Pre-Treatment Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy Using Intratumoral and Peritumoral Radiomics from T2-Weighted and Contrast-Enhanced T1-Weighted MRI
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
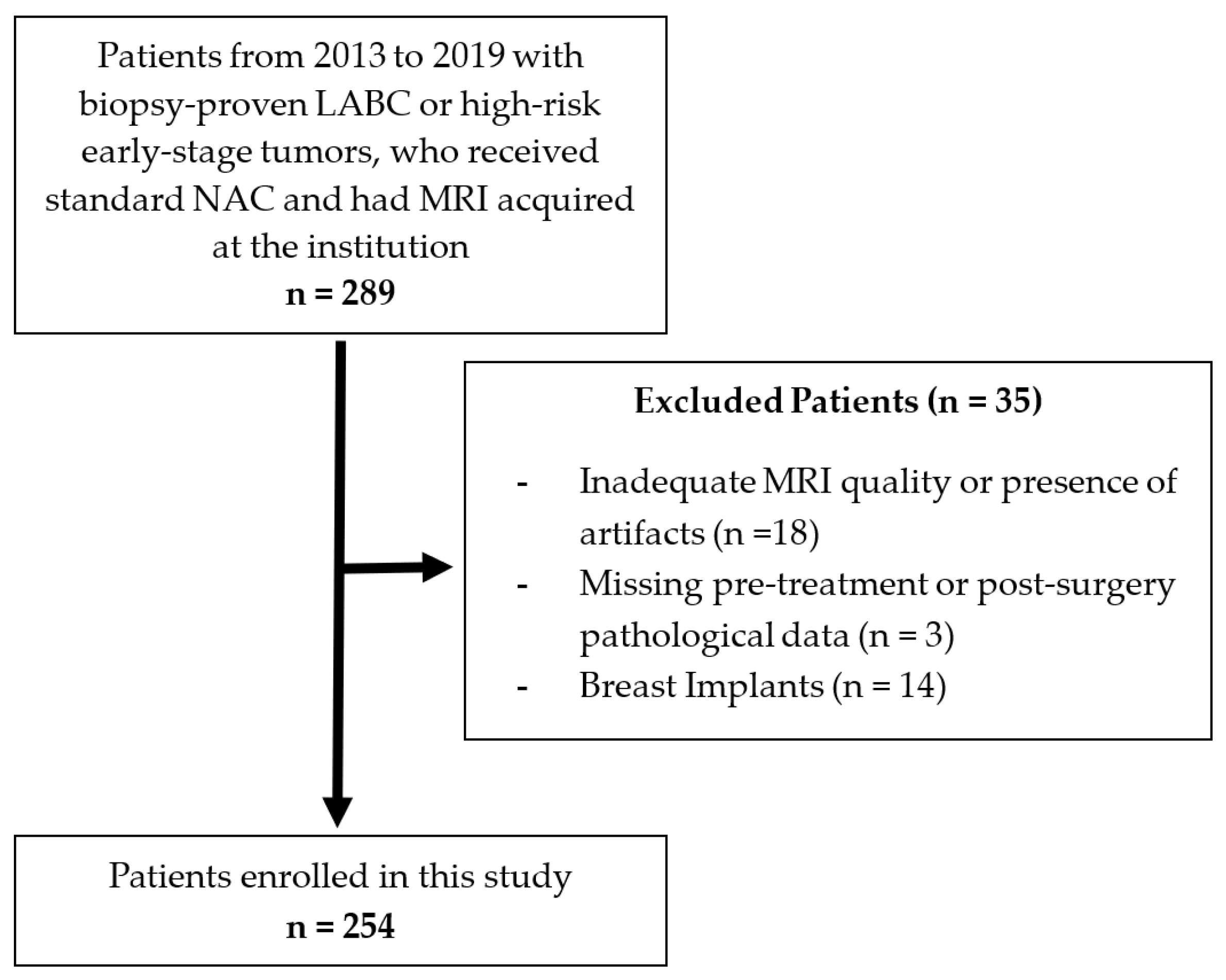
2.1. Patient Selection and Clinical Features
2.2. Response Evaluation
2.3. MRI Acquisition and Analysis
2.4. Radiomic Feature Extraction
2.5. Machine Learning Classification
2.6. Statistical Analysis
3. Results
3.1. Clinical Characteristics
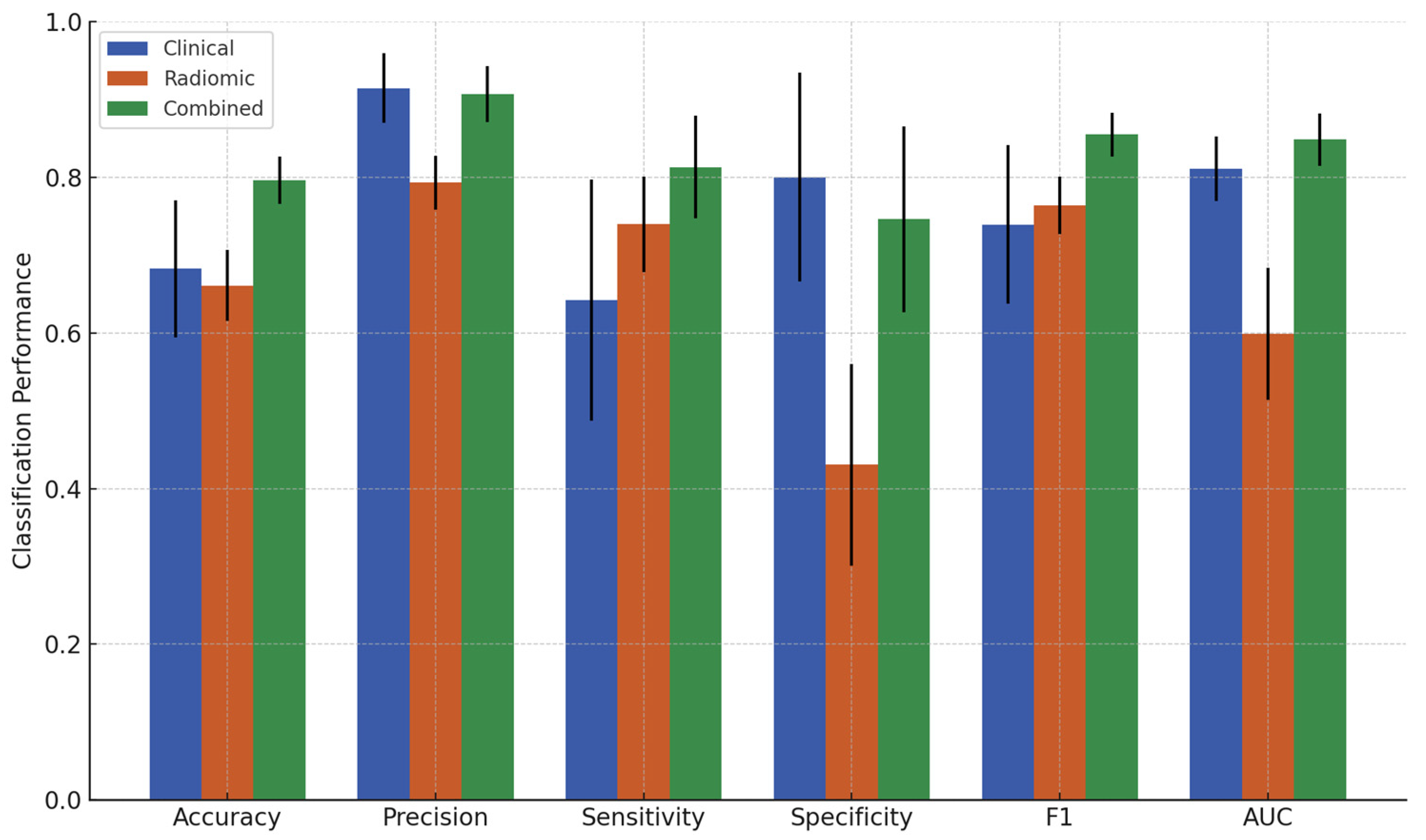
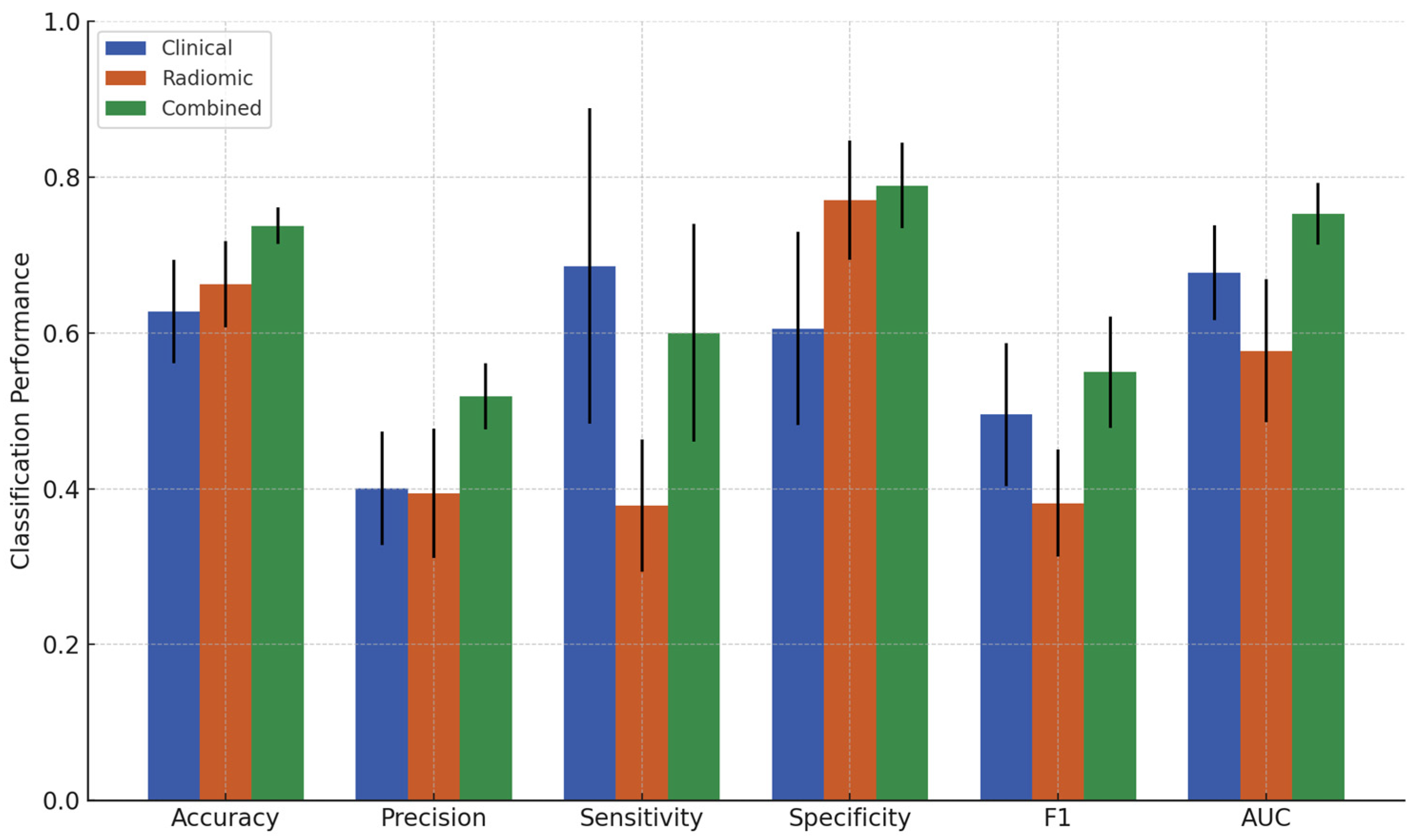
3.2. Classification Results
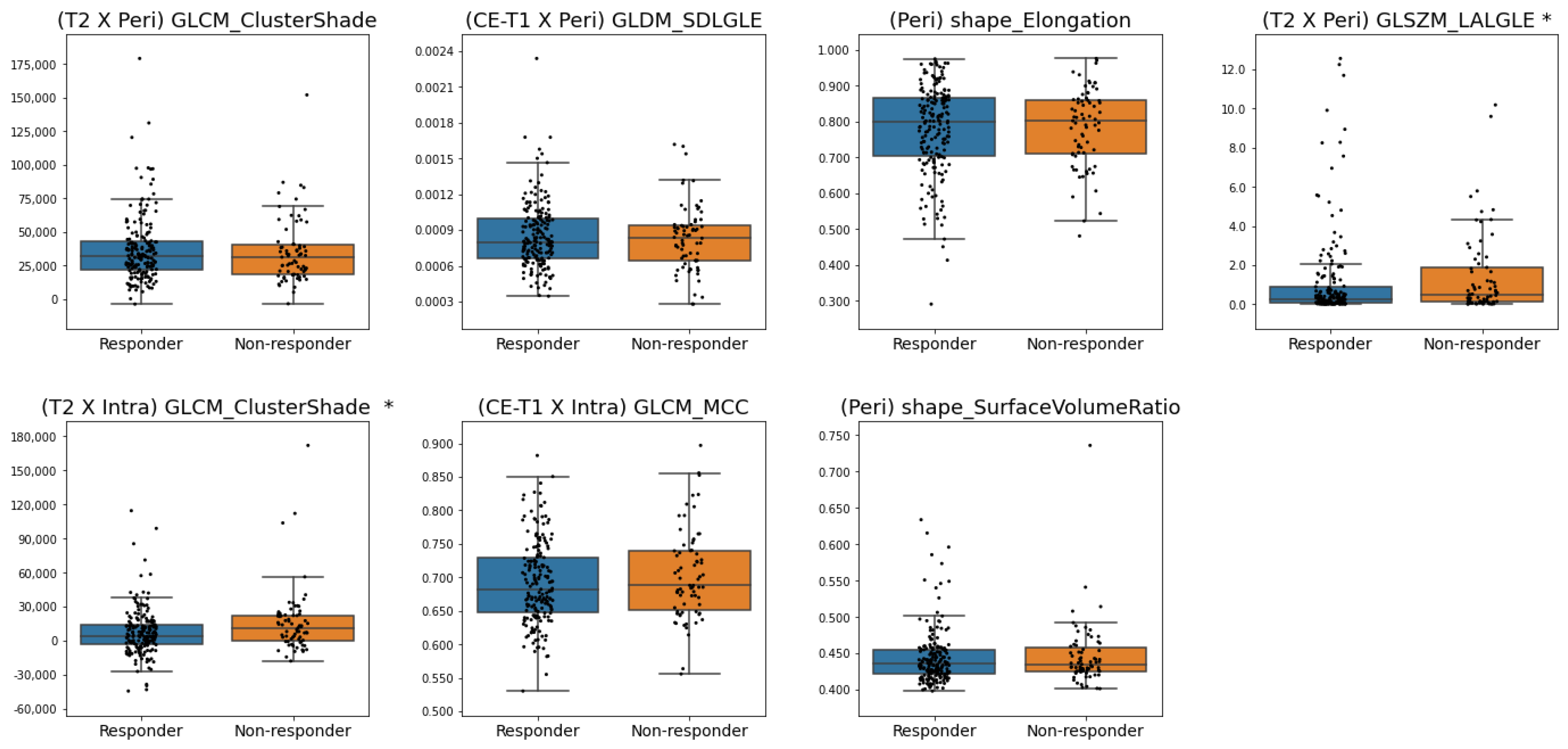
3.3. Features Selected
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Kim, J.; Harper, A.; McCormack, V.; Sung, H.; Houssami, N.; Morgan, E.; Mutebi, M.; Garvey, G.; Soerjomataram, I.; Fidler-Benaoudia, M.M. Global Patterns and Trends in Breast Cancer Incidence and Mortality across 185 Countries. Nat. Med. 2025, 31, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA—J. Am. Med. Assoc. 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Mieog, J.S.D.; Van De Velde, C.J.H. Neoadjuvant Chemotherapy for Early Breast Cancer. Expert Opin. Pharmacother. 2009, 10, 1423–1434. [Google Scholar] [CrossRef]
- Asaoka, M.; Gandhi, S.; Ishikawa, T.; Takabe, K. Neoadjuvant Chemotherapy for Breast Cancer: Past, Present, and Future. Breast Cancer 2020, 14, 1178223420980377. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Schott, A.F. Neoadjuvant Chemotherapy: What Are the Benefits for the Patient and for the Investigator? JNCI Monogr. 2015, 2015, 36–39. [Google Scholar] [CrossRef][Green Version]
- Cortazar, P.; Geyer, C.E. Pathological Complete Response in Neoadjuvant Treatment of Breast Cancer. Ann. Surg. Oncol. 2015, 22, 1441–1446. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and Impact of Pathologic Complete Response on Prognosis after Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef]
- Spanheimer, P.M.; Carr, J.C.; Thomas, A.; Sugg, S.L.; Scott-Conner, C.E.H.; Liao, J.; Weigel, R.J. The Response to Neoadjuvant Chemotherapy Predicts Clinical Outcome and Increases Breast Conservation in Advanced Breast Cancer. Am. J. Surg. 2013, 206, 2–7. [Google Scholar] [CrossRef][Green Version]
- Miller, M.; Ottesen, R.A.; Niland, J.C.; Kruper, L.; Chen, S.L.; Vito, C. Tumor Response Ratio Predicts Overall Survival in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Ann. Surg. Oncol. 2014, 21, 3317–3323. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Leung, V.W.Y.; Meterissian, S. MRI Performance in Detecting PCR After Neoadjuvant Chemotherapy by Molecular Subtype of Breast Cancer. World J. Surg. 2019, 43, 2254–2261. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.K.Y.; Tan, P.H. Liquid Biopsy in Breast Cancer: A Focused Review. Arch. Pathol. Lab. Med. 2021, 145, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Parekh, V.; Jacobs, M.A. Radiomics: A New Application from Established Techniques. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 207–226. [Google Scholar] [CrossRef]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in Breast Cancer Classification and Prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef]
- Reig, B.; Heacock, L.; Geras, K.J.; Moy, L. Machine Learning in Breast MRI. J. Magn. Reson. Imaging 2019, 52, 998–1018. [Google Scholar] [CrossRef]
- Valdora, F.; Houssami, N.; Rossi, F.; Calabrese, M.; Tagliafico, A.S. Rapid Review: Radiomics and Breast Cancer. Breast Cancer Res. Treat. 2018, 169, 217–229. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of Radiomics in Breast Cancer Diagnosis and Prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Sannachi, L.; Osapoetra, L.O.; DiCenzo, D.; Halstead, S.; Wright, F.; Look-Hong, N.; Slodkowska, E.; Gandhi, S.; Curpen, B.; Kolios, M.C.; et al. A Priori Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy Using Quantitative Ultrasound, Texture Derivative and Molecular Subtype. Sci. Rep. 2023, 13, 22687. [Google Scholar] [CrossRef] [PubMed]
- Osapoetra, L.O.; Sannachi, L.; Quiaoit, K.; Dasgupta, A.; DiCenzo, D.; Fatima, K.; Wright, F.; Dinniwell, R.; Trudeau, M.; Gandhi, S.; et al. A Priori Prediction of Response in Multicentre Locally Advanced Breast Cancer (LABC) Patients Using Quantitative Ultrasound and Derivative Texture Methods. Oncotarget 2021, 12, 81–94. [Google Scholar] [CrossRef]
- Moghadas-Dastjerdi, H.; Rahman, S.E.T.H.; Sannachi, L.; Wright, F.C.; Gandhi, S.; Trudeau, M.E.; Sadeghi-Naini, A.; Czarnota, G.J. Prediction of Chemotherapy Response in Breast Cancer Patients at Pre-Treatment Using Second Derivative Texture of CT Images and Machine Learning. Transl. Oncol. 2021, 14, 101183. [Google Scholar] [CrossRef]
- Kolios, C.; Sannachi, L.; Dasgupta, A.; Suraweera, H.; Dicenzo, D.; Stanisz, G.; Sahgal, A.; Wright, F.; Look-Hong, N.; Curpen, B.; et al. MRI Texture Features from Tumor Core and Margin in the Prediction of Response to Neoadjuvant Chemotherapy in Patients with Locally Advanced Breast Cancer. Oncotarget 2021, 12, 1354–1365. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef]
- Granzier, R.W.Y.; Ibrahim, A.; Primakov, S.P.; Samiei, S.; van Nijnatten, T.J.A.; de Boer, M.; Heuts, E.M.; Hulsmans, F.J.; Chatterjee, A.; Lambin, P.; et al. Mri-Based Radiomics Analysis for the Pretreatment Prediction of Pathologic Complete Tumor Response to Neoadjuvant Systemic Therapy in Breast Cancer Patients: A Multicenter Study. Cancers 2021, 13, 2447. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.M.; Wang, H.T.; Yu, T. The Application of Radiomics in Breast MRI: A Review. Technol. Cancer Res. Treat. 2020, 19, 1533033820916191. [Google Scholar] [CrossRef]
- Sannachi, L.; Gangeh, M.; Tadayyon, H.; Gandhi, S.; Wright, F.C.; Slodkowska, E.; Curpen, B.; Sadeghi-Naini, A.; Tran, W.; Czarnota, G.J. Breast Cancer Treatment Response Monitoring Using Quantitative Ultrasound and Texture Analysis: Comparative Analysis of Analytical Models. Transl. Oncol. 2019, 12, 1271–1281. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [PubMed]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Pletcha, D.; Madabhushi, A. Intratumoral and Peritumoral Radiomics for the Pretreatment Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy Based on Breast DCE-MRI. Breast Cancer Res. 2017, 19, 57. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Long, F.; Ding, C. Feature Selection Based on Mutual Information Criteria of Max-Dependency, Max-Relevance, and Min-Redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Tahmassebi, A.; Wengert, G.J.; Helbich, T.H.; Bago-Horvath, Z.; Alaei, S.; Bartsch, R.; Dubsky, P.; Baltzer, P.; Clauser, P.; Kapetas, P.; et al. Impact of Machine Learning with Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Investig. Radiol. 2019, 54, 110–117. [Google Scholar] [CrossRef]
- Vamvakas, A.; Tsivaka, D.; Logothetis, A.; Vassiou, K.; Tsougos, I. Breast Cancer Classification on Multiparametric MRI—Increased Performance of Boosting Ensemble Methods. Technol. Cancer Res. Treat. 2022, 21, 15330338221087828. [Google Scholar] [CrossRef]
- Meti, N.; Saednia, K.; Lagree, A.; Tabbarah, S.; Mohebpour, M.; Kiss, A.; Lu, F.-I.; Slodkowska, E.; Gandhi, S.; Joanna Jerzak, K.; et al. Machine Learning Frameworks to Predict Neoadjuvant Chemotherapy Response in Breast Cancer Using Clinical and Pathological Features. JCO Clin. Cancer Inf. 2021, 5, 66–80. [Google Scholar] [CrossRef]
- Lee, H.; Lee, J.H.; Lee, J.E.; Na, Y.M.; Park, M.H.; Lee, J.S.; Lim, H.S. Prediction of Early Clinical Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer: Incorporating Radiomics through Breast MRI. Sci. Rep. 2024, 14, 21691. [Google Scholar] [CrossRef]
- Lo, A.; Chernoff, H.; Zheng, T.; Lo, S.-H. Why Significant Variables Aren’t Automatically Good Predictors. Proc. Natl. Acad. Sci. USA 2015, 112, 13892–13897. [Google Scholar] [CrossRef]
- Kuhl, C.K. MRI of Breast Tumors. Eur. Radiol. 2000, 10, 46–58. [Google Scholar] [CrossRef]
- Jain, R.K. Normalization of Tumor Vasculature: An Emerging Concept in Antiangiogenic Therapy. Science 2005, 307, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Cho, N.; Moy, L. Breast MRI: State of the Art. Radiology 2019, 292, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.Y.; Hyun, C.L.; Jin, M.S.; Park, I.A.; Chung, Y.R.; Shim, B.; Lee, K.H.; Ryu, H.S. Histomorphological Factors Predicting the Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. J. Breast Cancer 2016, 19, 261–267. [Google Scholar] [CrossRef]
- Parikh, J.; Selmi, M.; Charles-Edwards, G.; Glendenning, J.; Ganeshan, B.; Verma, H.; Mansi, J.; Harries, M.; Tutt, A.; Goh, V. Changes in Primary Breast Cancer Heterogeneity May Augment Midtreatment MR Imaging Assessment of Response to Neoadjuvant Chemotherapy. Radiology 2014, 272, 100–112. [Google Scholar] [CrossRef]
- Braman, N.; Prasanna, P.; Whitney, J.; Singh, S.; Beig, N.; Etesami, M.; Bates, D.D.B.; Gallagher, K.; Bloch, B.N.; Vulchi, M.; et al. Association of Peritumoral Radiomics with Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2) -Positive Breast Cancer. JAMA Netw. Open 2019, 2, e192561. [Google Scholar] [CrossRef] [PubMed]
- Stanton, S.E.; Disis, M.L. Clinical Significance of Tumor-Infiltrating Lymphocytes in Breast Cancer. J. Immunother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef]
- Qi, Y.J.; Su, G.H.; You, C.; Zhang, X.; Xiao, Y.; Jiang, Y.Z.; Shao, Z.M. Radiomics in Breast Cancer: Current Advances and Future Directions. Cell Rep. Med. 2024, 5, 101719. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, Y.-H.; Zhu, T.; Zhang, Y.-M.; Zheng, X.-X.; Zhang, T.-F.; Lin, Y.-Y.; Wu, Z.-Y.; Liu, Z.-Y.; Lin, Y.; et al. Noninvasive Artificial Intelligence System for Early Predicting Residual Cancer Burden during Neoadjuvant Chemotherapy in Breast Cancer. Ann. Surg. 2024, 281, 645–654. [Google Scholar] [CrossRef]
- Tadayyon, H.; Sannachi, L.; Gangeh, M.J.; Kim, C.; Ghandi, S.; Trudeau, M.; Pritchard, K.; Tran, W.T.; Slodkowska, E.; Sadeghi-Naini, A.; et al. A Priori Prediction of Neoadjuvant Chemotherapy Response and Survival in Breast Cancer Patients Using Quantitative Ultrasound. Sci. Rep. 2017, 7, 457331. [Google Scholar] [CrossRef]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef]


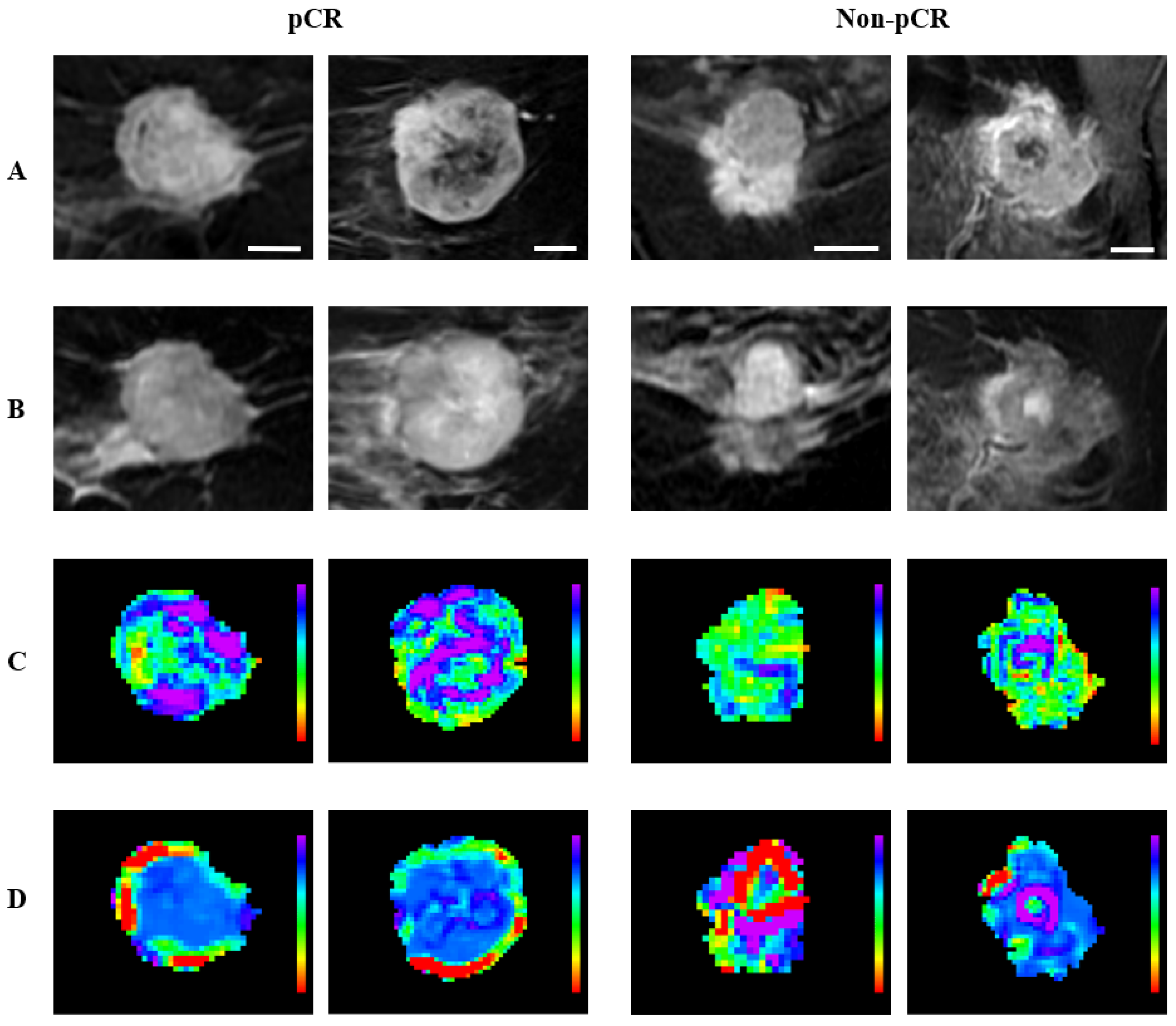
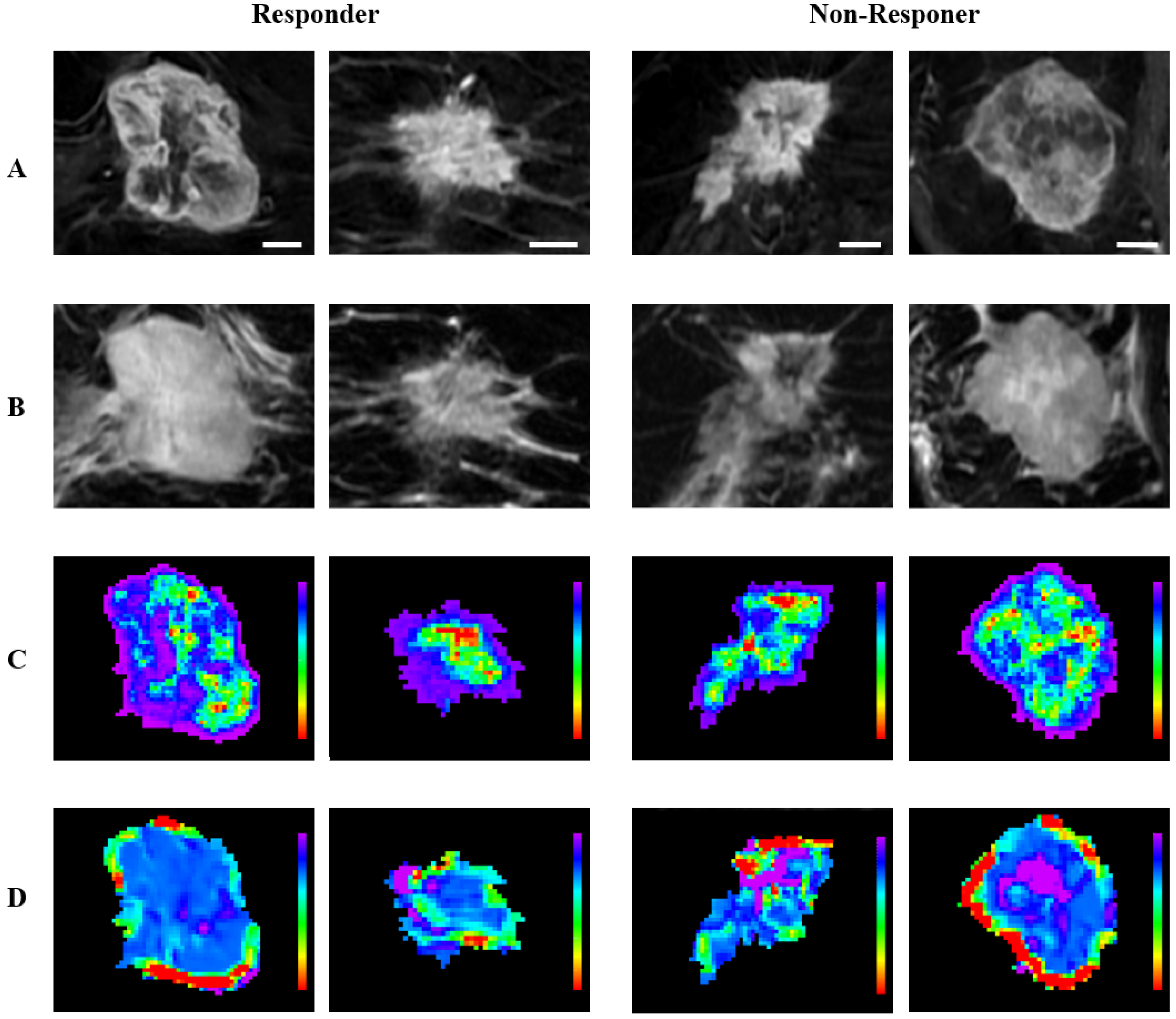
| Characteristics | pCR (n = 63) | Non-pCR (n = 191) | All (n = 254) | p Value |
|---|---|---|---|---|
| Age (year) | 50.2 ± 9.0 | 49.0 ± 11.1 | 49.3 ± 10.6 | 0.390 |
| Initial Tumor Size (mm) | 36.2 ± 17.4 | 43.9 ± 24.0 | 42.0 ± 22.8 | 0.012 |
| Histologic Grade | 0.001 | |||
| I (%) | 1 (1.6%) | 12 (6.3%) | 13 (5.1%) | |
| II (%) | 17 (27.0%) | 92 (48.2%) | 109 (42.9%) | |
| III (%) | 45 (71.4%) | 87 (45.5%) | 132 (52.0%) | |
| ER | <0.001 | |||
| Negative (%) | 41 (65.1%) | 56 (29.3%) | 97 (38.1%) | |
| Positive (%) | 22 (34.9%) | 135 (70.7%) | 157 (61.8%) | |
| PR | <0.001 | |||
| Negative (%) | 49 (77.8%) | 72 (37.7%) | 121 (47.6%) | |
| Positive (%) | 14 (22.2%) | 119 (62.3%) | 133 (52.3%) | |
| HER2 | <0.001 | |||
| Negative (%) | 18 (28.6%) | 140 (73.3%) | 158 (62.2%) | |
| Positive (%) | 45 (71.4%) | 51 (26.7%) | 96 (37.8%) | |
| Nodal Status | 0.011 | |||
| N0 (%) | 21 (33.3%) | 48 (25.1%) | 69 (27.2%) | |
| N1 (%) | 40 (63.5%) | 103 (53.9%) | 143 (56.3%) | |
| N2 (%) | 1 (1.6%) | 31 (16.2%) | 32 (12.6%) | |
| N3 (%) | 1 (1.6%) | 9 (4.7%) | 10 (3.9%) |
| Characteristics | Response (n = 183) | Non-Response (n = 71) | All (n = 254) | p Value |
|---|---|---|---|---|
| Age (year) | 48.5 ± 10.1 | 51.3 ± 11.8 | 49.3 ± 10.6 | 0.075 |
| Initial Tumor Size (mm) | 42.9 ± 24.1 | 39.5 ± 20.0 | 42.0 ± 22.8 | 0.561 |
| Histologic Grade | <0.001 | |||
| I (%) | 9 (4.9%) | 4 (5.6%) | 13 (5.1%) | |
| II (%) | 62 (33.9%) | 47 (66.2%) | 109 (42.9%) | |
| III (%) | 112 (61.2%) | 20 (28.2%) | 132 (52.0%) | |
| ER | <0.001 | |||
| Negative (%) | 86 (47.0%) | 11 (15.5%) | 97 (38.1%) | |
| Positive (%) | 97 (53.0%) | 60 (84.5%) | 157 (61.8%) | |
| PR | <0.001 | |||
| Negative (%) | 103 (56.3%) | 18 (25.4%) | 121 (47.6%) | |
| Positive (%) | 80 (43.7%) | 53 (74.6%) | 133 (52.3%) | |
| HER2 | <0.001 | |||
| Negative (%) | 101 (55.2%) | 57 (80.3%) | 158 (62.2%) | |
| Positive (%) | 82 (44.8%) | 14 (19.7%) | 96 (37.8%) | |
| Nodal Status | 0.317 | |||
| N0 (%) | 55 (30.1%) | 14 (19.7%) | 69 (27.2%) | |
| N1 (%) | 101 (55.2%) | 42 (59.2%) | 143 (56.3%) | |
| N2 (%) | 21 (11.5%) | 11 (15.5%) | 32 (12.6%) | |
| N3 (%) | 6 (3.3%) | 4 (5.6%) | 10 (3.9%) |
| Feature Set | Accuracy (%) ± SD | Precision (%) ± SD | Sensitivity (%) ± SD | Specificity (%) ± SD | F1 ± SD | AUC ± SD |
|---|---|---|---|---|---|---|
| Clinical | 68.2 ± 8.8 | 91.4 ± 4.5 | 64.2 ± 15.5 | 80.0 ± 13.4 | 0.739 ± 0.102 | 0.811 ± 0.042 |
| Radiomic | 66.1 ± 4.6 | 79.3 ± 3.4 | 73.9 ± 6.2 | 43.1 ± 13.0 | 0.764 ± 0.037 | 0.599 ± 0.085 |
| Combined | 79.6 ± 3.1 | 90.7 ± 3.6 | 81.3 ± 6.6 | 74.6 ± 11.9 | 0.855 ± 0.028 | 0.849 ± 0.034 |
| Feature Set | Accuracy (%) ± SD | Precision (%) ± SD | Sensitivity (%) ± SD | Specificity (%) ± SD | F1 ± SD | AUC ± SD |
|---|---|---|---|---|---|---|
| Clinical | 62.7 ± 6.6 | 40.1 ± 7.3 | 68.6 ± 20.3 | 60.5 ± 12.4 | 0.495 ± 0.092 | 0.677 ± 0.061 |
| Radiomic | 66.3 ± 5.5 | 39.4 ± 8.3 | 37.9 ± 8.5 | 77.0 ± 7.7 | 0.381 ± 0.069 | 0.576 ± 0.092 |
| Combined | 73.7 ± 2.4 | 51.8 ± 4.2 | 60.0 ± 14.0 | 78.9 ± 5.5 | 0.550 ± 0.071 | 0.752 ± 0.040 |
| Features | # |
|---|---|
| HER2 | 10 |
| PR | 10 |
| ER | 10 |
| Initial Tumor Size | 7 |
| (T2 × Intra) GLCM_ClusterShade | 6 |
| (CE-T1 × Peri) GLDM_SmallDependenceLowGrayLevelEmphasis | 6 |
| (T2 × Peri) GLCM_ClusterShade | 6 |
| (CE-T1 × Intra) GLCM_Correlation | 5 |
| (T2 × Peri) GLSZM_LargeAreaLowGrayLevelEmphasis | 5 |
| (Intra) Shape_Elongation | 5 |
| (Intra) Shape_Sphericity | 5 |
| Features | # |
|---|---|
| ER | 10 |
| Histologic Grade | 10 |
| Age | 10 |
| HER2 | 9 |
| PR | 9 |
| (T2 × Peri) GLCM_ClusterShade | 8 |
| (CE-T1 × Peri) GLDM_SmallDependenceLowGrayLevelEmphasis | 6 |
| (Peri) Shape_Elongation | 6 |
| (T2 × Peri) GLSZM_LargeAreaLowGrayLevelEmphasis | 6 |
| (T2 × Intra) GLCM_ClusterShade | 5 |
| (CE-T1 × Intra) GLCM_MCC | 5 |
| (Peri) Shape_SurfaceVolumeRatio | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, D.H.; Kolios, C.; Osapoetra, L.O.; Sannachi, L.; Curpen, B.; Pejović-Milić, A.; Czarnota, G.J. Pre-Treatment Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy Using Intratumoral and Peritumoral Radiomics from T2-Weighted and Contrast-Enhanced T1-Weighted MRI. Cancers 2025, 17, 1520. https://doi.org/10.3390/cancers17091520
Jang DH, Kolios C, Osapoetra LO, Sannachi L, Curpen B, Pejović-Milić A, Czarnota GJ. Pre-Treatment Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy Using Intratumoral and Peritumoral Radiomics from T2-Weighted and Contrast-Enhanced T1-Weighted MRI. Cancers. 2025; 17(9):1520. https://doi.org/10.3390/cancers17091520
Chicago/Turabian StyleJang, Deok Hyun, Christopher Kolios, Laurentius O. Osapoetra, Lakshmanan Sannachi, Belinda Curpen, Ana Pejović-Milić, and Gregory J. Czarnota. 2025. "Pre-Treatment Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy Using Intratumoral and Peritumoral Radiomics from T2-Weighted and Contrast-Enhanced T1-Weighted MRI" Cancers 17, no. 9: 1520. https://doi.org/10.3390/cancers17091520
APA StyleJang, D. H., Kolios, C., Osapoetra, L. O., Sannachi, L., Curpen, B., Pejović-Milić, A., & Czarnota, G. J. (2025). Pre-Treatment Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy Using Intratumoral and Peritumoral Radiomics from T2-Weighted and Contrast-Enhanced T1-Weighted MRI. Cancers, 17(9), 1520. https://doi.org/10.3390/cancers17091520







