Liquid and Tissue Biopsies for Lung Cancer: Algorithms and Perspectives
Abstract
Simple Summary
Abstract
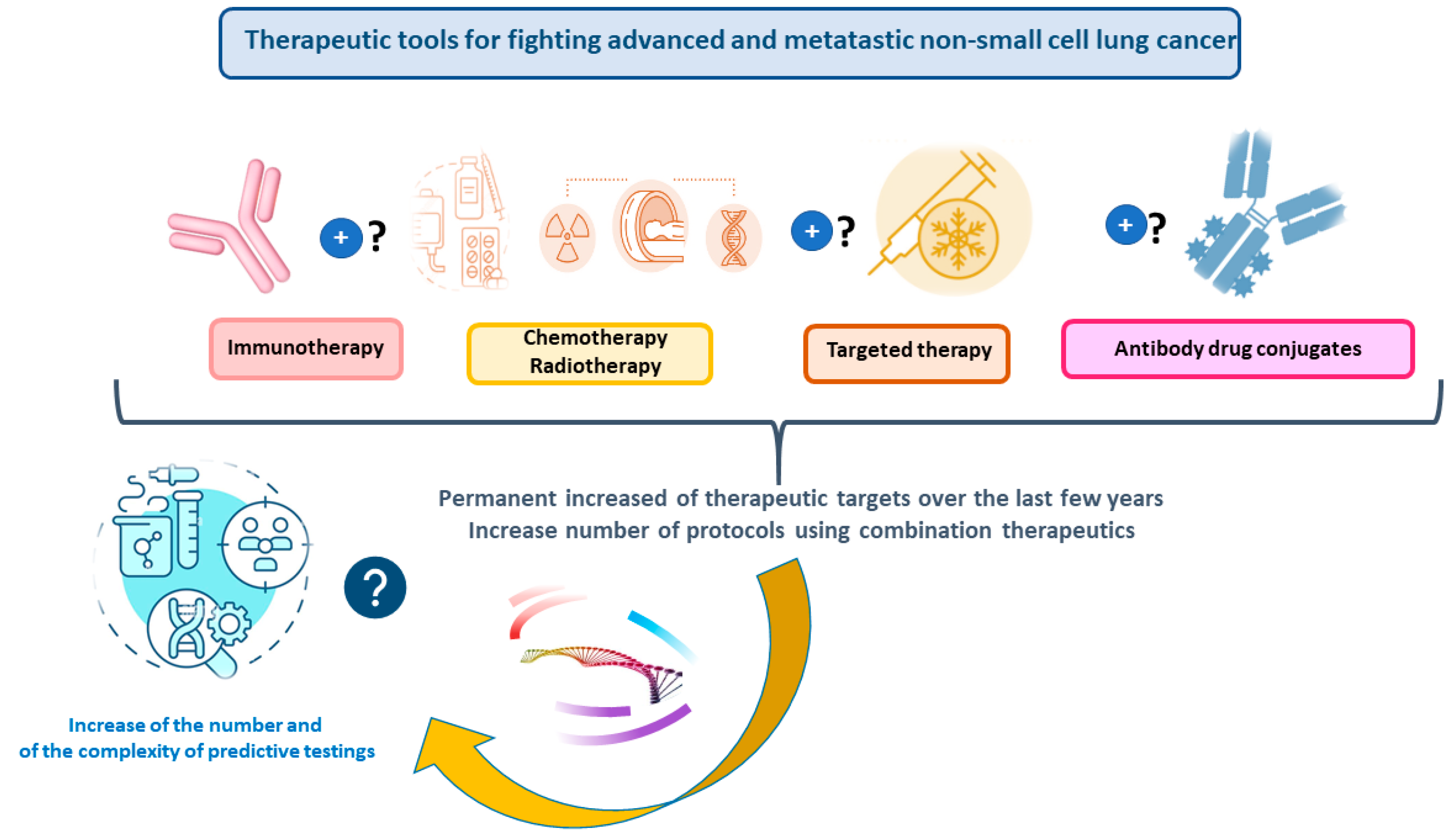
1. Introduction
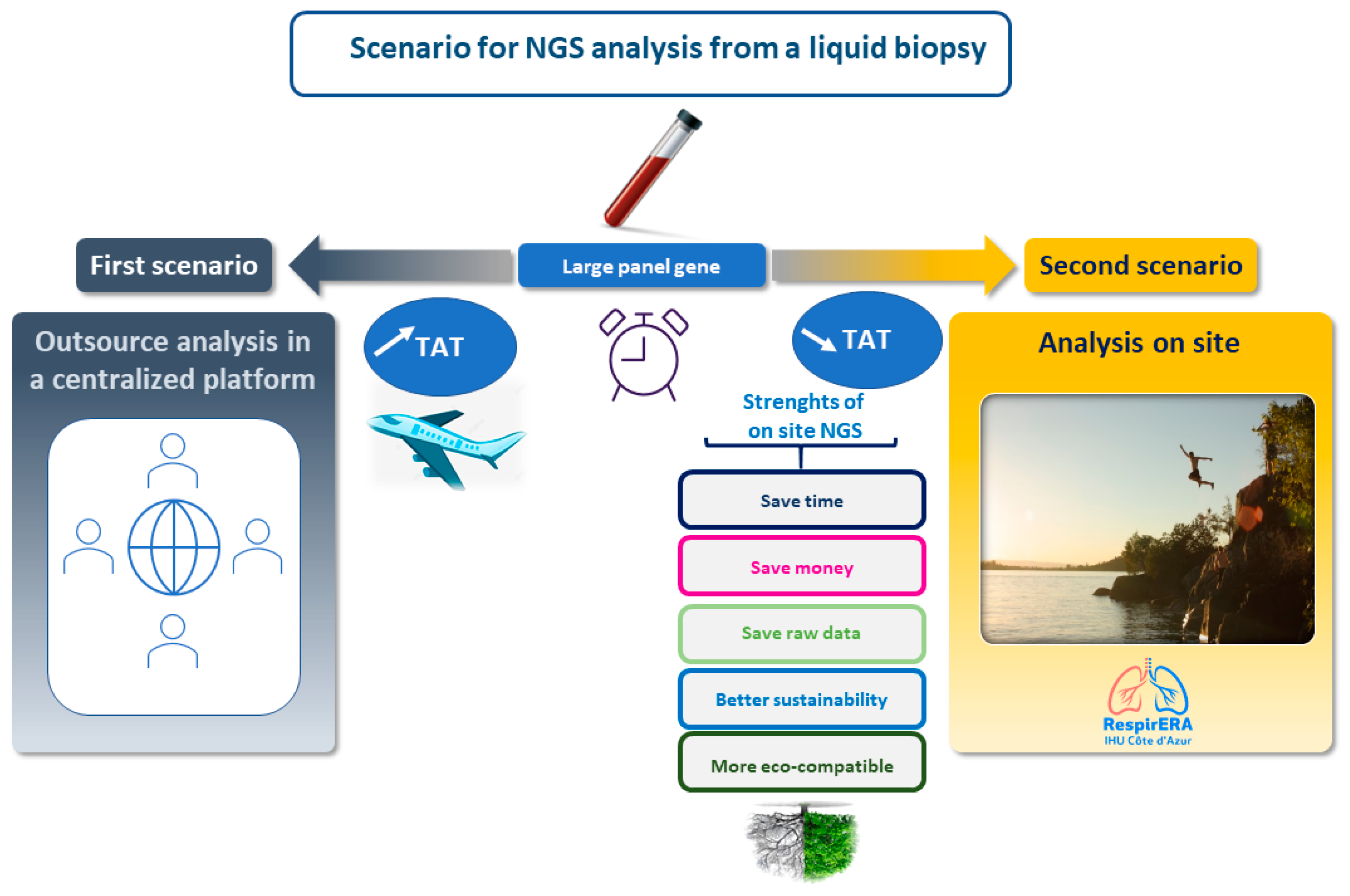
2. Liquid Biopsy for Nonsmall Cell Lung Cancer: One Gene Sequencing Testing versus NGS
| Company (City, Country) | Gene Panel (REF) | Number of Genes | Sequencing Approach | Sequencing Technology | System | Alternative NGS Compatibility |
|---|---|---|---|---|---|---|
| ThermoFisher Scientific (Waltham, MA, USA) | Oncomine™ Lung cfDNA Assay [31] | 11 | AmpliSeq | TFS: ion torrent: semiconductor technology | GeneStudio S5 | N/A |
| ThermoFisher Scientific (Waltham, MA, USA) | Oncomine Precision Assay [32] | 50 | AmpliSeq | TFS: ion torrent: technologie de semi-conducteur | Genexus | N/A |
| Pillar Biosciences (Natick, MA, USA) | OncoReveal Core and Fusion panel [33] | 20 | AmpliSeq | Illumina: sequencing by synthesis | NextSeq550Dx | N/A |
| Hedera Dx (Epalinges, Switzerland) | Hedera Profiling 2 ctDNA panel [34] | 32 | Hybrid Capture | Illumina: sequencing by synthesis | NextSeq550Dx and Higher | Element Aviti (sequencing-by-binding chemistry |
| Agilent (Santa Clara, CA, USA) | Beta test; Avida methyl 3400 DMR cancer [35,36] | 169 | Hybrid Capture | Illumina: sequencing by synthesis | NextSeq550Dx | Element Aviti (sequencing-by-binding chemistry) |
| Illumina (San Diego, CA, USA) | TruSight Oncology 500 [37] | 523 | Hybrid Capture | Illumina: sequencing by synthesis | NextSeq550Dx | No |
| Roche (Basel, Switzerland) | FoundationOne®Liquid CDx [38,39] | 324 | Hybrid Capture | Outsourced | N/A | |
| Guardant Health (Moorpark, CA, USA) | Guardant360 [40] | 739 | Hybrid Capture | Outsourced | ||
| SOPHIA GENETICS (Rolle, Switzerland) | MSK ACCESS powered by SOPHiA GENETICS [41] | 146 | Hybrid Capture | Illumina: sequencing by synthesis: Novaseq6000/NextSeq2000 | Element Aviti (sequencing-by-binding chemistry) | |
| Roche (Basel, Switzerland) | Avenio Expanded panel [42] | 77 | Hybrid Capture | Illumina: sequencing by synthesis | NextSeq550Dx | N/A |
3. Liquid Biopsy: A General View for a Complementary or an Alternative Approach to a Tissue Biopsy and Potential Limitations
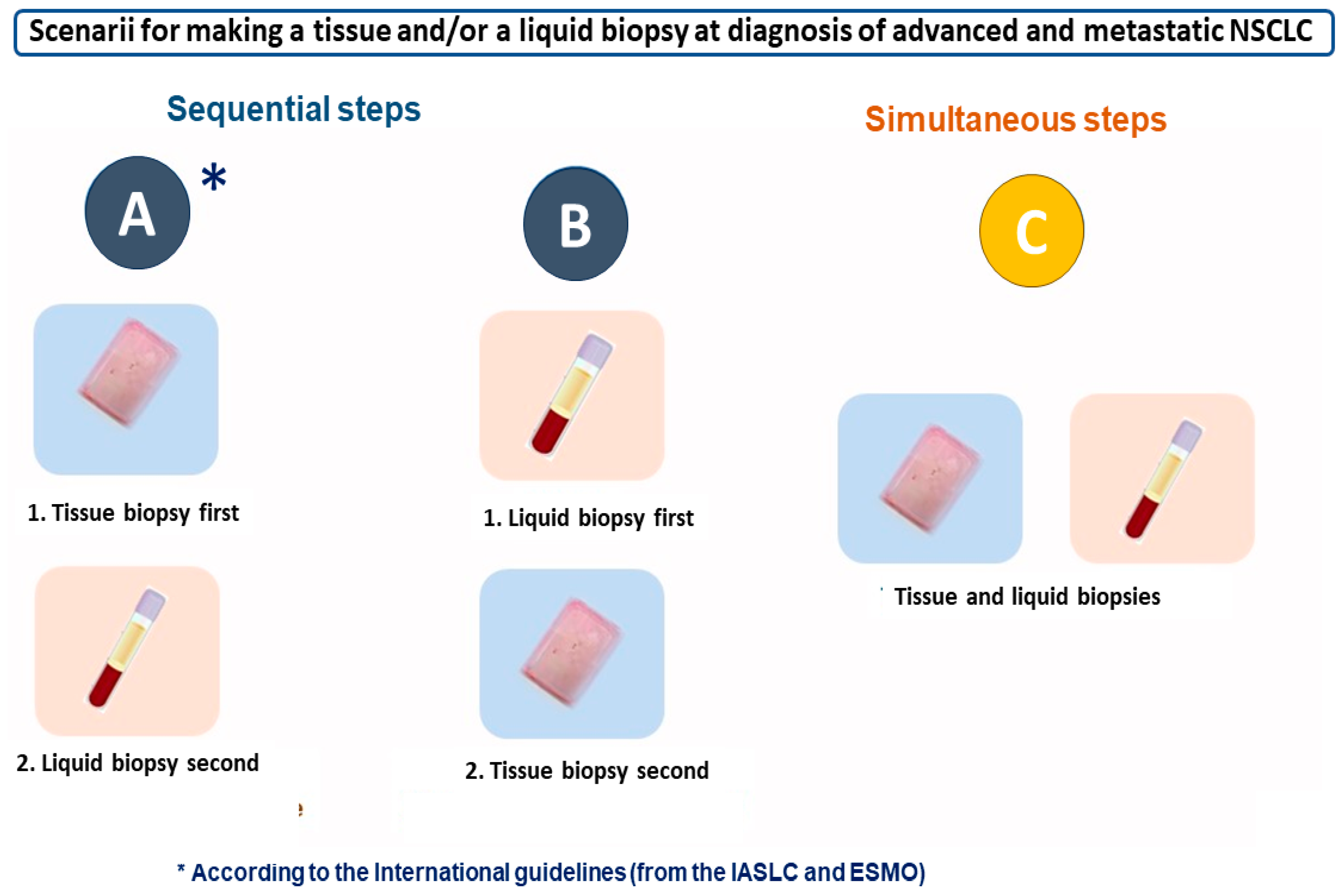
4. Liquid Biopsy and/or Tissue Biopsy: International Recommendations for Best Practice and Other Algorithms
5. Perspectives and Conclusions
Funding
Conflicts of Interest
References
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P.; Berezowska, S.; Kazdal, D.; Mograbi, B.; Ilié, M.; Stenzinger, A.; Hofman, V. Current challenges and practical aspects of molecular pathology for non-small cell lung cancers. Virchows Arch. 2024, 484, 233–246. [Google Scholar] [CrossRef] [PubMed]
- de Jager, V.D.; Timens, W.; Bayle, A.; Botling, J.; Brcic, L.; Büttner, R.; Fernandes, M.G.O.; Havel, L.; Hochmair, M.J.; Hofman, P.; et al. Developments in predictive biomarker testing and targeted therapy in advanced stage non-small cell lung cancer and their application across European countries. Lancet Reg. Health Eur. 2024, 38, 100838. [Google Scholar] [CrossRef] [PubMed]
- de Jager, V.D.; Timens, W.; Bayle, A.; Botling, J.; Brcic, L.; Büttner, R.; Fernandes, M.G.O.; Havel, L.; Hochmair, M.; Hofman, P.; et al. Future perspective for the application of predictive biomarker testing in advanced stage non-small cell lung cancer. Lancet Reg. Health Eur. 2024, 38, 100839. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Jänne, P.A.; Peters, S. Antibody-Drug Conjugates in Lung Cancer: Recent Advances and Implementing Strategies. J. Clin. Oncol. 2023, 41, 3747–3761. [Google Scholar] [CrossRef]
- Peters, S.; Loi, S.; André, F.; Chandarlapaty, S.; Felip, E.; Finn, S.P.; Jänne, P.A.; Kerr, K.M.; Munzone, E.; Passaro, A.; et al. Antibody-drug conjugates in lung and breast cancer: Current evidence and future directions-a position statement from the ETOP IBCSG Partners Foundation. Ann. Oncol. 2024, 35, 607–629. [Google Scholar] [CrossRef]
- Kerr, K.M.; Bubendorf, L.; Lopez-Rios, F.; Khalil, F.; Roy-Chowdhuri, S.; Joubert, P.; Hartmann, A.; Guerini-Rocco, E.; Yatabe, Y.; Hofman, P.; et al. Optimizing tissue stewardship in non-small cell lung cancer to support molecular characterization and treatment selection: Statement from a working group of thoracic pathologists. Histopathology 2024, 84, 429–439. [Google Scholar] [CrossRef]
- Hofman, P. The challenges of evaluating predictive biomarkers using small biopsy tissue samples and liquid biopsies from non-small cell lung cancer patients. J. Thorac. Dis. 2019, 11 (Suppl. S1), S57–S64. [Google Scholar] [CrossRef]
- Trisolini, R.; Bria, E.; Cetoretta, V.; Viscuso, M.; Malapelle, U. Seize the Opportunity with Small Tissue Samples: The Tailor Teaches! JTO Clin. Res. Rep. 2023, 4, 100507. [Google Scholar] [CrossRef]
- Mosele, M.F.; Westphalen, C.B.; Stenzinger, A.; Barlesi, F.; Bayle, A.; Bièche, I.; Bonastre, J.; Castro, E.; Dienstmann, R.; Krämer, A.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with advanced cancer in 2024: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2024, 35, 588–606. [Google Scholar] [CrossRef]
- Hofman, P.; Popper, H.H. Pathologists and liquid biopsies: To be or not to be? Virchows Arch. 2016, 469, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Al-Obeidi, E.; Riess, J.W.; Malapelle, U.; Rolfo, C.; Gandara, D.R. Convergence of Precision Oncology and Liquid Biopsy in Non-Small Cell Lung Cancer. Hematol. Oncol. Clin. N. Am. 2023, 37, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Malapelle, U.; André, F.; Paz-Ares, L.; Schuler, M.; Thomas, D.M.; Vainer, G.; Yoshino, T.; Rolfo, C. Practical Considerations for the Use of Circulating Tumor DNA in the Treatment of Patients with Cancer: A Narrative Review. JAMA Oncol. 2022, 8, 1830–1839. [Google Scholar] [CrossRef]
- García-Pardo, M.; Makarem, M.; Li, J.J.N.; Kelly, D.; Leighl, N.B. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: Opportunities and challenges. Br. J. Cancer. 2022, 127, 592–602. [Google Scholar] [CrossRef]
- Bayle, A.; Belcaid, L.; Aldea, M.; Vasseur, D.; Peyraud, F.; Nicotra, C.; Geraud, A.; Sakkal, M.; Seknazi, L.; Cerbone, L.; et al. Clinical utility of circulating tumor DNA sequencing with a large panel: A National Center for Precision Medicine (PRISM) study. Ann. Oncol. 2023, 34, 389–396. [Google Scholar] [CrossRef]
- Heeke, S.; Benzaquen, J.; Hofman, V.; Ilié, M.; Allegra, M.; Long-Mira, E.; Lassalle, S.; Tanga, V.; Salacroup, C.; Bonnetaud, C.; et al. Critical Assessment in Routine Clinical Practice of Liquid Biopsy for EGFR Status Testing in Non-Small-Cell Lung Cancer: A Single-Laboratory Experience (LPCE, Nice, France). Clin. Lung Cancer 2020, 21, 56–65.e8. [Google Scholar] [CrossRef]
- Heeke, S.; Benzaquen, J.; Vallee, A.; Allegra, M.; Mazieres, J.; Fayada, J.; Rajamani, J.; Lee, M.; Ordinario, E.; Tiotiu, A.; et al. Detection of ALK fusion transcripts in plasma of non-small cell lung cancer patients using a novel RT-PCR based assay. Ann. Transl. Med. 2021, 9, 922. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Hofman, V.; Benzaquen, J.; Otto, J.; Tanga, V.; Zahaf, K.; Allegra, M.; Long-Mira, E.; Lassalle, S.; Marquette, C.H.; et al. Detection of EGFR Mutations from Plasma of NSCLC Patients Using an Automatic Cartridge-Based PCR System. Front. Pharmacol. 2021, 14, 12:657743. [Google Scholar] [CrossRef]
- Malapelle, U.; Sirera, R.; Jantus-Lewintre, E.; Reclusa, P.; Calabuig-Fariñas, S.; Blasco, A.; Pisapia, P.; Rolfo, C.; Camps, C. Profile of the Roche cobas EGFR mutation test v2 for non-small cell lung cancer. Expert. Rev. Mol. Diagn. 2017, 17, 209–215. [Google Scholar] [CrossRef]
- Hofman, P. ALK Status Assessment with Liquid Biopsies of Lung Cancer Patients. Cancers 2017, 12, 106. [Google Scholar] [CrossRef]
- Hofman, P. Next-Generation Sequencing with Liquid Biopsies from Treatment-Naive Non-Small Cell Lung Carcinoma Patients. Cancers 2021, 13, 2049. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Circulating tumor DNA (ctDNA) as a biomarker for lung cancer: Early detection, monitoring and therapy prediction. Tumour Biol. 2024, 46, S283–S295. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, A.; Verzè, M.; Minari, R.; Perrone, F.; Gnetti, L.; Bordi, P.; Pluchino, M.; Nizzoli, R.; Azzoni, C.; Bottarelli, L.; et al. Resistance to osimertinib in advanced EGFR-mutated NSCLC: A prospective study of molecular genotyping on tissue and liquid biopsies. Br. J. Cancer 2024, 130, 135–142. [Google Scholar] [CrossRef]
- Ren, F.; Fei, Q.; Qiu, K.; Zhang, Y.; Zhang, H.; Sun, L. Liquid biopsy techniques and lung cancer: Diagnosis, monitoring and evaluation. J. Exp. Clin. Cancer Res. 2024, 43, 96. [Google Scholar] [CrossRef]
- Pezzuto, F.; Hofman, V.; Bontoux, C.; Fortarezza, F.; Lunardi, F.; Calabrese, F.; Hofman, P. The significance of co-mutations in EGFR-mutated non-small cell lung cancer: Optimizing the efficacy of targeted therapies? Lung Cancer 2023, 181, 107249. [Google Scholar] [CrossRef]
- Ricciuti, B.; Garassino, M.C. Precision Immunotherapy for STK11/KEAP1-Mutant NSCLC. J. Thorac. Oncol. 2024, 19, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Handorf, E.A.; Zhou, Y.; Borghaei, H.; Aggarwal, C.; Bauman, J. Outcomes in patients treated with frontline immune checkpoint inhibition (ICI) for advanced NSCLC with KRAS mutations and STK11/KEAP1 comutations across PD-L1 levels. Lung Cancer 2024, 190, 107510. [Google Scholar] [CrossRef]
- Cai, J.; Wang, W.; Zhang, W. A meta-analysis of liquid biopsy versus tumor histology for detecting EGFR mutations in non-small cell lung cancer. Transl. Oncol. 2024, 47, 102022. [Google Scholar] [CrossRef]
- Claus, J.; De Smet, D.; Breyne, J.; Wesolowski, J.; Himpe, U.; Demedts, I.; Martens, G.A. Patient-centric thresholding of Cobas EGFR mutation Test v2 for surveillance of EGFR-mutated metastatic non-small cell lung cancer. Sci. Rep. 2024, 14, 18191. [Google Scholar] [CrossRef]
- Ulivi, P.; Petracci, E.; Canale, M.; Priano, I.; Capelli, L.; Calistri, D.; Chiadini, E.; Cravero, P.; Rossi, A.; Delmonte, A.; et al. Liquid Biopsy for EGFR Mutation Analysis in Advanced Non-Small-Cell Lung Cancer Patients: Thoughts Drawn from a Real-Life Experience. Biomedicines 2021, 9, 1299. [Google Scholar] [CrossRef]
- Available online: https://www.thermofisher.com/fr/fr/home/life-science/cancer-research/cancer-genomics/liquid-biopsy-cancer-research-applications/oncomine-cell-free-dna-assays-liquid-biopsy-clinical-research.html (accessed on 16 September 2024).
- Available online: https://www.thermofisher.com/fr/fr/home/clinical/preclinical-companion-diagnostic-development/oncomine-oncology/oncomine-precision-assay.html (accessed on 16 September 2024).
- Available online: https://pillarbiosci.com (accessed on 15 September 2024).
- Available online: https://www.hederadx.com (accessed on 14 September 2024).
- Available online: https://www.agilent.com/en/product/next-generation-sequencing (accessed on 16 September 2024).
- Available online: https://www.agilent.com/en/product/next-generation-sequencing/ngs-library-prep-target-enrichment-reagents/methyl-seq-reagents/avida-duo-methyl-reagent-kit-4416759 (accessed on 15 September 2024).
- Available online: https://www.illumina.com/products/by-type/clinical-research-products/trusight-oncology-500-ctdna.html (accessed on 12 September 2024).
- Available online: https://www.foundationmedicine.fr/our-services/cdx.html (accessed on 16 September 2024).
- Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190032C.pdf (accessed on 15 September 2024).
- Available online: https://www.guardantcomplete.com/products/guardant360 (accessed on 12 September 2024).
- Available online: https://www.sophiagenetics.com/msk-access-powered-with-sophia-ddm (accessed on 17 September 2024).
- Available online: https://sequencing.roche.com/global/en/products/group/avenio-ctdna-expanded-kits.html (accessed on 15 September 2024).
- Hofman, P.; Calabrese, F.; Kern, I.; Adam, J.; Alarcão, A.; Alborelli, I.; Anton, N.T.; Arndt, A.; Avdalyan, A.; Barberis, M.; et al. Real-world EGFR testing practices for non-small-cell lung cancer by thoracic pathology laboratories across Europe. ESMO Open 2023, 8, 101628. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pardo, M.; Czarnecka, K.; Law, J.H.; Salvarrey, A.; Fernandes, R.; Fan, J.; Corke, L.; Waddell, T.K.; Yasufuku, K.; Donahoe, L.L.; et al. Plasma-first: Accelerating lung cancer diagnosis and molecular profiling through liquid biopsy. Ther. Adv. Med. Oncol. 2022, 14, 17588359221126151. [Google Scholar] [CrossRef] [PubMed]
- Jansen, J.P.; Ragavan, M.V.; Chen, C.; Douglas, M.P.; Phillips, K.A. The Health Inequality Impact of Liquid Biopsy to Inform First-Line Treatment of Advanced Non-Small Cell Lung Cancer: A Distributional Cost-Effectiveness Analysis. Value Health 2023, 26, 1697–1710. [Google Scholar] [CrossRef] [PubMed]
- Makarem, M.; Leighl, N.B. Molecular testing for lung adenocarcinoma: Is it time to adopt a “plasma-first” approach? Cancer 2020, 126, 3176–3180. [Google Scholar] [CrossRef] [PubMed]
- Raez, L.E.; Brice, K.; Dumais, K.; Lopez-Cohen, A.; Wietecha, D.; Izquierdo, P.A.; Santos, E.S.; Powery, H.W. Liquid Biopsy versus Tissue Biopsy to Determine Front Line Therapy in Metastatic Non-Small Cell Lung Cancer (NSCLC). Clin. Lung Cancer 2023, 24, 120–129. [Google Scholar] [CrossRef]
- Rathor, A.; Malik, P.S.; Tanwar, P.; Khurana, S.; Baskarane, H.; Pushpam, D.; Nambirajan, A.; Jain, D. ‘Plasma first’ approach for detecting epidermal growth factor receptor mutation in advanced non-small cell lung carcinoma. J. Cancer Res. Clin. Oncol. 2024, 150, 371. [Google Scholar] [CrossRef]
- Kurzrock, R.; Chaudhuri, A.A.; Feller-Kopman, D.; Florez, N.; Gorden, J.; Wistuba, I.I. Healthcare disparities, screening, and molecular testing in the changing landscape of non-small cell lung cancer in the United States: A review. Cancer Metastasis Rev. 2024, Online ahead of print. [Google Scholar] [CrossRef]
- Russo, A.; Lee, J.K.; Pasquina, L.W.; Del Re, M.; Dilks, H.H.; Murugesan, K.; Madison, R.W.; Lee, Y.; Schrock, A.B.; Comment, L.; et al. Liquid Biopsy of Lung Cancer before Pathological Diagnosis Is Associated with Shorter Time to Treatment. JCO Precis Oncol. 2024, 8, e2300535. [Google Scholar] [CrossRef]
- Esagian, S.M.; Grigoriadou, G.Ι.; Nikas, I.P.; Boikou, V.; Sadow, P.M.; Won, J.K.; Economopoulos, K.P. Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: A comprehensive systematic review. J. Cancer Res. Clin. Oncol. 2020, 146, 2051–2066. [Google Scholar] [CrossRef]
- Hofman, P. Matched tissue and liquid biopsies for advanced non-small cell lung cancer patients A potentially indispensable complementary approach. Transl. Oncol. 2023, 35, 101735. [Google Scholar] [CrossRef]
- Sugimoto, A.; Matsumoto, S.; Udagawa, H.; Itotani, R.; Usui, Y.; Umemura, S.; Nishino, K.; Nakachi, I.; Kuyama, S.; Daga, H.; et al. A Large-Scale Prospective Concordance Study of Plasma- and Tissue-Based Next-Generation Targeted Sequencing for Advanced Non-Small Cell Lung Cancer (LC-SCRUM-Liquid). Clin. Cancer Res. 2023, 29, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.T.; Ho, C.C.; Hsu, W.H.; Liao, W.Y.; Yang, C.Y.; Yu, C.J.; Tsai, T.H.; Yang, J.C.; Wu, S.G.; Hsu, C.L.; et al. Tissue or liquid rebiopsy? A prospective study for simultaneous tissue and liquid NGS after first-line EGFR inhibitor resistance in lung cancer. Cancer Med. 2024, 13, e6870. [Google Scholar] [CrossRef] [PubMed]
- Rotow, J.K.; Lee, J.K.; Madison, R.W.; Oxnard, G.R.; Jänne, P.A.; Schrock, A.B. Real-World Genomic Profile of EGFR Second-Site Mutations and Other Osimertinib Resistance Mechanisms and Clinical Landscape of NSCLC Post-Osimertinib. J. Thorac. Oncol. 2024, 19, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Scilla, K.A.; Mehra, R.; Gittens, A.; McCusker, M.G.; de Miguel-Perez, D.; Gomez, J.E.; Peleg, A.; Del Re, M.; Rolfo, C.D. Tracking Clonal Evolution of EGFR-Mutated Non-Small Cell Lung Cancer through Liquid Biopsy: Management of C797S Acquired Mutation. Clin. Lung Cancer 2023, 24, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Bayle, A.; Belcaid, L.; Palmieri, L.J.; Teysonneau, D.; Cousin, S.; Spalato-Ceruso, M.; Aldea, M.; Vasseur, D.; Alame, M.; Blouin, L.; et al. Circulating tumor DNA landscape and prognostic impact of acquired resistance to targeted therapies in cancer patients: A national center for precision medicine (PRISM) study. Mol. Cancer 2023, 22, 176. [Google Scholar] [CrossRef]
- Goksel, T.; Özgür, S.; Vardarlı, A.T.; Koç, A.; Karakuş, H.S.; Özdemir, T.R.; Erdoğan, K.M.; Aldağ, C.; Veral, A.; Komurcuoglu, B.; et al. Prognostic and predictive role of liquid biopsy in lung cancer patients. Front. Oncol. 2024, 13, 1275525. [Google Scholar] [CrossRef]
- Villa, M.; Malighetti, F.; Sala, E.; Sharma, G.G.; Arosio, G.; Gemelli, M.; Manfroni, C.; Fontana, D.; Cordani, N.; Meneveri, R.; et al. New pan-ALK inhibitor-resistant EML4::ALK mutations detected by liquid biopsy in lung cancer patients. NPJ Precis. Oncol. 2024, 8, 29. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, C.; Zhao, J.; Wang, Q.; Chu, X.; Li, J.; Zhou, F.; Ren, S.; Li, X.; Su, C.; et al. Re-biopsy and liquid biopsy for patients with non-small cell lung cancer after EGFR-tyrosine kinase inhibitor failure. Thorac. Cancer 2019, 10, 957–965. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Mack, P.C.; Banks, K.C.; Espenschied, C.R.; Burich, R.A.; Zill, O.A.; Lee, C.E.; Riess, J.W.; Mortimer, S.A.; Talasaz, A.; Lanman, R.B.; et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: Analysis of over 8000 cases. Cancer 2020, 126, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- García-Pardo, M.; Czarnecka-Kujawa, K.; Law, J.H.; Salvarrey, A.M.; Fernandes, R.; Fan, Z.J.; Waddell, T.K.; Yasufuku, K.; Liu, G.; Donahoe, L.L.; et al. Association of Circulating Tumor DNA Testing before Tissue Diagnosis with Time to Treatment Among Patients with Suspected Advanced Lung Cancer: The ACCELERATE Nonrandomized Clinical Trial. JAMA Netw. Open 2023, 6, e2325332. [Google Scholar] [CrossRef] [PubMed]
- Dziadziuszko, R.; Mok, T.; Peters, S.; Han, J.Y.; Alatorre-Alexander, J.; Leighl, N.; Sriuranpong, V.; Pérol, M.; de Castro Junior, G.; Nadal, E.; et al. J Blood First Assay Screening Trial (BFAST) in Treatment-Naive Advanced or Metastatic NSCLC: Initial Results of the Phase 2 ALK-Positive Cohort. J. Thorac. Oncol. 2021, 16, 2040–2050. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Herbst, R.S. BFAST but be smart: bTMB remains an exploratory biomarker in NSCLC. Nat. Rev. Clin. Oncol. 2023, 20, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Gadgeel, S.M.; Mok, T.; Nadal, E.; Kilickap, S.; Swalduz, A.; Cadranel, J.; Sugawara, S.; Chiu, C.H.; Yu, C.J.; et al. Entrectinib in ROS1-positive advanced non-small cell lung cancer: The phase 2/3 BFAST trial. Nat. Med. 2024, 30, 1923–1932. [Google Scholar] [CrossRef]
- Wang, H.Y.; Ho, C.C.; Lin, Y.T.; Liao, W.Y.; Chen, C.Y.; Shih, J.Y.; Yu, C.J. Comprehensive Genomic Analysis of Patients with Non-Small-Cell Lung Cancer Using Blood-Based Circulating Tumor DNA Assay: Findings from the BFAST Database of a Single Center in Taiwan. JCO Precis. Oncol. 2024, 8, e2300314. [Google Scholar] [CrossRef]
- Ernst, S.M.; van Marion, R.; Atmodimedjo, P.N.; de Jonge, E.; Mathijssen, R.H.J.; Paats, M.S.; de Bruijn, P.; Koolen, S.L.; von der Thüsen, J.H.; Aerts, J.G.J.V.; et al. Clinical Utility of Circulating Tumor DNA in Patients with Advanced KRASG12C-Mutated NSCLC Treated with Sotorasib. J. Thorac. Oncol. 2024, 19, 995–1006. [Google Scholar] [CrossRef]
- Sposito, M.; Belluomini, L.; Nocini, R.; Insolda, J.; Scaglione, I.M.; Menis, J.; Simbolo, M.; Lugini, A.; Buzzacchino, F.; Verderame, F.; et al. Tissue- and liquid-biopsy based NGS profiling in advanced non-small-cell lung cancer in a real-world setting: The IMMINENT study. Front. Oncol. 2024, 14, 1436588. [Google Scholar] [CrossRef]
- Rolfo, C.D.; Madison, R.W.; Pasquina, L.W.; Brown, D.W.; Huang, Y.; Hughes, J.D.; Graf, R.P.; Oxnard, G.R.; Husain, H. Measurement of ctDNA Tumor Fraction Identifies Informative Negative Liquid Biopsy Results and Informs Value of Tissue Confirmation. Clin. Cancer Res. 2024, 30, 2452–2460. [Google Scholar] [CrossRef]
- Budczies, J.; Kazdal, D.; Menzel, M.; Beck, S.; Kluck, K.; Altbürger, C.; Schwab, C.; Allgäuer, M.; Ahadova, A.; Kloor, M.; et al. Tumour mutational burden: Clinical utility, challenges and emerging improvements. Nat. Rev. Clin. Oncol. 2024, 21, 725–742. [Google Scholar] [CrossRef]
- Fridland, S.; Choi, J.; Nam, M.; Schellenberg, S.J.; Kim, E.; Lee, G.; Yoon, N.; Chae, Y.K. Assessing tumor heterogeneity: Integrating tissue and circulating tumor DNA (ctDNA) analysis in the era of immuno-oncology—Blood TMB is not the same as tissue TMB. J. Immunother. Cancer 2021, 9, e002551. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients with Non-Small Cell Lung Cancer with Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, J.; Wang, G.; He, X.; Mi, Y.; Cao, Y.; Yu, X. Predictive Efficacy of Blood-Based Tumor Mutation Burden Assay for Immune Checkpoint Inhibitors Therapy in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 795933. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; van den Broek, D.; Denis, M.G.; Hofman, P.; Hubank, M.; Mouliere, F.; Paz-Ares, L.; Schuuring, E.; Sültmann, H.; Vainer, G.; et al. Recommendations for a practical implementation of circulating tumor DNA mutation testing in metastatic non-small-cell lung cancer. ESMO Open 2022, 7, 100399. [Google Scholar] [CrossRef]
- Kasi, P.M.; Lee, J.K.; Pasquina, L.W.; Decker, B.; Vanden Borre, P.; Pavlick, D.C.; Allen, J.M.; Parachoniak, C.; Quintanilha, J.C.F.; Graf, R.P.; et al. Circulating Tumor DNA Enables Sensitive Detection of Actionable Gene Fusions and Rearrangements Across Cancer Types. Clin. Cancer Res. 2024, 30, 836–848. [Google Scholar] [CrossRef]
- Batool, S.M.; Hsia, T.; Beecroft, A.; Lewis, B.; Ekanayake, E.; Rosenfeld, Y.; Escobedo, A.K.; Gamblin, A.S.; Rawal, S.; Cote, R.J.; et al. Extrinsic and intrinsic preanalytical variables affecting liquid biopsy in cancer. Cell Rep. Med. 2023, 4, 101196. [Google Scholar] [CrossRef]
- Available online: https://www.biobank-cotedazur.fr (accessed on 15 September 2024).
- Bontoux, C.; Lespinet-Fabre, V.; Bordone, O.; Tanga, V.; Allegra, M.; Salah, M.; Lalvée, S.; Goffinet, S.; Benzaquen, J.; Marquette, C.H.; et al. Ultra-Fast Amplicon-Based Next-Generation Sequencing in Non-Squamous Non-Small Cell Lung Cancer. J. Vis. Exp. 2023, e65190. [Google Scholar] [CrossRef]
- Ilié, M.; Hofman, V.; Bontoux, C.; Heeke, S.; Lespinet-Fabre, V.; Bordone, O.; Lassalle, S.; Lalvée, S.; Tanga, V.; Allegra, M.; et al. Setting Up an Ultra-Fast Next-Generation Sequencing Approach as Reflex Testing at Diagnosis of Non-Squamous Non-Small Cell Lung Cancer; Experience of a Single Center (LPCE, Nice, France). Cancers 2022, 14, 2258. [Google Scholar] [CrossRef]
- Hofman, V.; Heeke, S.; Bontoux, C.; Chalabreysse, L.; Barritault, M.; Bringuier, P.P.; Fenouil, T.; Benzerdjeb, N.; Begueret, H.; Merlio, J.P.; et al. Ultrafast Gene Fusion Assessment for Nonsquamous NSCLC. JTO Clin. Res. Rep. 2022, 4, 100457. [Google Scholar] [CrossRef]
- Gosney, J.R.; Paz-Ares, L.; Jänne, P.; Kerr, K.M.; Leighl, N.B.; Lozano, M.D.; Malapelle, U.; Mok, T.; Sheffield, B.S.; Tufman, A.; et al. Pathologist-initiated reflex testing for biomarkers in non-small-cell lung cancer: Expert consensus on the rationale and considerations for implementation. ESMO Open 2023, 8, 101587. [Google Scholar] [CrossRef]
- Behel, V.; Chougule, A.; Noronha, V.; Patil, V.M.; Menon, N.; Singh, A.; Chopade, S.; Kumar, R.; Shah, S.; More, S.; et al. Clinical Utility of Liquid Biopsy (Cell-free DNA) Based EGFR Mutation Detection Post treatment Initiation as a Disease Monitoring Tool in Patients with Advanced EGFR-mutant NSCLC. Clin. Lung Cancer 2022, 23, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Fasano, R.; Serratì, S.; Rafaschieri, T.; Longo, V.; Di Fonte, R.; Porcelli, L.; Azzariti, A. Small-Cell Lung Cancer: Is Liquid Biopsy a New Tool Able to Predict the Efficacy of Immunotherapy? Biomolecules 2024, 14, 396. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Gay, C.M.; Estecio, M.R.; Tran, H.; Morris, B.B.; Zhang, B.; Tang, X.; Raso, M.G.; Rocha, P.; Lai, S.; et al. Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Cancer Cell 2024, 42, 225–237.e5. [Google Scholar] [CrossRef]
- Friedlaender, A.; Banna, G.; Malapelle, U.; Pisapia, P.; Addeo, A. Next Generation Sequencing and Genetic Alterations in Squamous Cell Lung Carcinoma: Where Are We Today? Front. Oncol. 2019, 9, 166. [Google Scholar] [CrossRef]
- Tan, A.C.; Lai, G.G.Y.; Saw, S.P.L.; Chua, K.L.M.; Takano, A.; Ong, B.H.; Koh, T.P.T.; Jain, A.; Tan, W.L.; Ng, Q.S.; et al. Detection of circulating tumor DNA with ultradeep sequencing of plasma cell-free DNA for monitoring minimal residual disease and early detection of recurrence in early-stage lung cancer. Cancer 2024, 130, 1758–1765. [Google Scholar] [CrossRef]
- Marinello, A.; Tagliamento, M.; Pagliaro, A.; Conci, N.; Cella, E.; Vasseur, D.; Remon, J.; Levy, A.; Dall’Olio, F.G.; Besse, B. Circulating tumor DNA to guide diagnosis and treatment of localized and locally advanced non-small cell lung cancer. Cancer Treat. Rev. 2024, 129, 102791. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Frankell, A.M.; Harrison, T.; Kisistok, J.; Garnett, A.; Johnson, L.; Veeriah, S.; Moreau, M.; Chesh, A.; Chaunzwa, T.L.; et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 2023, 616, 553–562. [Google Scholar] [CrossRef]
- Gristina, V.; La Mantia, M.; Peri, M.; Iacono, F.; Barraco, N.; Perez, A.; Viscardi, G.; Cutaia, S.; Russo, T.D.B.; Anwar, Z.; et al. Navigating the liquid biopsy Minimal Residual Disease (MRD) in non-small cell lung cancer: Making the invisible visible. Crit. Rev. Oncol. Hematol. 2023, 182, 103899. [Google Scholar] [CrossRef]
- Hofman, P.; Denis, M.G. The use of minimal residual disease in thoracic oncology: Gaps between promises and the on-the-ground reality of daily practice. Cytopathology 2024, 35, 7–15. [Google Scholar] [CrossRef]
- Pellini, B.; Chaudhuri, A.A. Circulating Tumor DNA Minimal Residual Disease Detection of Non-Small-Cell Lung Cancer Treated with Curative Intent. J. Clin. Oncol. 2022, 40, 567–575. [Google Scholar] [CrossRef]
- Rolfo, C.; Russo, A. Navigating into a stormy sea: Liquid biopsy enters peri-operative management in early-stage non-small cell lung cancer. Ann. Oncol. 2024, 35, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Heeke, S.; Sujit, S.; Vokes, N.; Zhang, J.; Aminu, M.; Lam, V.K.; Vaporciyan, A.; Swisher, S.G.; Godoy, M.C.B.; et al. Circulating tumor DNA and radiological tumor volume identify patients at risk for relapse with resected, early-stage non-small-cell lung cancer. Ann. Oncol. 2024, 35, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yang, F.; Shen, H.; Wang, C.; Li, X.; Chervova, O.; Wu, S.; Qiu, F.; Peng, D.; Zhu, X.; et al. Individualized tumor-informed circulating tumor DNA analysis for postoperative monitoring of non-small cell lung cancer. Cancer Cell 2023, 41, 1749–1762.e6. [Google Scholar] [CrossRef] [PubMed]
- Stetson, D.; Labrousse, P.; Russell, H.; Shera, D.; Abbosh, C.; Dougherty, B.; Barrett, J.C.; Hodgson, D.; Hadfield, J. Next-Generation Molecular Residual Disease Assays: Do We Have the Tools to Evaluate Them Properly? J. Clin. Oncol. 2024, 42, 2736–2740. [Google Scholar] [CrossRef]
- Berland, L.; Gabr, Z.; Chang, M.; Ilié, M.; Hofman, V.; Rignol, G.; Ghiringhelli, F.; Mograbi, B.; Rashidian, M.; Hofman, P. Further knowledge and developments in resistance mechanisms to immune checkpoint inhibitors. Front. Immunol. 2024, 15, 1384121. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P.; Heeke, S.; Alix-Panabières, C.; Pantel, K. Liquid biopsy in the era of immuno-oncology: Is it ready for prime-time use for cancer patients? Ann. Oncol. 2019, 30, 1448–1459. [Google Scholar] [CrossRef]
- Spagnolo, C.C.; Pepe, F.; Ciappina, G.; Nucera, F.; Ruggeri, P.; Squeri, A.; Speranza, D.; Silvestris, N.; Malapelle, U.; Santarpia, M. Circulating biomarkers as predictors of response to immune checkpoint inhibitors in NSCLC: Are we on the right path? Crit. Rev. Oncol. Hematol. 2024, 197, 104332. [Google Scholar] [CrossRef]
- Liu, S.; Graves, N.; Tan, A.C. The cost-effectiveness of including liquid biopsy into molecular profiling strategies for newly diagnosed advanced non-squamous non-small cell lung cancer in an Asian population. Lung Cancer 2024, 191, 07794. [Google Scholar] [CrossRef]
- Horgan, D.; Pesapane, F.; Van der Buckle, M.; de Maria, R.; Dube, F.; Singh, J.; Ługowska, I.; Bayle, A.; Hofman, P.; Malapelle, U.; et al. From theory to practice: Implementing next-generation sequencing and public health genomics in healthcare systems. Crit. Rev. Oncol. Hematol. 2024, 201, 104433. [Google Scholar] [CrossRef]
- Horgan, D.; Curigliano, G.; Rieß, O.; Hofman, P.; Büttner, R.; Conte, P.; Cufer, T.; Gallagher, W.M.; Georges, N.; Kerr, K.; et al. Identifying the Steps Required to Effectively Implement Next-Generation Sequencing in Oncology at a National Level in Europe. J. Pers. Med. 2022, 12, 72. [Google Scholar] [CrossRef]
- Horgan, D.; Van den Bulcke, M.; Malapelle, U.; Troncone, G.; Normanno, N.; Capoluongo, E.D.; Prelaj, A.; Rizzari, C.; Trapani, D.; Singh, J.; et al. Tackling the implementation gap for the uptake of NGS and advanced molecular diagnostics into healthcare systems. Heliyon 2023, 10, e23914. [Google Scholar] [CrossRef] [PubMed]
- Caputo, V.; De Falco, V.; Ventriglia, A.; Famiglietti, V.; Martinelli, E.; Morgillo, F.; Martini, G.; Corte, C.M.D.; Ciardiello, D.; Poliero, L.; et al. Comprehensive genome profiling by next generation sequencing of circulating tumor DNA in solid tumors: A single academic institution experience. Ther. Adv. Med. Oncol. 2022, 14, 17588359221096878. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.gustaveroussy.fr/fr/gustave-roussy-roche-et-foundation-medicine-annoncent-un-partenariat-unique-pour-etendre-lanalyse (accessed on 12 September 2024).
- Normanno, N.; De Luca, A.; Abate, R.E.; Morabito, A.; Milella, M.; Tabbò, F.; Curigliano, G.; Masini, C.; Marchetti, P.; Pruneri, G.; et al. Current practice of genomic profiling of patients with advanced solid tumours in Italy: The Italian Register of Actionable Mutations (RATIONAL) study. Eur. J. Cancer. 2023, 187, 174–184. [Google Scholar] [CrossRef]
- Porta, C.; Pradelli, L.; Sicari, E.; Castellani, S.; Sivakumar, S.; Sokol, E.; Montesion, M.; Wieland, T.; Rambichler, J.; Minari, R.; et al. Liquid Pbiopsy comprehensive genomic profiling of lung cancer in the Italian population: A real-world experience. Lung Cancer 2023, 185, 107359. [Google Scholar] [CrossRef] [PubMed]
- Wiedower, J.A.; Forbes, S.P.; Tsai, L.J.; Liao, J.; Raez, L.E. Real-world clinical and economic outcomes for patients with advanced non-small cell lung cancer enrolled in a clinical trial following comprehensive genomic profiling via liquid biopsy. J. Manag. Care Spec. Pharm. 2024, 30, 660–671. [Google Scholar] [CrossRef]
- Malapelle, U.; Leighl, N.; Addeo, A.; Hershkovitz, D.; Hochmair, M.J.; Khorshid, O.; Länger, F.; de Marinis, F.; Peled, N.; Sheffield, B.S.; et al. Recommendations for reporting tissue and circulating tumour (ct)DNA next-generation sequencing results in non-small cell lung cancer. Br. J. Cancer 2024, 131, 212–219. [Google Scholar] [CrossRef]
- Casolino, R.; Beer, P.A.; Chakravarty, D.; Davis, M.B.; Malapelle, U.; Mazzarella, L.; Normanno, N.; Pauli, C.; Subbiah, V.; Turnbull, C.; et al. Interpreting and integrating genomic tests results in clinical cancer care: Overview and practical guidance. CA. Cancer J. Clin. 2024, 74, 264–285. [Google Scholar] [CrossRef]
- Horgan, D.; Hofman, P.; Subbiah, V. Welcoming the future: Embracing novel technologies for a progressive health system. ESMO Open 2024, 9, 103656. [Google Scholar] [CrossRef]
- Venkataraman, V.; Martin-Giacalone, B.A.; Drake, B.F.; Salmi, L.; Claus, E.B.; Schuster, A.L.R.; Bridges, J.F.P.; Lenz, H.J.; Willman, C.L.; Diehl, D.; et al. Overcoming Systemic Barriers to Make Patient-Partnered Research a Reality. J. Clin. Oncol. 2024, Online ahead of print. [Google Scholar] [CrossRef]
- André, F.; Rassy, E.; Marabelle, A.; Michiels, S.; Besse, B. Forget lung, breast or prostate cancer: Why tumour naming needs to change. Nature 2024, 626, 26–29. [Google Scholar] [CrossRef]
- Haslam, A.; Olivier, T.; Tuia, J.; Prasad, V. A systematic review of basket and umbrella trials in oncology: The importance of tissue of origin and molecular target. Eur. J. Cancer 2023, 178, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Ilié, M.; Heeke, S.; Horgan, D.; Hofman, P. Navigating Change in Tumor Naming: Exploring the Complexities and Considerations of Shifting toward Molecular Classifications. J. Clin. Oncol. 2024, 42, 3183–3186. [Google Scholar] [CrossRef] [PubMed]
- Stenzinger, A.; Klauschen, F. Forget lung, breast or prostate cancer? Why we shouldn’t abandon tumour names yet. Nature 2024, 627, 38. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofman, P. Liquid and Tissue Biopsies for Lung Cancer: Algorithms and Perspectives. Cancers 2024, 16, 3340. https://doi.org/10.3390/cancers16193340
Hofman P. Liquid and Tissue Biopsies for Lung Cancer: Algorithms and Perspectives. Cancers. 2024; 16(19):3340. https://doi.org/10.3390/cancers16193340
Chicago/Turabian StyleHofman, Paul. 2024. "Liquid and Tissue Biopsies for Lung Cancer: Algorithms and Perspectives" Cancers 16, no. 19: 3340. https://doi.org/10.3390/cancers16193340
APA StyleHofman, P. (2024). Liquid and Tissue Biopsies for Lung Cancer: Algorithms and Perspectives. Cancers, 16(19), 3340. https://doi.org/10.3390/cancers16193340






