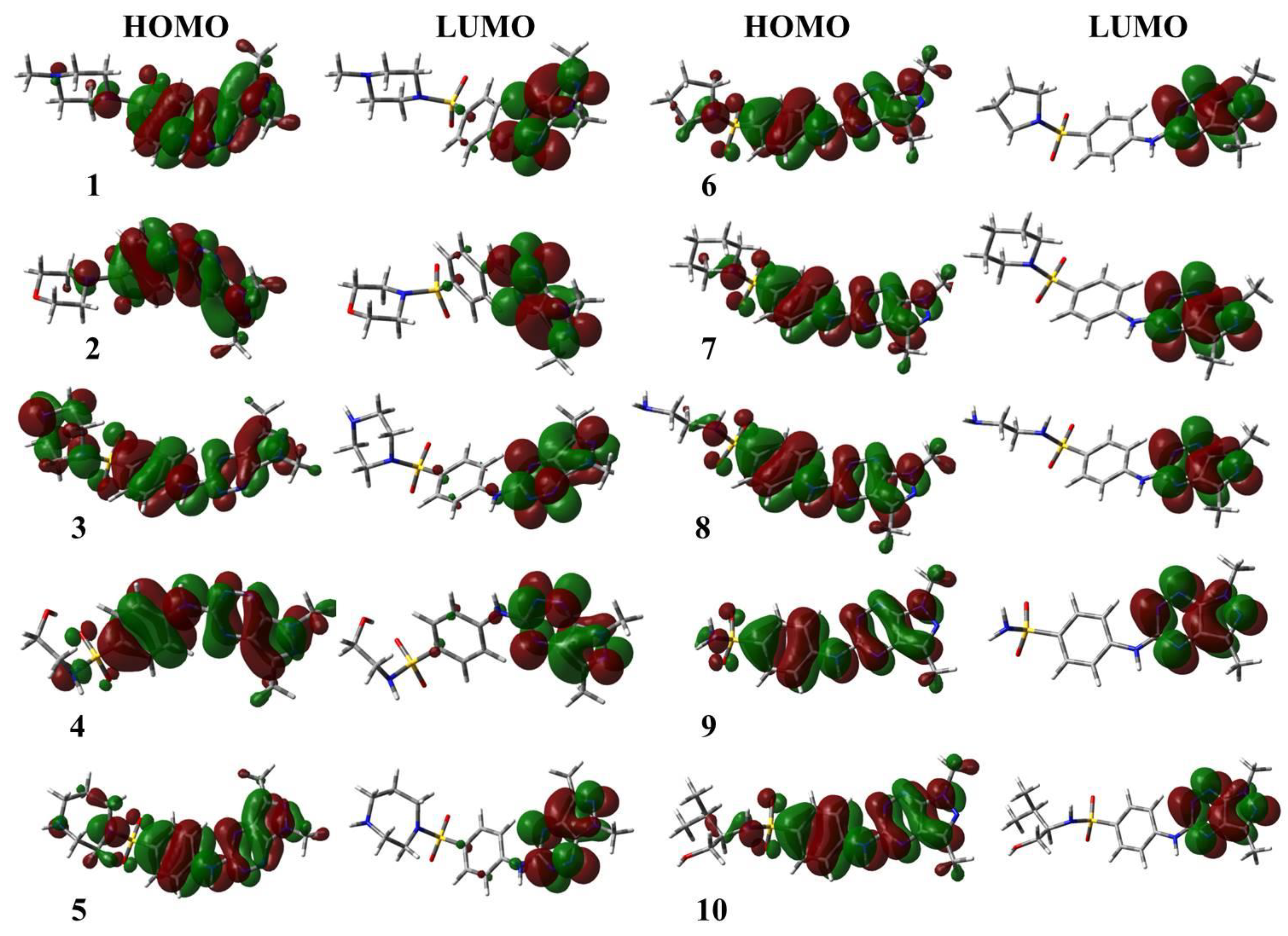


| Ligands | EHOMO (eV) | ELUMO (eV) | ΔEgap (eV) | IE (eV) | A (eV) | µ (eV) | χ (eV) | ƞ (eV) | S (eV) | ω (eV) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | −5.92 | −2.53 | 3.38 | 5.92 | 2.53 | −4.22 | 8.45 | 1.69 | 0.295 | 5.27 |
| 2 | −5.98 | −2.57 | 3.40 | 5.98 | 2.57 | −4.27 | 8.55 | 1.70 | 0.293 | 5.37 |
| 3 | −5.83 | −2.51 | 3.32 | 5.83 | 2.51 | −4.17 | 8.34 | 1.66 | 0.300 | 5.23 |
| 4 | −6.00 | −2.58 | 3.41 | 6.00 | 2.58 | −4.29 | 8.58 | 1.70 | 0.292 | 5.39 |
| 5 | −5.81 | −2.50 | 3.30 | 5.81 | 2.50 | −4.16 | 8.32 | 1.65 | 0.302 | 5.23 |
| 6 | −5.79 | −2.54 | 3.25 | 5.79 | 2.54 | −4.17 | 8.34 | 1.62 | 0.307 | 5.34 |
| 7 | −5.86 | −2.57 | 3.28 | 5.86 | 2.57 | −4.21 | 8.43 | 1.64 | 0.304 | 5.41 |
| 8 | −5.874 | −2.57 | 3.29 | 5.87 | 2.578 | −4.22 | 8.45 | 1.64 | 0.303 | 5.41 |
| 9 | −5.88 | −2.57 | 3.30 | 5.88 | 2.57 | −4.23 | 8.46 | 1.65 | 0.302 | 5.41 |
| 10 | −5.87 | −2.58 | 3.28 | 5.87 | 2.58 | −4.22 | 8.45 | 1.64 | 0.304 | 5.43 |
| Ligand | 3RHK | 5GTY | 6PL2 | 7JXH | ||||
|---|---|---|---|---|---|---|---|---|
| Docking Score kcal/mol | ∆G Energy kcal/mol | Docking Score kcal/mol | ∆G Energy kcal/mol | Docking Score kcal/mol | ∆G Energy kcal/mol | Docking Score kcal/mol | ∆G Energy kcal/mol | |
| 1 | −3.101 | −29.511 | −8.204 | −50.933 | −3.884 | −35.62 | −5.655 | −46.97 |
| 2 | −4.374 | −33.526 | −8.572 | −52.125 | −2.287 | −27.867 | −6.634 | −46.897 |
| 3 | −3.238 | −32.331 | −9.03 | −56.017 | −4.835 | −37.531 | −6.124 | −49.091 |
| 4 | −4.181 | −37.568 | −8.449 | −53.274 | −4.075 | −34.647 | −6.258 | −50.734 |
| 5 | −2.261 | −22.686 | −9.099 | −56.537 | −4.522 | −39.877 | −6.587 | −46.836 |
| 6 | −4.224 | −32.129 | −8.608 | −52.009 | −2.998 | −33.2 | −6.025 | −47.675 |
| 7 | −3.081 | −27.479 | −8.727 | −54.31 | −2.864 | −30.155 | −6.143 | −46.758 |
| 8 | −2.765 | −25 | −7.791 | −53.477 | −5.146 | −36.581 | −6.462 | −50.689 |
| 9 | −3.963 | −32.622 | −7.666 | −46.033 | −3.989 | −30.021 | −6.491 | −44.485 |
| 10 | −5.277 | −37.388 | −9.498 | −61.971 | −3.26 | −35.013 | −6.617 | −51.322 |
| Erlotinib | −4.143 | −32.362 | −7.629 | −54.808 | −2.279 | −31.124 | −6.327 | −52.283 |
| Neratinib | −2.894 | −36.451 | −5.674 | −58.645 | −0.676 | −28.228 | −4.009 | −52.789 |
| Tepotinib | −2.811 | −36.912 | −9.029 | −66.287 | −3.339 | −47.78 | −7.204 | −59.378 |
| Compd | Mol MW | HBD | HBA | QPlog Po/w | QPlogS | QPPCaco | metab | QPlog Khsa | Human Oral Absorption | Percent Human Oral Absorption | Rule of Five | Rule of Three |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 402.473 | 1 | 11 | 1.051 | −3.145 | 87.847 | 2 | −0.387 | 3 | 67.89 | 0 | 0 |
| 2 | 389.431 | 1 | 10.7 | 1.205 | −3.703 | 366.53 | 2 | −0.529 | 3 | 79.892 | 0 | 0 |
| 3 | 388.446 | 2 | 10.5 | 0.653 | −3.1 | 42.575 | 1 | −0.336 | 2 | 59.928 | 0 | 0 |
| 4 | 363.393 | 3 | 10.7 | 0.242 | −3.397 | 60.53 | 2 | −0.639 | 3 | 60.254 | 0 | 0 |
| 5 | 402.473 | 2 | 10.5 | 0.754 | −2.831 | 48.159 | 1 | −0.288 | 3 | 61.479 | 0 | 0 |
| 6 | 373.432 | 1 | 9 | 1.745 | −4.325 | 280.231 | 1 | −0.221 | 3 | 80.971 | 0 | 0 |
| 7 | 387.459 | 1 | 9 | 2.138 | −4.774 | 329.24 | 1 | −0.081 | 3 | 84.525 | 0 | 0 |
| 8 | 362.409 | 4 | 10 | −0.052 | −2.073 | 25.439 | 3 | −0.541 | 2 | 51.799 | 0 | 0 |
| 9 | 319.34 | 3 | 9 | 0.068 | −3.242 | 60.275 | 1 | −0.543 | 2 | 59.203 | 0 | 0 |
| 10 | 405.474 | 3 | 10.7 | 1.249 | −4.018 | 119.707 | 2 | −0.375 | 3 | 71.454 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutahir, S.; Khan, M.A.; Naglah, A.M.; Al-Omar, M.A.; Almehizia, A.A.; Huwaimel, B.; Abouzied, A.S.; Alharbi, A.S.; Refat, M.S. Structural Characterization and Molecular Docking Screening of Most Potent 1,2,4-Triazine Sulfonamide Derivatives as Anti-Cancer Agents. Crystals 2023, 13, 767. https://doi.org/10.3390/cryst13050767
Mutahir S, Khan MA, Naglah AM, Al-Omar MA, Almehizia AA, Huwaimel B, Abouzied AS, Alharbi AS, Refat MS. Structural Characterization and Molecular Docking Screening of Most Potent 1,2,4-Triazine Sulfonamide Derivatives as Anti-Cancer Agents. Crystals. 2023; 13(5):767. https://doi.org/10.3390/cryst13050767
Chicago/Turabian StyleMutahir, Sadaf, Muhammad Asim Khan, Ahmed M. Naglah, Mohamed A. Al-Omar, Abdulrahman A. Almehizia, Bader Huwaimel, Amr S. Abouzied, Amirah Senaitan Alharbi, and Moamen S. Refat. 2023. "Structural Characterization and Molecular Docking Screening of Most Potent 1,2,4-Triazine Sulfonamide Derivatives as Anti-Cancer Agents" Crystals 13, no. 5: 767. https://doi.org/10.3390/cryst13050767
APA StyleMutahir, S., Khan, M. A., Naglah, A. M., Al-Omar, M. A., Almehizia, A. A., Huwaimel, B., Abouzied, A. S., Alharbi, A. S., & Refat, M. S. (2023). Structural Characterization and Molecular Docking Screening of Most Potent 1,2,4-Triazine Sulfonamide Derivatives as Anti-Cancer Agents. Crystals, 13(5), 767. https://doi.org/10.3390/cryst13050767











