Simple Summary
Mutations in PIK3CA occur in approximately one-third of hormone receptor-positive (HR+)/HER2-negative (HER2−) breast cancer (BC) and represent a target for specific inhibitors. The current study established the frequency and spectrum of PIK3CA alterations in HR+/HER2− BC from Russian patients. We conclude that the overall prevalence and characteristics of PIK3CA-positive cases are consistent with previously published data and do not depend on the patient’s ethnic background. We also show that PIK3CA testing should not be limited to hot-spot variants, as commercially available kits may miss up to 12% of PIK3CA-mutated cases.
Abstract
Background: Several targeted drugs have been recently approved for the treatment of PIK3CA-mutated hormone receptor-positive (HR+)/HER2-negative (HER2−) breast cancer (BC). This study aimed at a comprehensive evaluation of the spectrum of PIK3CA alterations in Russian BC patients. Methods: The tumor material from 1872 patients with ER+/HER2− BC was tested by a combination of PCR-based methods. Results: Mutations were detected in 693/1872 (37%) cases, including 46 BC with two PIK3CA lesions. The three most common substitutions (E542K, E545K, and H1047R) were identified in 542/693 (78%) PIK3CA-mutated cases, while as many as 5.5–12% of identified mutations were not potentially detectable by common commercial kits. The study included patients of Slavic and non-Slavic ethnicities residing in regions with different climate conditions, however, these factors did not influence the distribution of PIK3CA mutations. The presence of PIK3CA variants was associated with older patient age at diagnosis (p = 0.0002), smaller tumor size (p = 0.005), lower grade (p = 0.005), Ki67 <20% (p = 0.0001) and progesterone receptor-positive status (p = 0.002) at the initial disease diagnosis, and fewer distant metastases at the time of the detection of BC spread (p = 0.0001). In a subgroup of 413 BC patients who received adjuvant tamoxifen or aromatase inhibitors, PIK3CA mutations were not associated with resistance to either type of treatment. Conclusions: The results of this study highlight the need to extend the PIK3CA testing beyond the hotspot regions of this gene. Although PIK3CA alterations contribute to the pathogenesis of HR+/HER2− BC and represent a target for several novel drugs, they are not intrinsically associated with unfavorable clinical characteristics of this subtype of cancer disease.
1. Introduction
Mutations affecting PIK3CA, the gene encoding the catalytic subunit of p110-alpha kinase PI3K, represent one of the most common genetic driver events in human tumors [1]. The attempts to develop inhibitors of the PI3K/AKT/mTOR signaling cascade emerged some time ago, but it was not until recently that the balance between clinical efficacy and drug tolerability could be achieved. Monotherapy of solid neoplasms with PI3K inhibitors proved ineffective, however, several compounds have already been approved for the treatment of PIK3CA-mutated hormone-receptor (HR)-positive/HER2-negative breast cancer (BC) in combination with other agents. Combined use of a selective inhibitor of the PI3K-alpha alpelisib and estrogen degrader fulvestrant received approval for patients progressing on or after prior endocrine-based therapy [2,3]. Similar approval was granted to AKT inhibitor capivasertib; however, in addition to PIK3CA mutations, the indication of this drug includes nucleotide sequence alterations in AKT1 and PTEN genes [4]. Another AKT inhibitor, ipatasertib, is currently under investigation in breast cancer [5]. Inavolisib has the ability to drive the degradation of mutated p110-alpha while sparing the wild-type isoform of this protein; it has been approved for clinical use in combination with fulvestrant and CDK4/6 inhibitor palbociclib [6]. Several other PI3K inhibitors have failed to achieve a significant improvement in the progression-free survival or objective response rate, and their use has been associated with considerable toxicity (e.g., buparlisib, pictilisib, and taselisib). A number of novel compounds, including inhibitors that selectively target mutant PIK3CA isoforms, have shown promising results in preclinical and early-phase clinical studies [5]. There are also ongoing trials utilizing combinations of PI3K inhibitors with other drugs and involving cancer types other than BC [7,8,9,10].
PIK3CA mutations affect a variety of biological properties of tumor cells, including some essential metabolic features, profiles of secreted immunocytokines, regulation of the cell cycle, formation of tolerance to polyploidy and chromosomal instability, etc. [11,12,13]. Activating PIK3CA lesions were shown to have a prognostic value in breast cancer, however, their role may differ in early- and late-stage tumors. In particular, PIK3CA mutations appear to be associated with a favorable prognosis in early BC, while in metastatic HR+ and/or HER2+ carcinomas, they may be enriched in cases with unfavorable disease course and resistance to standard therapy schemes [14,15,16]. The presence of PIK3CA alterations negatively affects the results of targeted therapy in HER2-positive BC [17,18,19,20].
There are several hotspot PIK3CA regions most frequently affected by somatic alterations. About 70% of identified mutations are located in codons 542, 545 (exon 9, helical domain) and 1047 (exon 20, kinase domain). Substitutions in the helical domain initiate transmission of a signal through the RAS-MAPK molecular cascade independently of the regulatory PI3K subunit, p85. In contrast, kinase domain mutations are associated with the requirement for interaction with p85, but the activation of signaling does not depend on RAS-GTP binding [21]. Recently, a phenomenon of multiple (usually two) co-occurring PIK3CA mutations was described in breast cancer and other tumor types: these mutations occur mainly in cis, are mutually exclusive with other driver genetic alterations, and confer increased sensitivity to PI3K inhibition [22,23]. PIK3CA H1047R substitution was shown to be associated with a low proportion of complete responses to anthracycline and taxane therapy in triple-negative breast cancer, and with resistance to anti-HER2 therapy in HER2-positive tumors [24,25]. Mutations in the helical domain occur more frequently in well-differentiated neoplasms of the luminal A expression subtype, as well as in lobular BC [26,27]. Despite the functional differences, the predictive role of exon 9 and exon 20 mutations in relation to alpelisib seems to be similar [2]. Less frequently, somatic mutations are observed in the C2 domain of PI3K (exon 7) and in the other gene parts. The clinical and biological significance of atypical mutations in the helical and kinase domains, as well as of rare alterations in PIK3CA, have not yet been fully characterized [28], although the evidence for correlation of several types of rare variants with clinical benefit from PI3K inhibition continues to emerge [29,30].
There is some evidence that the occurrence of PIK3CA mutations in breast cancer may have interethnic differences [31,32,33]. The aim of the current study was to analyze the frequency, spectrum, and clinical associations of PIK3CA lesions in Russian patients with HR+/HER2− breast cancer.
2. Materials and Methods
The study included 1872 patients with ER+/HER2− breast cancer diagnosed between 1980 and 2022 and treated in more than 50 cities of Russia. All patients were referred to PIK3CA mutation analysis in 2020–2023, with about 85% of them suffering from the recurrent or metastatic disease at the time of genetic testing. Approximately half of patients underwent PIK3CA testing in the first 4 years after the disease manifestation. The study cohort consisted of 1861 women and 11 men with breast tumors. The median age of the patients was 51 years (range: 20–88 years). DNA for genetic analysis was extracted from formalin-fixed paraffin-embedded (FFPE) primary tumor tissues (≈70%) or FFPE material from tumors obtained from tumor recurrence/metastasis (≈30%). Testing for PIK3CA mutations was performed by a combination of high-resolution melting (HRM) analysis, allelic discrimination TaqMan PCR (AD-PCR), digital droplet PCR, and pyrosequencing. The presence of alterations in exons 2 (codons 64–117), 7 (codons 418–455), 10 (codons 526–552), 20 (codons 1027–1057) was determined by HRM of PCR products; mutations G118D, N345K, E726K were investigated by AD-PCR. Cases showing abnormal melting patterns were tested for hot-spot variants by the corresponding AD-PCR assays (exon 2: codons 81, 88, 108, 110, 111; exon 7: codons 420, 453; exon 10: codons 542, 545, 546; exon 20: codons 1043, 1044, 1047, 1049). Tumor samples with equivocal results were further analyzed by digital droplet PCR assays for the mutations E542K, E545K, H1047R, and H1047L. Cases with abnormal melting curves, which were negative for the hot-spot variants, were subsequently subjected to pyrosequencing. The list of primers and TaqMan probes, assay conditions, and utilized equipment is given in Table S1.
Associations between PIK3CA alterations and clinicopathological parameters were estimated using the chi-squared test. The Mann–Whitney U test was used to compare age in patients with different PIK3CA status. Event-free survival (EFS) was determined as the interval between diagnosis and detection of disease recurrence or distant metastasis in stage I-III cases. Data on the time of disease progression were extracted from available medical records. Kaplan–Meier method and log-rank test were used to construct and compare survival curves to determine the parameters affecting EFS. The Cox proportional hazards regression model was used to assess the prognostic significance of multiple parameters simultaneously. Variables were entered into the Cox regression model using a stepwise method. p < 0.05 was considered statistically significant. Statistical analyses were performed using MedCalc software (Version 19).
3. Results
PIK3CA mutations were identified in 693/1872 (37.0%) patients (687/1861 (36.9%) female and 6/11 (54.5%) male BC cases). Forty-six out of 693 tumors (6.6%) harbored two PIK3CA alterations simultaneously (Table S2). At least one mutation in exon 10 or 20 was identified in the majority of PIK3CA-positive BC (624/693, 90%). The most frequent substitution types were known major hotspots: p.H1047R in exon 20 (310/739, 42.0%), p.E545K (153/739, 20.7%), and p.E542K (84/739, 11.4%) in exon 10. These three alterations were identified in 542/693 (78.2%) PIK3CA-mutated cases. Several other missense variants occurred more than 10 times: p.H1047L (n = 30), p.N345K (n = 28), p.E726K (n = 23), p.C420R (n = 17), p.Q546K (n = 15), p.E453K (n = 12) (Figure 1). P.E726K substitution (exon 14) was predominantly observed in double-mutated cases (18/23, 78.3%). There was a trend towards a higher mutation detection rate in primary tumor samples than in metastatic material (39.0% vs. 34.3%, p = 0.107).
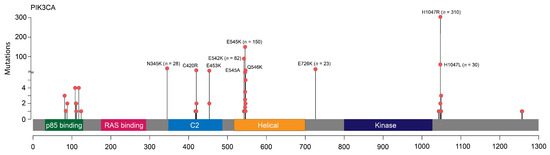
Figure 1.
Schematic representation of the spectrum of PIK3CA mutations.
The median age at BC diagnosis was higher in PIK3CA-mutated cases (53.0 vs. 50.0 years, p = 0.0002); PIK3CA variants were detected in 43.9% of patients older than 60 years compared to 33.9% in younger individuals (p = 0.0002). PIK3CA alterations were associated with a number of favorable prognostic features: smaller tumor size, lower grade, Ki67 <20%, and positive PR status (p ≤ 0.005 for all comparisons) (Figure 2, Table 1). In our case series, about 85% of patients had metastatic disease at the time of PIK3CA testing. PIK3CA mutations were found less frequently in cases with multiple metastases than in patients with only one metastasis-affected zone (32.8% vs. 43.1%, p = 0.001). Mutations also tended to be underrepresented in patients with distant metastases to soft tissues, skin, and lymph nodes (Table 1).
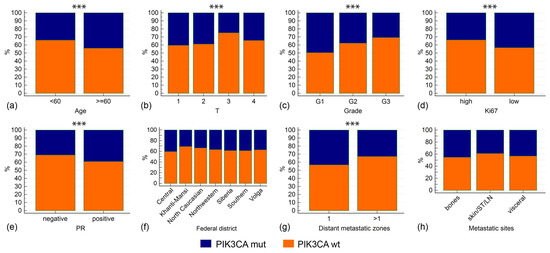
Figure 2.
Associations between PIK3CA mutations and clinicopathological characteristics. Asterisks note the comparisons with p < 0.01. Clinicopathological characteristics include age at diagnosis (a), tumor size (b), tumor grade (c), Ki67 proliferation index (d), PR status (e), Federal district (f), number of metastatic zones (g), types of metastatic sites (h). In (f), only Federal districts with >100 analyzed cases are shown. In (h): ST—soft tissues, LN—lymph nodes.

Table 1.
Clinical and pathological characteristics of the studied BC samples in relation to PIK3CA mutations.
Our study sample included BC patients residing in different parts of Russia. Women living in the North Caucasus belong mostly to non-Slavic ethnic groups (Kabardins, Ossetins, Chechens, Karachays, Avars, Ingush) and are generally characterized by earlier age at first delivery and a higher number of childbirths when compared to Western-style communities. For example, the total fertility rate in 2023 was 1.24 in North Western Russia and 1.72 in the North Caucasus [34]. The above factors significantly affect BC epidemiology: BC incidence per 100,000 women in the North Caucasus is among the lowest in Russia [35]. However, patients from this region had no specific features with regard to the spectrum and frequency of PIK3CA mutations. Other regions with predominantly non-Slavic populations, e.g., the Republics of Tatarstan and Bashkortostan, also had distribution of PIK3CA alterations similar to the general sample. Women residing in an extremely cold climate (e.g., Khanti-Mansi region) had essentially the PIK3CA mutation rate as patients living in Southern Russia (Table 2, Figure 2).

Table 2.
PIK3CA mutation frequency in different regions of Russia.
Median event-free survival (EFS) in stage I–III tumors was 3 years. In univariable analysis, longer EFS was associated with patients’ age less than 60 years, lower tumor stage, absence of nodal involvement, and smaller tumor size. PIK3CA mutations had no effect on EFS (Table 3, Figure 3). In Cox regression analysis, including age, T, and N categories, these variables retained statistical significance in relation to EFS.

Table 3.
Univariable and multivariable analysis of the factors associated with event-free survival.
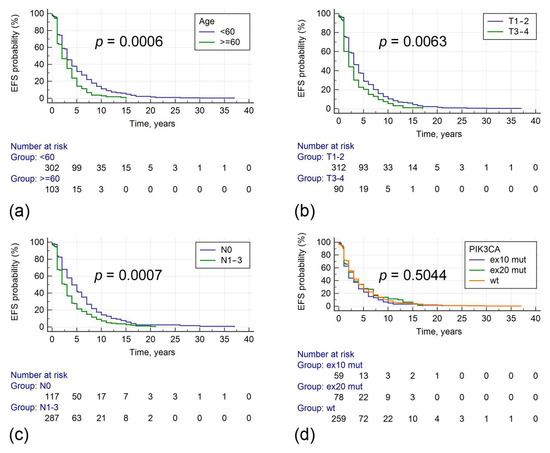
Figure 3.
Associations between clinical parameters, PIK3CA mutations, and event-free survival. The evaluated parameters include age at diagnosis (a), tumor size (b), regional lymph node status (c), PIK3CA mutations (d).
We were able to obtain the data on sensitivity to adjuvant hormonal therapy in 413 BC cases: 331 patients received tamoxifen and 82 were treated by aromatase inhibitors (anastrozole or letrozole). Patients were categorized as having primary endocrine resistance (a relapse within the first two years of adjuvant therapy), secondary resistance (defined as disease progression detected after the first two years of adjuvant therapy or within a year after treatment completion), or sensitivity (disease relapse diagnosed in more than a year after completion of adjuvant treatment or later) [36]. PIK3CA mutation prevalence did not significantly differ between cases with sensitivity or resistance to aromatase inhibitors or tamoxifen (Table 4).

Table 4.
Frequency of PIK3CA mutations in patients with various sensitivities to adjuvant endocrine therapy.
4. Discussion
This study analyzed a large sample of Russian patients with HR+/HER2− BC who were referred for PIK3CA genetic testing between 2020 and 2023. The spectrum of the mutations identified was generally consistent with the published data [37,38], although the incidence of PIK3CA alterations was lower (37%) compared to that in European patients with ER+/HER2− BC (7706/17,687, 44%) [33]. The most plausible explanation for these differences is the distinct age distribution in the patient series. Indeed, our BC group had a relatively low median age at diagnosis (51 years) compared to the other large BC cohorts (58–61 years) [33,39]. Importantly, in our study, the frequency of PIK3CA mutations in subjects older than 60 years approached 44%. Similar to previous observations, the three main hotspot variants (p.H1047R, p.E545K, p.E542K) constituted 78% of all PIK3CA mutations [40].
Double mutations account for 12–15% of all BC cases with PIK3CA alterations and confer increased benefit from alpelisib therapy [22,23]. Our cohort contained 6,6% of co-occurring PIK3CA alterations. Most of the double mutations (40/46, 87%) included variants in major hotspot codons E542, E545, and H1047. Substitution E726K was enriched in cases with multiple mutations: it was observed as a single variant in only 5/23 BC (21.7%) and was one of the co-occurring variants in 18/46 (39%) double-mutated cases. The second common minor mutation in double-mutated BC was E453K/Q, which is consistent with previous findings [22].
Currently, there are a number of commercially available PCR-based PIK3CA mutation kits aimed at detecting the most common missense variants (e.g., therascreen PIK3CA RGQ PCR Kit (QIAGEN Manchester, Manchester, UK; cobas PIK3CA Mutation Test (Roche Diagnostics, Mannheim, Germany); AmoyDx PIK3CA Mutation Detection Kit (Amoy Diagnostics, Xiamen, China); Idylla PIK3CA-AKT1 Mutation Assay (BioCartis Idylla, Mechelen, Belgium, etc.)). These kits detect 11–17 PIK3CA variants. Using our stepwise PCR-based approach, we identified 43 unique mutation types, including 39 missense variants and 4 indels. Twenty-five of these variants were identified more than once, 10 of which could not be targeted by any of the commercial assays mentioned. Notably, neither of the above commercial assays included testing for the E453K and E726K variants, which were detected 12 and 23 times, respectively, in our dataset. Compared to our approach, Cobas, Idylla, Therascreen, and AmoyDx PIK3CA assays would miss 38/693 (5.5%), 42/693 (6.1%), 83/693 (12%), and 83/693 (12%) PIK3CA-mutated cases, respectively.
Most of the identified PIK3CA alterations (37 out of 43 mutation types and 731 out of a total of 739 mutations) have been reported previously. However, we could not find any mention of several of the identified variants (p.P124S, p.H419_C420delinsR, p.H419_P421>QT, p.C420_P421del, p.N1044H, and p.H1048A) in the literature. There is experimental evidence of the activating role of the majority of the found alterations (30/43 mutation types; 723/739 individual mutations) [28,41,42,43,44,45,46]. Nevertheless, the functional significance of the newly identified mutation types remains to be evaluated.
PIK3CA mutation status may be discordant between primary and metastatic tumor lesions in up to 10% of BC, with most of the discrepancy attributed to the loss of PIK3CA in metastatic tissues [47]. In accordance with this, we observed a trend towards lower PIK3CA occurrence in metastatic material.
Accumulating evidence suggests that the impact of PIK3CA mutations on tumor behavior and prognosis depends on the BC subtype. In HR+/HER2− BC, PIK3CA has been related to improved clinical outcome in operable primary cases, and to poor prognosis in metastatic disease [14,15,48,49,50]. We could not demonstrate an association between PIK3CA alterations and event-free survival in stage I-III BC, although their presence was strongly associated with smaller tumor size, lower grade, Ki67<20%, and PR protein expression. We also found that PIK3CA variants were more prevalent in patients with lower metastatic burden (43% vs. 33%, p = 0.0001). Our findings on the clinical associations of PIK3CA alterations generally replicate the data from several previous studies involving at least 400–500 BC cases [15,42,51,52,53,54,55]. At the same time, in most of these works, authors did not distinguish between individual BC subtypes, and the associations were described for the entire study sample. Our study included a large number of cases and focused on a relatively homogenous category of BC, i.e., patients with ER+/HER2− tumors.
ER and PI3K/AKT/mTOR signaling pathways are characterized by a complex interplay in breast tumors, and their coordinated activation supports the survival of ER-positive BC cells [56]. It is generally accepted that the activated PI3K/AKT/mTOR molecular cascade is implicated in endocrine resistance [57,58]; however, PIK3CA mutations were not associated with reduced benefit from adjuvant endocrine therapy in most of the previous studies. In fact, they were not significantly associated with outcomes of adjuvant tamoxifen therapy [55,59,60,61] and showed conflicting associations in relation to aromatase inhibitors. Ramirez-Ardila et al. [60] demonstrated an increased benefit of aromatase inhibitors in PIK3CA-mutated cases, while a recent study reported an opposite effect [55]. However, the authors of the latter work failed to validate their findings in an independent cohort [62]. We did not find an enrichment of PIK3CA mutations in patients with primary or secondary resistance to adjuvant tamoxifen or aromatase inhibitors. It was suggested that PIK3CA alterations do not confer pronounced resistance to endocrine therapy because they render only weak activation of the canonical PI3K pathway and probably exert their oncogenic potential through alternative molecular mechanisms [59,63].
5. Conclusions
In conclusion, our results highlight the importance of expanded testing of the PIK3CA gene, not limited to hotspot mutations. Although PIK3CA alterations contribute to the pathogenesis of HR+/HER2− BC and represent a target for several novel drugs, they are not intrinsically associated with unfavorable features of this subtype of cancer disease.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers17111833/s1, Table S1: Description of methods for detection of PIK3CA mutations; Table S2: List of double mutations in the PIK3CA gene.
Author Contributions
Conceptualization, T.N.S. and E.N.I.; methodology, A.S.N., A.S.M., Y.V.B. and Y.S.Z.; formal analysis, T.M.S., Y.S.Z., A.P.C. and S.N.A.; investigation, L.M.M., Z.K.K., V.V.K., A.I.N., N.G.R., D.Y.Y., D.V.R., Y.A.T., A.Y.G., M.I.G., R.A.Z., Y.N.M., A.V.S. (Aleksandr V. Sultanbaev), E.N.V., L.D.O., M.D.C., Y.I.B., I.O.S., Z.M.K., L.V.B., V.I.V., K.S.S., S.A.T., V.L.C., A.A.G., A.S.S., P.I.K., A.G.K., I.A.B., M.B.B., Y.A.U., A.V.S. (Aleksandr V. Shkradyuk), S.G.P., T.A.K., V.N.D., I.K.A., A.V.F., E.I.R., A.A.V., A.E.V., E.L.S., I.S.S., K.T.A., O.L.P., K.S.M., A.I.S., A.V.Z., A.S.A., V.N.P., A.N.L., A.E.K., A.V.A., D.D.G., N.A.K., D.A.M., G.G.R., S.P.E., V.S.B., A.V.V., N.O.P., S.V.O., K.A.S., V.E.G., M.V.K., K.V.M., I.M.K., D.V.K., A.M.D., A.A.E., S.Y.B., C.K.G., Y.V.M., E.A.B. and M.A.M.; data curation, R.V.O.; writing—original draft preparation, T.N.S., G.A.Y. and A.G.I.; writing—review and editing, E.N.I. and S.N.A.; visualization, P.R.K. and J.O.B. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by Novartis Pharma (agreement number 2545, dated 27 July 2020, between FSBI “N.N. Petrov National Medical Research Center of Oncology” and Novartis Pharma). The sponsors partially financed the PIK3CA laboratory testing and the analysis of clinical data. The sponsors did not influence the study design, selection of patients for the study, interpretation of the data, preparation of the paper, or decision to submit the report for publication.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the NMRC of Oncology named after N.N. Petrov (protocol 20/25 approved 23 January 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further enquiries can be directed to the corresponding author.
Conflicts of Interest
Author Svetlana V. Odintsova was employed by the company EuroCityClinic LLC, The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.M.; Juric, D.; Loibl, S.; Campone, M.; Mayer, I.A.; Rubovszky, G.; Yamashita, T.; Kaufman, B.; Lu, Y.S.; et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: Final overall survival results from SOLAR-1. Ann. Oncol. 2021, 32, 208–217. [Google Scholar] [CrossRef]
- Turner, N.C.; Oliveira, M.; Howell, S.J.; Dalenc, F.; Cortes, J.; Gomez Moreno, H.L.; Hu, X.; Jhaveri, K.; Krivorotko, P.; Loibl, S.; et al. Capivasertib in Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2023, 388, 2058–2070. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Wei, Y.; Meng, W.; Zhang, J.; Yang, X. PI3K/AKT/mTOR inhibitors for hormone receptor-positive advanced breast cancer. Cancer Treat. Rev. 2025, 132, 102861. [Google Scholar] [CrossRef]
- Jhaveri, K.L.; Accordino, M.K.; Bedard, P.L.; Cervantes, A.; Gambardella, V.; Hamilton, E.; Italiano, A.; Kalinsky, K.; Krop, I.E.; Oliveira, M.; et al. Phase I/Ib Trial of Inavolisib Plus Palbociclib and Endocrine Therapy for PIK3CA-Mutated, Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced or Metastatic Breast Cancer. J. Clin. Oncol. 2024, 42, 3947–3956. [Google Scholar] [CrossRef]
- Ye, Y.; Huang, Z.; Zhang, M.; Li, J.; Zhang, Y.; Lou, C. Synergistic therapeutic potential of alpelisib in cancers (excluding breast cancer): Preclinical and clinical evidences. Biomed. Pharmacother. 2023, 159, 114183. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Gonzalez-Martin, A.; Cruz, F.M.; Friedlander, M.; Glasspool, R.; Lorusso, D.; Marth, C.; Monk, B.J.; Kim, J.W.; Hinson, P.; et al. EPIK-O/ENGOT-OV61: Alpelisib plus olaparib vs cytotoxic chemotherapy in high-grade serous ovarian cancer (phase III study). Future Oncol. 2022, 18, 3481–3492. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Morita, T.Y.; Ohashi, A.; Haeno, H.; Hakozaki, Y.; Fujii, M.; Kashima, Y.; Kobayashi, S.S.; Mukohara, T. Combination treatment with a PI3K/Akt/mTOR pathway inhibitor overcomes resistance to anti-HER2 therapy in PIK3CA-mutant HER2-positive breast cancer cells. Sci. Rep. 2020, 10, 21762. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Abramson, V.G.; Sanders, M.E.; Mayer, E.L.; Haddad, T.C.; Nanda, R.; Van Poznak, C.; Storniolo, A.M.; Nangia, J.R.; Gonzalez-Ericsson, P.I.; et al. TBCRC 032 IB/II Multicenter Study: Molecular Insights to AR Antagonist and PI3K Inhibitor Efficacy in Patients with AR(+) Metastatic Triple-Negative Breast Cancer. Clin. Cancer Res. 2020, 26, 2111–2123. [Google Scholar] [CrossRef]
- Berenjeno, I.M.; Piñeiro, R.; Castillo, S.D.; Pearce, W.; McGranahan, N.; Dewhurst, S.M.; Meniel, V.; Birkbak, N.J.; Lau, E.; Sansregret, L.; et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat. Commun. 2017, 8, 1773. [Google Scholar] [CrossRef] [PubMed]
- Sobral-Leite, M.; Salomon, I.; Opdam, M.; Kruger, D.T.; Beelen, K.J.; van der Noort, V.; van Vlierberghe, R.L.P.; Blok, E.J.; Giardiello, D.; Sanders, J.; et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019, 21, 90. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Karali, E.; Tripp, A.; Valle, A.; Inglese, P.; Perry, N.J.S.; Magee, D.J.; Anjomani Virmouni, S.; Elder, G.A.; Tyson, A.L.; et al. Metabolic Fingerprinting Links Oncogenic PIK3CA with Enhanced Arachidonic Acid-Derived Eicosanoids. Cell 2020, 181, 1596–1611.e27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.R.; Jiang, Y.Z.; Zuo, W.J.; Yu, K.D.; Shao, Z.M. PIK3CA mutations define favorable prognostic biomarkers in operable breast cancer: A systematic review and meta-analysis. Onco Targets Ther. 2014, 7, 543–552. [Google Scholar] [CrossRef]
- Mollon, L.E.; Anderson, E.J.; Dean, J.L.; Warholak, T.L.; Aizer, A.; Platt, E.A.; Tang, D.H.; Davis, L.E. A Systematic Literature Review of the Prognostic and Predictive Value of PIK3CA Mutations in HR+/HER2− Metastatic Breast Cancer. Clin. Breast Cancer 2020, 20, e232–e243. [Google Scholar] [CrossRef]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Tran Dien, A.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef]
- Shi, Q.; Xuhong, J.; Tian, H.; Qu, M.; Zhang, Y.; Jiang, J.; Qi, X. Predictive and prognostic value of PIK3CA mutations in HER2-positive breast cancer treated with tyrosine kinase inhibitors: A systematic review and meta-analysis. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188847. [Google Scholar] [CrossRef]
- Loibl, S.; Majewski, I.; Guarneri, V.; Nekljudova, V.; Holmes, E.; Bria, E.; Denkert, C.; Schem, C.; Sotiriou, C.; Loi, S.; et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: Pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann. Oncol. 2016, 27, 1519–1525. [Google Scholar] [CrossRef]
- Fan, H.; Li, C.; Xiang, Q.; Xu, L.; Zhang, Z.; Liu, Q.; Zhang, T.; Zhou, Y.; Zhao, X.; Cui, Y. PIK3CA mutations and their response to neoadjuvant treatment in early breast cancer: A systematic review and meta-analysis. Thorac. Cancer 2018, 9, 571–579. [Google Scholar] [CrossRef]
- Kim, J.W.; Lim, A.R.; You, J.Y.; Lee, J.H.; Song, S.E.; Lee, N.K.; Jung, S.P.; Cho, K.R.; Kim, C.Y.; Park, K.H. PIK3CA Mutation is Associated with Poor Response to HER2-Targeted Therapy in Breast Cancer Patients. Cancer Res. Treat. 2023, 55, 531–541. [Google Scholar] [CrossRef]
- Zhao, L.; Vogt, P.K. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): Differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 2010, 9, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Razavi, P.; Johnson, J.L.; Shao, H.; Shah, H.; Antoine, A.; Ladewig, E.; Gorelick, A.; Lin, T.Y.; Toska, E.; et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 2019, 366, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S.; Jin, D.X.; Rathod, R.; Ross, J.; Cantley, L.C.; Scaltriti, M.; Chen, J.W.; Hutchinson, K.E.; Wilson, T.R.; Sokol, E.S.; et al. Genetic Heterogeneity and Tissue-specific Patterns of Tumors with Multiple PIK3CA Mutations. Clin. Cancer Res. 2023, 29, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Garay, J.P.; Smith, R.; Devlin, K.; Hollern, D.P.; Liby, T.; Liu, M.; Boddapati, S.; Watson, S.S.; Esch, A.; Zheng, T.; et al. Sensitivity to targeted therapy differs between HER2-amplified breast cancer cells harboring kinase and helical domain mutations in PIK3CA. Breast Cancer Res. 2021, 23, 81. [Google Scholar] [CrossRef]
- Guo, S.; Loibl, S.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Denkert, C. PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane-Based Neoadjuvant Chemotherapy. Cancer Res. Treat. 2020, 52, 689–696. [Google Scholar] [CrossRef]
- Barbareschi, M.; Buttitta, F.; Felicioni, L.; Cotrupi, S.; Barassi, F.; Del Grammastro, M.; Ferro, A.; Dalla Palma, P.; Galligioni, E.; Marchetti, A. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin. Cancer Res. 2007, 13, 6064–6069. [Google Scholar] [CrossRef]
- Papaxoinis, G.; Kotoula, V.; Alexopoulou, Z.; Kalogeras, K.T.; Zagouri, F.; Timotheadou, E.; Gogas, H.; Pentheroudakis, G.; Christodoulou, C.; Koutras, A.; et al. Significance of PIK3CA Mutations in Patients with Early Breast Cancer Treated with Adjuvant Chemotherapy: A Hellenic Cooperative Oncology Group (HeCOG) Study. PLoS ONE 2015, 10, e0140293. [Google Scholar] [CrossRef][Green Version]
- Dogruluk, T.; Tsang, Y.H.; Espitia, M.; Chen, F.; Chen, T.; Chong, Z.; Appadurai, V.; Dogruluk, A.; Eterovic, A.K.; Bonnen, P.E.; et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 2015, 75, 5341–5354. [Google Scholar] [CrossRef]
- Spangle, J.M.; Von, T.; Pavlick, D.C.; Khotimsky, A.; Zhao, J.J.; Roberts, T.M. PIK3CA C-terminal frameshift mutations are novel oncogenic events that sensitize tumors to PI3K-α inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 24427–24433. [Google Scholar] [CrossRef]
- Rugo, H.S.; Raskina, K.; Schrock, A.B.; Madison, R.W.; Graf, R.P.; Sokol, E.S.; Sivakumar, S.; Lee, J.K.; Fisher, V.; Oxnard, G.R.; et al. Biology and Targetability of the Extended Spectrum of PIK3CA Mutations Detected in Breast Carcinoma. Clin. Cancer Res. 2023, 29, 1056–1067. [Google Scholar] [CrossRef]
- Ademuyiwa, F.O.; Tao, Y.; Luo, J.; Weilbaecher, K.; Ma, C.X. Differences in the mutational landscape of triple-negative breast cancer in African Americans and Caucasians. Breast Cancer Res. Treat. 2017, 161, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Omilian, A.R.; Wei, L.; Hong, C.C.; Bandera, E.V.; Liu, S.; Khoury, T.; Ambrosone, C.B.; Yao, S. Somatic mutations of triple-negative breast cancer: A comparison between Black and White women. Breast Cancer Res. Treat. 2020, 182, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Murugesan, K.; Newberg, J.Y.; Sokol, E.S.; Savage, H.M.; Stout, T.J.; Maund, S.L.; Hutchinson, K.E. Comparison of PIK3CA Mutation Prevalence in Breast Cancer Across Predicted Ancestry Populations. JCO Precis. Oncol. 2022, 6, e2200341. [Google Scholar] [CrossRef] [PubMed]
- Russian Unified Interdepartmental Information and Statistical System. Available online: https://www.fedstat.ru/indicator/31517 (accessed on 5 February 2025).
- Kaprin, A.D.; Starinsky, V.V.; Shakhzadova, A.O. Malignant Tumors in Russia in 2023 (Morbidity and Mortality); P.A. Gertsen Moscow Research Institute of Oncology—Branch of the Federal State Budgetary Institution «Scientific Medical Research Centre of Radiology» of the Ministry of Health of Russia: Moscow, Russia, 2024; 276p. (In Russian) [Google Scholar]
- Lambertini, M.; Blondeaux, E.; Bisagni, G.; Mura, S.; De Placido, S.; De Laurentiis, M.; Fabi, A.; Rimanti, A.; Michelotti, A.; Mansutti, M.; et al. Prognostic and clinical impact of the endocrine resistance/sensitivity classification according to international consensus guidelines for advanced breast cancer: An individual patient-level analysis from the Mammella InterGruppo (MIG) and Gruppo Italiano Mammella (GIM) studies. EClinicalMedicine 2023, 59, 101931. [Google Scholar] [CrossRef]
- Anderson, E.J.; Mollon, L.E.; Dean, J.L.; Warholak, T.L.; Aizer, A.; Platt, E.A.; Tang, D.H.; Davis, L.E. A Systematic Review of the Prevalence and Diagnostic Workup of PIK3CA Mutations in HR+/HER2− Metastatic Breast Cancer. Int. J. Breast Cancer 2020, 2020, 3759179. [Google Scholar] [CrossRef]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Rajadurai, P.; Semiglazova, T.; Hegmane, A.; Karak, F.; Chiu, J.; Gupta, S.; Azim, H.; Kitzen, J.; Arnaud, A.; Haftchenary, S.; et al. PIK3CA Registry: A Non-interventional, Descriptive, Retrospective Cohort Study of PIK3CA Mutations in Patients With Hormone Receptor-Positive (HR+), Human Epidermal Growth Factor Receptor 2-Negative (HER2−) Advanced Breast Cancer (ABC). Cancer Res. 2022, 82, 5–13. [Google Scholar] [CrossRef]
- Gymnopoulos, M.; Elsliger, M.A.; Vogt, P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA 2007, 104, 5569–5574. [Google Scholar] [CrossRef]
- Jin, N.; Keam, B.; Cho, J.; Lee, M.J.; Kim, H.R.; Torosyan, H.; Jura, N.; Ng, P.K.; Mills, G.B.; Li, H.; et al. Therapeutic implications of activating noncanonical PIK3CA mutations in head and neck squamous cell carcinoma. J. Clin. Investig. 2021, 131, e150335. [Google Scholar] [CrossRef]
- Ikenoue, T.; Kanai, F.; Hikiba, Y.; Obata, T.; Tanaka, Y.; Imamura, J.; Ohta, M.; Jazag, A.; Guleng, B.; Tateishi, K.; et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005, 65, 4562–4567. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.K.; Li, J.; Jeong, K.J.; Shao, S.; Chen, H.; Tsang, Y.H.; Sengupta, S.; Wang, Z.; Bhavana, V.H.; Tran, R.; et al. Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer Cell 2018, 33, 450–462.e10. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Okada, J.; Timmerman, L.; Rodriguez-Viciana, P.; Stokoe, D.; Shoji, K.; Taketani, Y.; Kuramoto, H.; Knight, Z.A.; Shokat, K.M.; et al. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008, 68, 8127–8136. [Google Scholar] [CrossRef] [PubMed]
- Miled, N.; Yan, Y.; Hon, W.C.; Perisic, O.; Zvelebil, M.; Inbar, Y.; Schneidman-Duhovny, D.; Wolfson, H.J.; Backer, J.M.; Williams, R.L. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 2007, 317, 239–242. [Google Scholar] [CrossRef]
- Rosin, J.; Svegrup, E.; Valachis, A.; Zerdes, I. Discordance of PIK3CA mutational status between primary and metastatic breast cancer: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2023, 201, 161–169. [Google Scholar] [CrossRef]
- Dumont, A.G.; Dumont, S.N.; Trent, J.C. The favorable impact of PIK3CA mutations on survival: An analysis of 2587 patients with breast cancer. Chin. J. Cancer 2012, 31, 327–334. [Google Scholar] [CrossRef]
- Zardavas, D.; Te Marvelde, L.; Milne, R.L.; Fumagalli, D.; Fountzilas, G.; Kotoula, V.; Razis, E.; Papaxoinis, G.; Joensuu, H.; Moynahan, M.E.; et al. Tumor PIK3CA Genotype and Prognosis in Early-Stage Breast Cancer: A Pooled Analysis of Individual Patient Data. J. Clin. Oncol. 2018, 36, 981–990. [Google Scholar] [CrossRef]
- Fillbrunn, M.; Signorovitch, J.; André, F.; Wang, I.; Lorenzo, I.; Ridolfi, A.; Park, J.; Dua, A.; Rugo, H.S. PIK3CA mutation status, progression and survival in advanced HR+/HER2− breast cancer: A meta-analysis of published clinical trials. BMC Cancer 2022, 22, 1002. [Google Scholar] [CrossRef]
- Cizkova, M.; Susini, A.; Vacher, S.; Cizeron-Clairac, G.; Andrieu, C.; Driouch, K.; Fourme, E.; Lidereau, R.; Bièche, I. PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res. 2012, 14, R28. [Google Scholar] [CrossRef]
- Kalinsky, K.; Jacks, L.M.; Heguy, A.; Patil, S.; Drobnjak, M.; Bhanot, U.K.; Hedvat, C.V.; Traina, T.A.; Solit, D.; Gerald, W.; et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin. Cancer Res. 2009, 15, 5049–5059. [Google Scholar] [CrossRef]
- Wu, H.; Wang, W.; Du, J.; Li, H.; Wang, H.; Huang, L.; Xiang, H.; Xie, J.; Liu, X.; Li, H.; et al. The distinct clinicopathological and prognostic implications of PIK3CA mutations in breast cancer patients from Central China. Cancer Manag. Res. 2019, 11, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Du, C.; Zhang, Y.; Wu, F.; Jin, Y.; Chen, X.; Liu, X.; Feng, C.; Ma, X.; Zhang, S. Clinicopathological characteristics and prognostic analysis of PIK3CA mutation in breast cancer patients in Northwest China. Pathol. Res. Pract. 2022, 238, 154063. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, K.; Stückrath, K.; Hartung, C.; Kaufhold, S.; Uleer, C.; Hanf, V.; Lantzsch, T.; Peschel, S.; John, J.; Pöhler, M.; et al. PIK3CA-mutations in breast cancer. Breast Cancer Res. Treat. 2022, 196, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Toska, E.; Scaltriti, M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann. Oncol. 2019, 30 (Suppl. 10), x3–x11. [Google Scholar] [CrossRef]
- Beelen, K.; Hoefnagel, L.D.; Opdam, M.; Wesseling, J.; Sanders, J.; Vincent, A.D.; van Diest, P.J.; Linn, S.C. PI3K/AKT/mTOR pathway activation in primary and corresponding metastatic breast tumors after adjuvant endocrine therapy. Int. J. Cancer 2014, 135, 1257–1263. [Google Scholar] [CrossRef]
- Presti, D.; Quaquarini, E. The PI3K/AKT/mTOR and CDK4/6 Pathways in Endocrine Resistant HR+/HER2− Metastatic Breast Cancer: Biological Mechanisms and New Treatments. Cancers 2019, 11, 1242. [Google Scholar] [CrossRef]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef]
- Ramirez-Ardila, D.E.; Helmijr, J.C.; Look, M.P.; Lurkin, I.; Ruigrok-Ritstier, K.; van Laere, S.; Dirix, L.; Sweep, F.C.; Span, P.N.; Linn, S.C.; et al. Hotspot mutations in PIK3CA associate with first-line treatment outcome for aromatase inhibitors but not for tamoxifen. Breast Cancer Res. Treat. 2013, 139, 39–49. [Google Scholar] [CrossRef]
- Beelen, K.; Opdam, M.; Severson, T.M.; Koornstra, R.H.; Vincent, A.D.; Wesseling, J.; Muris, J.J.; Berns, E.M.; Vermorken, J.B.; van Diest, P.J.; et al. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res. 2014, 16, R13. [Google Scholar] [CrossRef]
- Reinhardt, K.; Vetter, M.; Kaufhold, S.; Kantelhardt, E.; Thomssen, C. Adjuvant aromatase inhibitors in patients with PIK3CA mutation early breast cancer. Ann. Oncol. 2023, 34 (Suppl. 2), S278–S324. [Google Scholar] [CrossRef]
- Vasudevan, K.M.; Barbie, D.A.; Davies, M.A.; Rabinovsky, R.; McNear, C.J.; Kim, J.J.; Hennessy, B.T.; Tseng, H.; Pochanard, P.; Kim, S.Y.; et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 2009, 16, 21–32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).