Genomic and Demographic Characteristics of Angiosarcoma as Described in the AACR Project GENIE Registry
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Patient Demographics
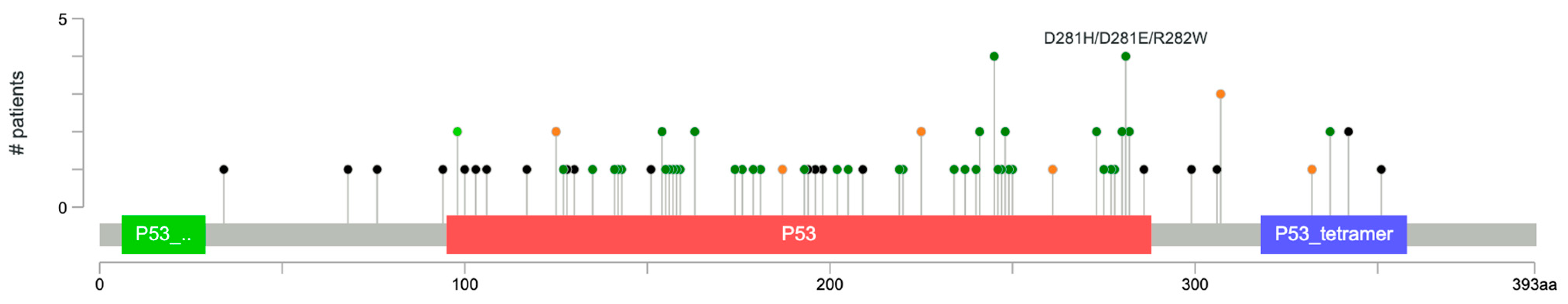
3.2. Somatic Mutations and Copy Number Alterations
3.3. Mutational Landscapes by Sex
3.4. Mutational Landscapes by Race
3.5. Co-Occurrence and Mutual Exclusivity
3.6. Metastatic vs. Primary Mutations
3.7. Breast Angiosarcoma vs. Liver Angiosarcoma
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WHO | World Health Organization |
| TMB | tumor mutation burden |
| AACR GENIE | American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange |
| CNAs | copy number alterations |
| SD | standard deviation |
| FDR | false discovery rate |
| VAF | variant allele frequency |
| VUS | variants of unknown significance |
| MAF | mutation annotation format |
| TKI | tyrosine kinase inhibitor |
References
- Spiker, A.M.; Mangla, A.; Ramsey, M.L. Angiosarcoma. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Kim, W.J.; Kim, H.K. Current Understanding of Angiosarcoma: Disease Biology and Evolving Treatment. Arch. Craniofacial Surg. 2023, 24, 203–210. [Google Scholar] [CrossRef]
- Young, R.J.; Brown, N.J.; Reed, M.W.; Hughes, D.; Woll, P.J. Angiosarcoma. Lancet Oncol. 2010, 11, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, J.; He, C.; Fang, M. Angiosarcoma: A Review of Diagnosis and Current Treatment. Am. J. Cancer Res. 2019, 9, 2303–2313. [Google Scholar] [PubMed]
- Liu, S.; Bellamkonda, V.; Phung, T. Angiosarcoma. Available online: https://www.pathologyoutlines.com/topic/softtissueangiosarcoma.html (accessed on 12 September 2025).
- Gaballah, A.H.; Jensen, C.T.; Palmquist, S.; Pickhardt, P.J.; Duran, A.; Broering, G.; Elsayes, K.M. Angiosarcoma: Clinical and Imaging Features from Head to Toe. Br. J. Radiol. 2017, 90, 20170039. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.J.; Ravi, V.; Schaub, S.K.; Kim, E.Y.; Sharib, J.; Mogal, H.; Park, M.; Tsai, M.; Duarte-Bateman, D.; Tufaro, A.; et al. Incidence and Presenting Characteristics of Angiosarcoma in the US, 2001–2020. JAMA Netw. Open 2024, 7, e246235. [Google Scholar] [CrossRef]
- Colas, M.; Gérazime, A.; Popescu, D.; Puzenat, E.; Chaigneau, L.; Woronoff, A.S.; Dupond, A.S.; Nardin, C.; Aubin, F. Angiosarcoma: A Population-Based Cancer Registry Descriptive Study of 45 Consecutive Cases Diagnosed between 1979 and 2016. Rare Tumors 2020, 12, 2036361320979216. [Google Scholar] [CrossRef]
- Takemori, T.; Ogura, K.; Morizane, C.; Satake, T.; Iwata, S.; Toda, Y.; Muramatsu, S.; Kondo, H.; Kobayashi, E.; Higashi, T.; et al. Incidence and Site Specific Characteristics of Angiosarcoma in Japan Using a Population-Based National Cancer Registry from 2016 to 2019. Sci. Rep. 2025, 15, 9960. [Google Scholar] [CrossRef]
- Friedrich, A.-K.U.; Reisenbichler, E.S.; Heller, D.R.; LeBlanc, J.M.; Park, T.S.; Killelea, B.K.; Lannin, D.R. Characteristics and Long-Term Risk of Breast Angiosarcoma. Ann. Surg. Oncol. 2021, 28, 5112–5118. [Google Scholar] [CrossRef]
- Fury, M.G.; Antonescu, C.R.; Van Zee, K.J.; Brennan, M.E.; Maki, R.G. A 14-Year Retrospective Review of Angiosarcoma: Clinical Characteristics, Prognostic Factors, and Treatment Outcomes with Surgery and Chemotherapy. Cancer J. 2005, 11, 241. [Google Scholar] [CrossRef]
- Veiga, L.H.; Vo, J.B.; Curtis, R.E.; Mille, M.M.; Lee, C.; Ramin, C.; Bodelon, C.; Aiello Bowles, E.J.; Buist, D.S.; Weinmann, S.; et al. Treatment-Related Thoracic Soft Tissue Sarcomas in US Breast Cancer Survivors: A Retrospective Cohort Study. Lancet Oncol. 2022, 23, 1451–1464. [Google Scholar] [CrossRef]
- Rosenbaum, E.; Antonescu, C.R.; Smith, S.; Bradic, M.; Kashani, D.; Richards, A.L.; Donoghue, M.; Kelly, C.M.; Nacev, B.; Chan, J.E.; et al. Clinical, Genomic, and Transcriptomic Correlates of Response to Immune Checkpoint Blockade-Based Therapy in a Cohort of Patients with Angiosarcoma Treated at a Single Center. J. Immunother. Cancer 2022, 10, e004149. [Google Scholar] [CrossRef]
- Chen, T.W.-W. Angiosarcoma: Role of Immunotherapy. Curr. Treat. Options Oncol. 2025, 26, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Espejo-Freire, A.; Elliott, A.; Rosenberg, A.; Costa, P.A.; Barreto-Coelho, P.; Jonczak, E.; D’Amato, G.; Subhawong, T.; Arshad, J.; Diaz-Perez, J.A.; et al. Genomic Landscape of Angiosarcoma: A Targeted and Immunotherapy Biomarker Analysis. Cancers 2021, 13, 4816. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- The AACR Project GENIE Consortium; André, F.; Arnedos, M.; Baras, A.S.; Baselga, J.; Bedard, P.L.; Berger, M.F.; Bierkens, M.; Calvo, F.; Cerami, E.; et al. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef]
- Caplan, U.C. U.S. Older Population Grew from 2010 to 2020 at Fastest Rate Since 1880 to 1890. Available online: https://www.census.gov/library/stories/2023/05/2020-census-united-states-older-population-grew.html (accessed on 13 September 2025).
- Toro, J.R.; Travis, L.B.; Wu, H.J.; Zhu, K.; Fletcher, C.D.M.; Devesa, S.S. Incidence Patterns of Soft Tissue Sarcomas, Regardless of Primary Site, in the Surveillance, Epidemiology and End Results Program, 1978–2001: An Analysis of 26,758 Cases. Int. J. Cancer 2006, 119, 2922–2930. [Google Scholar] [CrossRef]
- Wang, L.; Lao, I.W.; Yu, L.; Wang, J. Clinicopathological Features and Prognostic Factors in Angiosarcoma: A Retrospective Analysis of 200 Patients from a Single Chinese Medical Institute. Oncol. Lett. 2017, 14, 5370–5378. [Google Scholar] [CrossRef]
- Wali, A.; Robinson, J.; Iqbal, A.; Yasinzai, A.Q.K.; Sohail, A.H.; Jain, H.; Fadhil, N.; Khan, M.; Khan, I.; Karki, N.R.; et al. Demographics, Prognostic Factors, and Survival Outcomes in Hepatic Angiosarcoma: A Retrospective Analysis. J. Gastrointest. Cancer 2024, 56, 33. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, G.; Liu, Z.; Xu, Y.; Lin, F.; Baklaushev, V.P.; Chekhonin, V.P.; Peltzer, K.; Mao, M.; Wang, X.; et al. Epidemiology, Tumor Characteristics and Survival in Patients with Angiosarcoma in the United States: A Population-Based Study of 4537 Cases. Jpn. J. Clin. Oncol. 2019, 49, 1092–1099. [Google Scholar] [CrossRef]
- Albores-Saavedra, J.; Schwartz, A.M.; Henson, D.E.; Kostun, L.; Hart, A.; Angeles-Albores, D.; Chablé-Montero, F. Cutaneous Angiosarcoma. Analysis of 434 Cases from the Surveillance, Epidemiology, and End Results Program, 1973–2007. Ann. Diagn. Pathol. 2011, 15, 93–97. [Google Scholar] [CrossRef]
- U.S. Census Bureau. Hispanic or Latino, and not Hispanic or Latino by Race. U.S. Census Bureau Decennial Census; 2020. Available online: https://data.census.gov/table?q=p2 (accessed on 13 September 2025).
- Benton, A.; Liu, B.; Gartenhaus, L.E.; Hanna, J.A. Genomic Landscape and Preclinical Models of Angiosarcoma. Mol. Oncol. 2024, 19, 965–983. [Google Scholar] [CrossRef]
- Chen, Z.G.; Saba, N.F.; Teng, Y. The Diverse Functions of FAT1 in Cancer Progression: Good, Bad, or Ugly? J. Exp. Clin. Cancer Res. 2022, 41, 248. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Medarde, A.; De Las Rivas, J.; Santos, E. 40 Years of RAS—A Historic Overview. Genes 2021, 12, 681. [Google Scholar] [CrossRef] [PubMed]
- van Ravensteijn, S.G.; Versleijen-Jonkers, Y.M.H.; Hillebrandt-Roeffen, M.H.S.; Weidema, M.E.; Nederkoorn, M.J.L.; Bol, K.F.; Gorris, M.A.J.; Verrijp, K.; Kroeze, L.I.; de Bitter, T.J.J.; et al. Immunological and Genomic Analysis Reveals Clinically Relevant Distinctions between Angiosarcoma Subgroups. Cancers 2022, 14, 5938. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.S.; Lee, M.G. Cancer-Epigenetic Function of the Histone Methyltransferase KMT2D and Therapeutic Opportunities for the Treatment of KMT2D-Deficient Tumors. Oncotarget 2021, 12, 1296–1308. [Google Scholar] [CrossRef]
- Accardo, M.-L.; Osborne, J.; Else, T. POT1 Tumor Predisposition. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 1993. [Google Scholar]
- Panse, G.; Chrisinger, J.S.; Leung, C.H.; Ingram, D.R.; Khan, S.; Wani, K.; Lin, H.; Lazar, A.J.; Wang, W.-L. Clinicopathological Analysis of ATRX, DAXX and NOTCH Receptor Expression in Angiosarcomas. Histopathology 2018, 72, 239–247. [Google Scholar] [CrossRef]
- Fontana, B.; Gallerani, G.; Salamon, I.; Pace, I.; Roncarati, R.; Ferracin, M. ARID1A in Cancer: Friend or Foe? Front. Oncol. 2023, 13, 1136248. [Google Scholar] [CrossRef]
- Ueno, S.; Sudo, T.; Hirasawa, A. ATM: Functions of ATM Kinase and Its Relevance to Hereditary Tumors. Int. J. Mol. Sci. 2022, 23, 523. [Google Scholar] [CrossRef]
- Cozzi, S.; Finocchi Ghersi, S.; Tava, F.; Bardoscia, L.; Najafi, M.; Ruggieri, M.P.; Serre, A.-A.; Roukoz, C.; Gutierrez Miguelez, C.; Lazrek, A.; et al. Radiation-Associated Angiosarcoma of the Breast: The State of the Art of a Rare and Aggressive Disease. J. Pers. Med. 2024, 14, 859. [Google Scholar] [CrossRef]
- Nishikawa, S.; Iwakuma, T. Drugs Targeting P53 Mutations with FDA Approval and in Clinical Trials. Cancers 2023, 15, 429. [Google Scholar] [CrossRef]
- Painter, C.A.; Jain, E.; Tomson, B.N.; Dunphy, M.; Stoddard, R.E.; Thomas, B.S.; Damon, A.L.; Shah, S.; Kim, D.; Gómez Tejeda Zañudo, J.; et al. The Angiosarcoma Project: Enabling Genomic and Clinical Discoveries in a Rare Cancer through Patient-Partnered Research. Nat. Med. 2020, 26, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Kim, M.-J.; Kumar, A.; Lee, H.-W.; Yang, Y.; Kim, Y. Vascular Endothelial Growth Factor Signaling in Health and Disease: From Molecular Mechanisms to Therapeutic Perspectives. Signal Transduct. Target. Ther. 2025, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Zhang, L.; Chang, N.-E.; Singer, S.; Maki, R.G.; Antonescu, C.R. Consistent MYC and FLT4 Gene Amplification in Radiation-Induced Angiosarcoma but Not in Other Radiation-Associated Atypical Vascular Lesions. Genes Chromosomes Cancer 2011, 50, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.L.; Ranga-Prasad, H.; Parson, M.A.H.; Harris, N.J.; Rathinaswamy, M.K.; Burke, J.E. Oncogenic Mutations of PIK3CA Lead to Increased Membrane Recruitment Driven by Reorientation of the ABD, P85 and C-Terminus. Nat. Commun. 2023, 14, 181. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Davoudi, Z.; Coussens, N.P.; Silvers, T.; Morris, J.; Takebe, N.; Said, R.; Moscow, J.A.; Doroshow, J.H.; Teicher, B.A. Combinatorial Screen of Targeted Agents with the PI3K Inhibitors Inavolisib, Alpelisib, Duvelisib, and Copanlisib in Multi-Cell Type Tumor Spheroids. SLAS Discov. Adv. Life Sci. R D 2025, 32, 100222. [Google Scholar] [CrossRef]
- Gulve, N.; Su, C.; Deng, Z.; Soldan, S.S.; Vladimirova, O.; Wickramasinghe, J.; Zheng, H.; Kossenkov, A.V.; Lieberman, P.M. DAXX-ATRX Regulation of P53 Chromatin Binding and DNA Damage Response. Nat. Commun. 2022, 13, 5033. [Google Scholar] [CrossRef]
- Wang, T.; Li, J.; Du, J.; Zhou, W.; Lu, G. Recent Advances in the Role of Atypical Cadherin FAT1 in Tumorigenesis (Review). Oncol. Lett. 2024, 29, 110. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, X.; Gao, L.; Xu, Y.; Yi, C. Differences in Potential Key Genes and Pathways between Primary and Radiation-Associated Angiosarcoma of the Breast. Transl. Oncol. 2022, 19, 101385. [Google Scholar] [CrossRef]
- Chang, C.-J.; Chao, C.-H.; Xia, W.; Yang, J.-Y.; Xiong, Y.; Li, C.-W.; Yu, W.-H.; Rehman, S.K.; Hsu, J.L.; Lee, H.-H.; et al. P53 Regulates Epithelial-Mesenchymal Transition (EMT) and Stem Cell Properties through Modulating miRNAs. Nat. Cell Biol. 2011, 13, 317–323. [Google Scholar] [CrossRef]
- Peng, Z.; Gong, Y.; Liang, X. Role of FAT1 in Health and Disease. Oncol. Lett. 2021, 21, 398. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q. The Roles of EZH2 in Cancer and Its Inhibitors. Med. Oncol. Northwood Lond. Engl. 2023, 40, 167. [Google Scholar] [CrossRef]
- Golde, T.E.; Koo, E.H.; Felsenstein, K.M.; Osborne, B.A.; Miele, L. γ-Secretase Inhibitors and Modulators. Biochim. Biophys. Acta 2013, 1828, 2898–2907. [Google Scholar] [CrossRef]
- Dai, J.; Wang, T.; Wang, W.; Zhang, S.; Liao, Y.; Chen, J. Role of MAPK7 in Cell Proliferation and Metastasis in Ovarian Cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10444–10451. [Google Scholar] [PubMed]
- Green, D.; Eyre, H.; Singh, A.; Taylor, J.T.; Chu, J.; Jeys, L.; Sumathi, V.; Coonar, A.; Rassl, D.; Babur, M.; et al. Targeting the MAPK7/MMP9 Axis for Metastasis in Primary Bone Cancer. Oncogene 2020, 39, 5553–5569. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.; Xu, P.-P.; Xu, Y.-H.; Guo, Y. A Bioinformatics Analysis: ZFHX4 Is Associated with Metastasis and Poor Survival in Ovarian Cancer. J. Ovarian Res. 2022, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Sun, J.; Ma, Y.; Chen, H.; Tian, C.; Dong, M. MSI2 Regulates NLK-Mediated EMT and PI3K/AKT/mTOR Pathway to Promote Pancreatic Cancer Progression. Cancer Cell Int. 2024, 24, 273. [Google Scholar] [CrossRef] [PubMed]
- Abelman, R.O.; Wu, B.; Barnes, H.; Medford, A.; Norden, B.; Putur, A.; Bitman, E.; Thant, W.; Liu, T.; Weipert, C.; et al. TOP1 Mutations and Cross-Resistance to Antibody–Drug Conjugates in Patients with Metastatic Breast Cancer. Clin. Cancer Res. 2025, 31, 1966–1974. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Y.; Xie, Y.; Yang, D.; Sun, Y.; Yuan, Y.; Chen, H.; Zhang, Y.; Huang, K.; Zheng, L. Histone H1.2 Promotes Hepatocarcinogenesis by Regulating Signal Transducer and Activator of Transcription 3 Signaling. Cancer Sci. 2022, 113, 1679–1692. [Google Scholar] [CrossRef]
- Miura, T.; Fujita, M.; Kawano, M.; Imadome, K.; Yasuda, T.; Nishihara, S.; Imamura, T.; Masuzawa, M.; Imai, T.; Nakayama, F. Strong Radioprotective FGF1 Signaling Down-Regulates Proliferative and Metastatic Capabilities of the Angiosarcoma Cell Line, ISOS-1, through the Dual Inhibition of EGFR and VEGFR Pathways. Clin. Transl. Radiat. Oncol. 2017, 7, 83–90. [Google Scholar] [CrossRef] [PubMed]


| Demographics | Category | n (%) |
|---|---|---|
| Sex | Male | 141 (40.8%) |
| Female | 194 (56.1%) | |
| Unknown | 11 (3.2%) | |
| Age category | Adult | 355 (98.9%) |
| Pediatric | 4 (1.1%) | |
| Ethnicity | Non-Hispanic | 264 (76.3%) |
| Unknown/Not Collected | 51 (14.7%) | |
| Hispanic | 31 (9.0%) | |
| Race | Asian | 24 (6.9%) |
| White | 239 (69.1%) | |
| Black | 20 (5.8%) | |
| Other | 28 (8.1%) | |
| Pacific Islander | 1 (0.3%) | |
| Not collected | 4 (1.2%) | |
| Unknown | 30 (8.7%) | |
| Sample Type | Primary | 252 (70.2%) |
| Metastasis | 79 (22.0%) | |
| Not Collected | 16 (4.5%) | |
| Unspecified | 12 (3.3%) |
| Gene (Chi-Squared) | Asian, n (%) | Non-Asian, n (%) | p Value |
|---|---|---|---|
| CHEK1 | 2 (8.33% | 0 (0.00%) | 7.950 × 10−3 |
| RBM10 | 2 (8.33%) | 0 (0.00%) | 8.197 × 10−3 |
| CBL | 3 (12.50%) | 3 (1.13%) | 8.580 × 10−3 |
| EXT1 | 1 (100%) | 0 (0.00%) | 0.0115 |
| FGF3 | 2 (8.70%) | 0 (0.00%) | 0.0116 |
| BRAF | 4 (16.00%) | 8 (2.95%) | 0.0124 |
| SETD2 | 3 (12.50% | 5 (1.91%) | 0.0221 |
| Gene (Chi-Squared) | White, n (%) | Non-White, n (%) | p Value |
| MYC | 4 (8.89%) | 64 (26.56%) | 0.0121 |
| GLI3 | 0 (0.00%) | 3 (13.64%) | 0.0307 |
| MAPK7 | 0 (0.00%) | 1 (50.00%) | 0.0417 |
| Gene (Chi-Squared) | Black, n (%) | Non-Black, n (%) | p Value |
| PRPF40B | 2 (100%) | 1 (2.50%) | 3.484 × 10−3 |
| CREBBP | 3 (15.00%) | 3 (1.13%) | 5.147 × 10−3 |
| AURKA | 2 (10.00%) | 1 (0.38%) | 0.0135 |
| KDM6A | 2 (10.00%) | 2 (0.75%) | 0.0258 |
| PARP1 | 2 (14.29%) | 2 (1.12%) | 0.0271 |
| STAT3 | 2 (10.00%) | 2 (0.79%) | 0.0279 |
| PPM1D | 2 (11.11%) | 3 (1.36%) | 0.0472 |
| Gene (Chi-Squared) | Pacific-Islander n (%) | Non-Pacific Islander, n (%) | p Value |
| PMS1 | 1 (100%) | 1 (0.35%) | 7.042 × 10−3 |
| BORCS8-MEF2B | 1 (100%) | 2 (0.68%) | 0.0101 |
| FOXA1 | 1 (100%) | 3 (1.16%) | 0.0154 |
| CYLD | 1 (100%) | 3 (1.23%) | 0.0163 |
| CALR | 1 (100%) | 5 (2.02%) | 0.0241 |
| PIK3CB | 1 (100%) | 5 (2.55%) | 0.030 |
| ARID1B | 1 (100%) | 10 (3.57%) | 0.0391 |
| Gene (Chi-Squared) | Male, n (%) | Female, n (%) | p Value |
| MYC | 8 (5.84%) | 66 (34.02%) | 2.14 × 10−10 |
| POT1 | 20 (27.78%) | 6 (6.52%) | 3.870 × 10−4 |
| NTRK2 | 1 (0.52%) | 8 (5.93%) | 4.232 × 10−3 |
| FGFR4 | 0 (0.00%) | 6 (4.35%) | 4.578 × 10−3 |
| PIK3C2B | 0 (0.00%) | 6 (12.50%) | 8.582 × 10−3 |
| FAT1 | 19 (14.84%) | 10 (5.71%) | 9.757 × 10−3 |
| HRAS | 14 (6.86%) | 3 (2.08%) | 0.0456 |
| Gene | Number of Samples by Mutation | Total Breast Samples | Percentage |
|---|---|---|---|
| MYC | 35 | 73 | 47.90% |
| KDR | 17 | 73 | 23.30% |
| FLT4 | 11 | 73 | 15.10% |
| PIK3CA | 9 | 73 | 12.30% |
| KMT2D | 10 | 73 | 13.70% |
| TP53 | 4 | 73 | 5.50% |
| Gene Pair | p Value | Impact of Mutation | Therapeutic Implications |
|---|---|---|---|
| TP53-ATRX | <0.001 | Dysregulation of telomere maintenance and DNA repair | Sensitivity to CHK1/2 or WEE1 inhibition |
| ARID1A-NOTCH1 | <0.001 | Dysregulation of chromatin remodeling via chromatin–Notch–Hippo/Wnt crosstalk | Sensitivity to EZH2 or γ-secretase inhibitors |
| ARID1A-NOTCH2 | 0.002 | Dysregulation of chromatin remodeling via chromatin–Notch–Hippo/Wnt crosstalk | Sensitivity to EZH2 or γ-secretase inhibitors |
| KDR-FLT4 | 0.022 | Dysregulation of VEGFR-driven angiogenesis | Sensitivity to VEGFR-targeted TKIs |
| TP53-FAT1 | <0.001 | Dysregulation of telomere maintenance, DNA repair, epithelial–mesenchymal transition | Sensitivity to CHK1/2, WEE1, or Wnt/β-catenin inhibition |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leach, E.; Jafari, A.; Torbenson, E.; Hsia, B.; Tauseef, A. Genomic and Demographic Characteristics of Angiosarcoma as Described in the AACR Project GENIE Registry. Cancers 2025, 17, 3663. https://doi.org/10.3390/cancers17223663
Leach E, Jafari A, Torbenson E, Hsia B, Tauseef A. Genomic and Demographic Characteristics of Angiosarcoma as Described in the AACR Project GENIE Registry. Cancers. 2025; 17(22):3663. https://doi.org/10.3390/cancers17223663
Chicago/Turabian StyleLeach, Eileen, Amir Jafari, Elijah Torbenson, Beau Hsia, and Abubakar Tauseef. 2025. "Genomic and Demographic Characteristics of Angiosarcoma as Described in the AACR Project GENIE Registry" Cancers 17, no. 22: 3663. https://doi.org/10.3390/cancers17223663
APA StyleLeach, E., Jafari, A., Torbenson, E., Hsia, B., & Tauseef, A. (2025). Genomic and Demographic Characteristics of Angiosarcoma as Described in the AACR Project GENIE Registry. Cancers, 17(22), 3663. https://doi.org/10.3390/cancers17223663






