Simple Summary
Colorectal cancer (CRC) is the second-leading cause of cancer-related deaths worldwide. In this review article, we examine the role of gut microbiota in the development and progression of CRC. We also examine the use of gut microbiota as a biomarker to predict CRC and its possible therapeutic outcome.
Abstract
Colorectal cancer (CRC) is the second-leading cause of cancer-related deaths worldwide. While CRC is thought to be an interplay between genetic and environmental factors, several lines of evidence suggest the involvement of gut microbiota in promoting inflammation and tumor progression. Gut microbiota refer to the ~40 trillion microorganisms that inhabit the human gut. Advances in next-generation sequencing technologies and metagenomics have provided new insights into the gut microbial ecology and have helped in linking gut microbiota to CRC. Many studies carried out in humans and animal models have emphasized the role of certain gut bacteria, such as Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, and colibactin-producing Escherichia coli, in the onset and progression of CRC. Metagenomic studies have opened up new avenues for the application of gut microbiota in the diagnosis, prevention, and treatment of CRC. This review article summarizes the role of gut microbiota in CRC development and its use as a biomarker to predict the disease and its potential therapeutic applications.
1. Introduction
Colorectal cancer (CRC) is one of the major causes of cancer mortality in humans, accounting for 9.4% of deaths worldwide in 2020 [1]. It is the third most common cancer in males and the second most common cancer in females. The incidence of CRC has increased in recent years. In 2020, there were 1.9 million new cases and 0.9 million deaths worldwide [1,2]. It has been estimated that by 2030, there would be about 2.2 million cases and 1.1 million CRC deaths per year [3]. The incidence of CRC has been rising rapidly in developing countries, whereas stabilizing or decreasing trends have been observed in developed countries where rates remain high [4]. Mutations in many genes have been identified that are associated with CRC, and some with high frequencies of occurence are listed in Table 1. The first gene mutation identified was the adenomatous polyposis coli (APC) gene [5]. The initial formation of polyps occurs in response to mutations in tumor-suppressor genes, such as APC, which is a component of the Wnt/β-catenin pathway and controls cell proliferation. Additionally, mutations may also take place in genes involved in DNA repair, such as hMSH2, contributing to colorectal tumorigenesis. Some of these genetic alterations are also inherited. The majority of CRC cases are sporadic [6], and heritable CRC accounts for only 12–35% of cases [7].

Table 1.
Genes associated with familial CRC.
CRC development is multifactorial and influenced by host genetic and environmental factors. CRC development usually takes decades, during which mutations are accumulated in oncogenes and tumor suppressor genes [8]. The sequence of genetic changes leading to CRC is commonly referred to as the adenoma-carcinoma sequence [9]. Tumors are more frequent in the distal region than in the proximal region of the large intestine [10] reflecting differences in the luminal environments of these two regions [11].
In this review, we discuss the relationship between gut microbiota and CRC development. We restrict our discussion mainly to bacteria, which have gained much attention in recent years due to their involvement in human health and diseases. We also discuss the potential role of gut microbiota in the diagnosis, treatment, and prevention of CRC.
2. Human Gut Microbiota
The human gut contains about 40 trillion microorganisms that constitute the gastrointestinal microbiota [12]. The number of microorganisms in the human gut is about three times greater than the total number of cells in the human body [12]. The human gut microbiome is sometimes referred to as the “forgotten organ” [13,14]. The human colon is considered to be one of the most densely populated microbial ecosystems [12,15]. The colorectum harbors about 30 trillion bacteria that constantly crosstalk with the intestinal epithelium, immunological cells, and mucosal barrier [16,17]. The human gut microbiome represents a complex microecosystem, the composition of which varies between individuals. The human gut microbial genome (microbiome) is at least two orders larger than the human genome [16]. The gut microbiota is acquired during the initial stages of life from the mother. The colonization of the gut by microorganisms is affected by the type of delivery [18] and the type of diet [19]. Babies born through vaginal delivery are exposed to vaginal microbes, such as Lactobacillus, while babies born through C-section are exposed to skin microbes, such as Staphylococcus and Corynebacterium. Breast milk and formula milk also affect the types of microorganisms that colonize the gut. According to a study published in 2018, the composition and structure of gut microbiota are predominantly shaped by environmental factors and only 1.9% of the gut microbiome is heritable [20].
The composition of gut microbiota shows variations between individuals [21]. but it is relatively stable within an individual [22]. The composition of gut microbiota in adults is influenced by diet, age, geographic location, race, external environmental microorganisms, use of antibiotics, infectious diarrhea, or international immigration [23,24,25,26,27,28].
The gut microbiota is critical for normal gut physiology, including digestion, biosynthesis of vitamins, generation of heat, gut immunity, and maintenance of gut homeostasis [15,29,30,31,32,33,34]. It is estimated that microbial metabolism results in the generation of about 70% of body heat at rest [35,36], which is important for maintaining a stable gut and body temperature. The composition of the gut microbiota influences individual variations in immunity [37,38,39] and is necessary for the development of the immune system [40,41]. Certain gut symbiotic Gram-negative bacteria can induce an IgG response that promotes the phagocytosis of pathogenic E. coli and Salmonella [42]. Germ-free mice are prone to harbor deficiencies in the development of gut-associated lymphoid tissues [43], suggesting the role of gut microbiota in the development of the immune system.
Study of the composition and diversity of the human gut microbiome is made possible by recent advances in next-generation sequencing technologies. Every individual has a unique microbiota, and the exact numbers of bacterial phyla and species vary among individuals. Humans share 40% of the core microbiota, and the remaining 60% of the microbiota is variable and depends on various host factors [16,44,45]. It has been estimated that normal human commensal gut microbiota comprises more than 50 bacterial phyla and 1000 bacterial species [16,45,46,47]. The number and diversity of gut bacterial species depend on the lifestyle, type of diet, and genotype of the host [48,49]. The microbiota constantly evolves during the lifetime of an individual. Though there is no consensus on an average intestinal microbiota, studies suggest that the dominant bacterial phyla in the human gut are Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Fusobacteria [50]. The gut microbial composition shows diversity at the genus and species levels. Besides bacteria, the gut microbiota also includes a variety of viruses, archaea, protozoa, and fungi. The composition of the gut microbiota also varies in different parts of the gut. Bacteroidetes (Bacteroidota) and Actinobacteria (Actinomycetota) are the dominant phyla (representing more than 90% of bacterial phyla) in the colon, while Firmicutes is the dominant phylum (representing 40% of bacterial phyla) in the small intestine [27,51]. The proximal gut shows a relatively low number of microbes (108 cells/mL), which mostly belong to Bacteroidetes (Bacteroidota) and Clostridiales. The large intestine shows a thousand-fold higher number of microbes (1011 cells/mL), which belong to Bacteroidetes (Bacteroidota), Firmicutes (Bacillota), Proteobacteria (Pseudomonadota), Actinobacteria (Actinomycetota), and Verrucomicrobia (Verrucomicrobiota) [52]. The Firmicutes/Bacteroidetes ratio (Bacillota/Bacteroidota) has been used as a critical parameter for gut health [53] and is critical for CRC progression [54]. A change in this ratio is associated with inflammatory bowel diseases (IBDs) [55,56,57], which are risk factors for CRC.
3. Gut Microbiota and Colorectal Cancer
In recent years, much attention has been paid to the role of microbes in cancer development. It has been found that microbes are involved in 20% of cancers [58], including CRC [59]. The first study that showed the effect of gut microbiota in mediating the carcinogenic effects of cycasin in germ-free mice was published in 1967 [60]. Several studies have found a link between dysbiosis of the gut microbiota and the development of CRC [61,62,63,64,65,66,67]. Mounting evidence suggests that altering the gut microbiota affects CRC progression [68,69,70].
Gut dysbiosis refers to the compositional and functional alterations caused by imbalance between symbiotic and opportunistic microbes [71]. Dysbiosis is categorized into three types: (i) loss of beneficial microbes, (ii) expansion of pathogenic microbes, and (iii) loss of microbial diversity [72]. Dysbiosis contributes to many pathological conditions, such as diabetes [73,74]. obesity [75,76], neurogenerative diseases [77], and cancers [78,79,80].
Studies have suggested a link between environmental factors and dysbiosis in the gut microbiota with CRC carcinogenesis and prognosis [81,82,83,84]. Various factors, such as lack of exercise, antibiotics, western diet, aging, and obesity, may cause a shift in the gut microbiota to a pro-inflammatory type [85]. Long-term antibiotic use is associated with an increased risk of CRC, linking gut microbiota with CRC [86]. With advancing age, there is a loss of CD4 T-cells and a shift in the microbiota to a pro-inflammatory type. This decreases the ability of immune cells to suppress inflammation in the colon [45]. There is also a reduction in butyrate-producing bacteria, increasing the intracolonic pH, which along with dysbiosis and inflammation contributes to CRC [81]. Smoking also alters gut microbiota composition and induces CRC in a mouse model [87].
A study published in 2017 showed that gavage of fecal samples from patients with CRC to germ-free and conventional mice promoted intestinal carcinogenesis [88]. Studies carried out on germ-free mouse and rat models of CRC have indicated a reduced tumor load as compared to those reared under conventional conditions [89,90]. Alterations in the microbiota are not only restricted to the tumor site but are also seen in surrounding healthy tissues that show the same microbiota composition as tumor tissue [91]. Alterations in gut microbiota are also linked to other cancers, including hepatocellular carcinomas [92,93], pancreatic cancer [94], and breast cancer [95].
Studies on humans have shown that the gut microbiota of patients with CRC differ from the microbiota of healthy subjects, with a lower abundance of commensal bacteria and a higher abundance of procarcinogenic bacteria [96,97]. Studies have also found differences in the fecal and mucosal microbiota of CRC patients [98]. Touchefeu et al. [99] found that F. prausnitzii, Barnesiella intestinihominis, Alistipes finegoldii, Bacteroides eggerthii, and Eubacterium siraeum were significantly decreased in CRC patients compared with controls. Several studies have tried to dive deep into the microbiota composition associated with CRC in fecal [96,97,100,101,102,103] and mucosal samples [84,104,105,106,107]. These studies have found a global compositional shift in the CRC microbiota. Studies conducted across different populations have shown the association of certain bacterial species with CRC [108,109,110] (Table 2). Many studies have found a lower bacterial diversity and an increase in certain pro-tumorigenic bacteria in CRC [65,84,111,112,113,114]. The bacterial species commonly associated with colorectal carcinogenesis include Fusobacterium nucleatum [33,115], E. coli [68,116], Bacteroides fragilis [61,117,118,119], Streptococcus bovis/gallolyticus [120], Clostridium septicum [121,122], Enterococcus faecalis [123,124], and Peptostreptococcus anaerobius [125,126] (Figure 1). A meta-analysis of 536 fecal shotgun metagenomes identified a core set of seven bacterial species enriched in CRC. These were Fusobacterium nucleatum, Bacteroides fragilis, Parvimonas micra, Porphyromonas asaccharolytica, Prevotella intermedia, Alistipes finegoldii, and Thermanaerovibrio acidaminovorans [109]. A study published in 2019 identified 29 core species enriched in CRC across eight different geographical regions [102]. Wang et al. [124] reported that Bacteroides fragilis, Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus, and Peptostreptococcus were significantly more abundant in the gut microbiota of CRC patients, while Roseburia and other butyrate-producing bacteria of the family Lachnospiraceae were less abundant. Studies have suggested that certain bacteria, including E. coli, Bacteroides fragilis, and Peptostreptococcus anaerobius, are associated with colorectal carcinogenesis through activating Th17 cell response [127] and inducing DNA damage [68,128].

Table 2.
Putative CRC-associated gut microbes reported to have a mechanistic role in carcinogenesis.
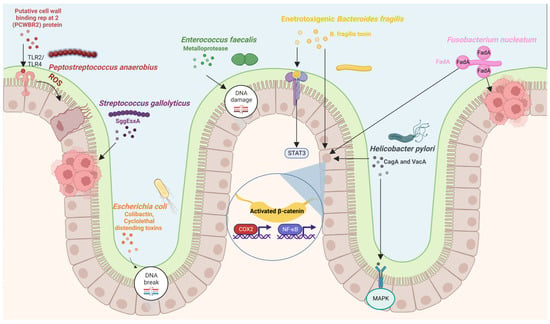
Figure 1.
Diagram showing putative bacteria implicated in colorectal carcinogenesis and their molecular mechanisms. Figure created with BioRender.com.
Apart from bacteria, many viruses have been identified in human CRC samples, including human papillomavirus [129] and cytomegalovirus [130]. A study published in 2018 found alterations in enteric virome profiles that were associated with survival outcomes of CRC patients [131]. A few studies have found changes in the mycobiome in CRC samples. One study found an increase in the Ascomycota/Basidiomycota ratio, with increased proportions of Trichosporon and Malassezia [132], while the other study found an increase in the Basidiomycota/Ascomycota ratio, with an increase in Malasseziomycetes and a decrease in Saccharomycetes and Pneumocystidomycetes [133]. Wang et al. [134] reported a significant increase in Candida albicans in CRC patients. A recent study found alterations in mycobiota in CRC with enrichment of Aspergillus rambellii, Cordyceps sp. RAO-2017, Erysiphe pulchra, Moniliophthora perniciosa, Sphaerulina musiva, and Phytophthora capsici [135]. Another recent study found significant enrichment of Phanerochaete chrysosporium, Lachancea waltii, and Aspergillus rambellii in CRC [136]. Coker et al. [137] reported alterations in the archaeomes of patients with CRC, with enrichment of halophiles and depletion of methanogens.
These results indicate that transkingdom crosstalk may be required for colorectal carcinogenesis. A recent study [138] found four kingdom microbiota alterations in samples from eight distinct geographical cohorts. This study identified 16 multi-kingdom markers (11 bacterial, 4 fungal, and 1 archaeal) that could help in diagnosing patients with CRC. Many studies have also reported associations between CRC and oral microbiota [139,140].
The role of microorganisms in CRC development has been explained by the ‘bacterial driver-passenger’ model [141,142]. According to this model, pathogenic intestinal bacteria called “drivers” produce genotoxins that induce DNA damage, causing genome instability and CRC initiation. The CRC microenvironment promotes the proliferation of specific opportunistic bacteria called “passenger bacteria” that not only have growth advantages but also show carcinogenic effects. These passenger bacteria can outgrow other bacteria due to competitive advantage. However, it is still unknown whether dysbiosis is a cause or a consequence of CRC.
3.1. Fusobacterium nucleatum
Fusobacterium nucleatum is a Gram-positive anaerobic bacteria that has been found to be enriched in CRC [33,34,143,144,145,146,147]. Though a normal constituent of the human oral cavity, F. nucleatum is less commonly detected in the gut microbiota of healthy individuals [148]. Since F. nucleatum is an important organizer of biofilms in the oral cavity, it is thought to be a pioneer organism that is responsible for creating a microenvironment conducive to other pathogenic microorganisms.
Studies have confirmed increased colonization of F. nucleatum in adenomas, which may be 400 times higher than the adjacent normal tissues [33,149,150]. High colonization is mostly seen in advanced stage III-IV CRC [151,152]. A high abundance of F. nucleatum is associated with increased expression of β-catenin, NF-kB, and tumor necrosis factor (TNF)-β [153]. Besides, F. nucleatum has been found to be associated with the CpG island methylator phenotype (CIMP) and microsatellite instability (MSI) in CRC [151,154]. An abundance of Fusobacterium is positively associated with lymph node metastasis [143], lower T cell infiltration [155], and poor patient survival [146,156]. The presence of F. nucleatum in distant metastatic lesions suggests its role in CRC metastasis [157]. Recent studies have supported the role of F. nucleatum in promoting liver metastasis [158,159]. Kostic et al. [33], while working with Apcmin/+ mice, found that exposure to F. nucleatum was sufficient to promote small intestinal adenocarcinoma development. They also found that F. nucleatum induced carcinogenesis by selectively recruiting tumor-infiltrating myeloid cells. F. nucleatum is also associated with a decrease in antitumor M1 macrophages and an increase in protumor M2 macrophages [160].
F. nucleatum uses FadA adhesin for adhesion and invasion into epithelial cells. FadA binds to E-cadherin, stimulates the Wnt/β-catenin pathway [115] and increases the permeability of endothelial cells, which allows F. nucleatum to cross cell-cell junctions [161]. The synthetic peptides that prevent the binding of F. nucleatum to E-cadherin suppress CRC development. Another protein, Fap2, can inhibit natural killer (NK) cells by associating with TIGIT, an inhibitory receptor on NK cells [162]. F. nucleatum can induce carcinogenesis through the inflammatory NF-κb signaling pathway and downregulates CD3+-T-cell-mediated adaptive immunity [33,155]. A significant association between F. nucleatum and patient outcome suggests that F. nucleatum may be used as a prognostic biomarker for CRC [146,156]. F. nucleatum can also activate TLR4 signaling in mice to promote tumor development [163,164]. A recent study found that F. nucleatum activates YAP signaling, inhibits FOXD3 expression, and reduces METTL3 transcription, thus reducing m6A modifications in CRC cells [165]. Formate, a metabolite produced by F. nucleatum, increases tumor incidence and Th17 cell expansion, promoting CRC development [166].
3.2. Escherichia coli
Escherichia coli is a commensal Gram-negative and facultative anaerobic bacterium that is classified into four phylogenetic groups (A, B1, B2, and D) that are predominant in several human populations [167,168]. Various studies have confirmed a link between E. coli, particularly from the B2 phylogroup, and CRC [68,70,116,169,170]. Most pathogenic strains of E. coli are involved in inflammatory bowel diseases, such as Crohn’s disease, which are risk factors for CRC [116,171,172]. Moreover, an increase in the colonization of colon mucosa by mucosa-associated E. coli has been reported in patients with CRC [68,70,116,173,174], suggesting the role of E. coli in CRC. Many pathogenic strains of E. coli produce toxins called cyclomodulins, such as colibactin, cytolethal distending toxins (CDTs), cycle inhibiting factors, and cytotoxic necrotizing factors (CNFs) [174,175,176].
Colibactin is encoded by pks island in E. coli. It alkylates DNA on adenine residues [177], induces double-stranded breaks in DNA [175], and interferes with the cell cycle [128]. Studies have shown that colibactin-producing E. coli induces inflammation pathways and thus has a pro-carcinogenic effect [68,70,178]. Colibactin-producing E. coli is found to be prevalent in patients with advanced stages of CRC [70]. CDTs damage DNA [179] and have been shown to be potent carcinogens [180,181]. Pleguezuelos-Manzano et al. [182] identified a distinct mutational signature from human intestinal organoids exposed to genotoxic pks+ E. coli. This signature has also been found in patients with CRC, suggesting that CRC results from past exposure to E. coli harboring a colibactin-producing pks pathogenicity island. A recent study found that colibactin-producing E. coli induced colorectal carcinogenesis in a CRC mouse model lacking genetic susceptibility [183].
3.3. Bacteroides fragilis
Bacteroides fragilis is a commensal Gram-negative anaerobic bacterium that represents less than 1% of the gut microbiota [184,185]. There are two subtypes of B. fragilis: non-toxigenic and enterotoxigenic [185,186]. Enterotoxigenic B. fragilis (ETBF) is responsible for diarrhea in children [187]. Studies have indicated increased colonic colonization in CRC patients [61,64] and the inflammatory potential of ETBF [188], suggesting a link between B. fragilis and CRC. Most ETBF strains have a bft gene that codes for B. fragilis toxin (BFT or fragilysin), which is responsible for their toxigenicity [186,189]. BFT is a zinc-dependent metalloprotease that directly affects signaling pathways, such as the Wnt, NF-κB, and mitogen-activated protein kinase (MAPK) pathways, leading to increased cell proliferation and production of pro-inflammatory mediators [117,118,119,190]. ETBF also activates the Stat3 transcription factor in the colon of Apcmin/+ mice [127]. Studies have found that infection by ETBF increases Th17 and T regulatory cells (Treg) [191], with a crucial role of BFT in triggering carcinogenesis through inflammation pathways [186,192]. High levels of B. fragilis are correlated with increased expression of cyclooxygenase 2 (COX-2) and NF-kB [153]. A recent study found a correlation between B. fragilis and the levels of inflammatory cytokines [193]. This study also reported that B. fragilis in polyps were bft-negative and enriched in genes associated with LPS biosynthesis. High abundances of F. nucleatum and B. fragilis have ben found to be indicators of poor survival of CRC patients [153].
3.4. Enterococcus faecalis
Enterococcus faecalis is a commensal Gram-positive, facultative anaerobic bacterium. Studies have confirmed an increased abundance of E. faecalis in the feces of patients with CRC [123,124]. However, the role of E. faecalis in CRC development remains controversial [194]. Many studies have suggested its ability to generate reactive oxygen species (ROS) and extracellular superoxide that can damage colonic epithelial cell DNA, leading to mutations and CRC [124,195,196,197]. E. faecalis also produces metalloprotease, which can affect the intestinal epithelial barrier and induce inflammation [198]. Contrary to this, other studies indicate that E. faecalis is an important probiotic microorganism [199] and may have a role in CRC prevention [200,201,202].
3.5. Streptococcus bovis/gallolyticus
Streptococcus bovis/gallolyticus is a Gram-positive bacterium associated with endocarditis [203]. The first case of endocarditis-associated CRC was reported in 1951 [204]. Studies have confirmed the association of S. bovis/gallolyticus infection with CRC [205,206,207,208,209] and the prevalence of S. bovis/gallolyticus in CRC tissues [210]. The exact mechanism by which S. bovis/gallolyticus induces carcinogenesis is still to be characterized, but studies have indicated its involvement during the early stages of colorectal carcinogenesis [205,206,208,211], and therefore, it may serve as an early marker for CRC screening. S. gallolyticus uses Pil3 pilus for adhesion to and translocation across colonic epithelial cells [212,213]. Additionally, bacterial type VII secretion systems (T7SS) may mediate the interactions of S. gallolyticus with host cells and are important for its virulence [214]. S. gallolyticus probably caused CRC development by upregulating β-catenin levels and inducing inflammation via IL-1, IL-8, and COX-2 [210]. Oral gavage with S. bovis/gallolyticus has been found to increase the tumor burden in azoxymethane (AOM) and dextran sulfate sodium (DSS) mouse models of tumorigenesis [215,216].
3.6. Helicobacter pylori
Helicobacter pylori is a Gram-negative, microaerophilic bacterium that is found in the stomach and is responsible for peptic ulcers, chronic gastritis, mucosa-associated lymphoid tissue lymphoma, and gastric adenocarcinoma [217,218,219]. The eradication of H. pylori can, therefore, prevent gastric cancer [220]. H. pylori was designated as a Group 1 human carcinogen by the International Agency for Research on Cancer (IARC) in 1994. H. pylori promotes carcinogenesis by activating the β-catenin signaling pathway [221]. In gastric epithelial cells, H. pylori and IL-22 induce matrix metalloproteinase-10 (MMP-10) through the extracellular signal-regulated kinase (ERK) pathway. MMP-10 induces inflammation and damages gastric mucosa by inhibiting tight junction proteins [222]. Studies have suggested increased risk of CRC in patients with H. pylori infection [223,224,225]. Moreover, increased colonization of colonic mucosa by H. pylori has also been reported in adenomas and adenocarcinomas [226]. The pathogenicity islands in some H. pylori strains code for virulence factors, such as cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA) [227], which may induce inflammation pathways and cell proliferation [228]. CagA+ and VacA+ H. pyroli strains are associated with an increased risk of gastric cancer [229,230] and CRC [231,232,233]. Infection with Cag+ H. pyroli strains inactivates tumor suppressor pathways with induced P53 mutations [234,235]. A study on human gastric organoids confirmed that CagA protein bound and phosphorylated c-Met, stimulating epithelial cell proliferation [236]. VacA promotes cell vacuolation [237], upregulates MAP kinase and ERK1/2 expression [238] and the Wnt/β-catenin signaling pathway [239], and activates vascular endothelial growth factor [240], thereby inducing epithelial cell proliferation. H. pylori infections are also associated with methylations on CpG islands [241], but the direct role of H. pylori in hypermethylation remains controversial.
H. pylori infections are associated with an increase in Proteobacteria, Acidobacteria, and Spirochaetes and a decrease in Firmicutes, Bacteriodetes, and Actinobacteria [242], suggesting that H. pylori infection is associated with dysbiosis [243]. Studies have also found an inverse relationship between H. pylori abundance and bacterial diversity [244,245].
3.7. Peptostreptococcus anaerobius
Peptostreptococcus anaerobius is a Gram-negative anaerobic bacterium commonly found in the oral cavity and the gut. It promotes carcinogenesis by modulating immune cells and interacting with toll-like receptors, TLR-2 and TLR-4, on colon cells to induce ROS formation [125,126]. The binding of P. anaerobius to colon cancer cells is mediated by its surface protein, putative cell wall binding repeat 2 (PCWBR2), which interacts with α2/β1 integrins on colon cells. The binding of P. anaerobius was found to activate the PI3K-Akt pathway, stimulating inflammation and cell proliferation [126].
3.8. Parvimonas micra
Several oral diseases, including endodontic abscesses, odontogenic infections, periodontitis lesions, and other infections, are frequently associated with Parvimonas micra [246]. A recent study using an isolated strain from a CRC patient showed that it could promote coloncyte proliferation via the enhancement of Th17 cell infiltration and the oncogenic Wnt signalling pathway [247].
4. Biomarkers for CRC Screening
For diagnosis of CRC, the identification of novel biomarkers, which are reliable and non-invasive, is desirable. The identification of microbial biomarkers is helpful for designing non-invasive tools for CRC diagnosis. It has been estimated that the use of accurate tests for screening average-risk individuals can reduce the incidence and mortality associated with CRC [248]. Current clinical diagnostic procedures, such as the fecal occult blood test (FOBT), have limited sensitivity for detecting CRC [249].
Though fecal immunochemical testing (FIT) has high sensitivity for the detection of CRC [250], it can detect CRC with a sensitivity of 79% [249] and colorectal adenomas with a sensitivity of 25–27% [251].
CRC-enriched bacteria may serve as potential diagnostic bacterial markers [102,109]. Large-scale studies on fecal metagenomes have identified microbial signatures that can predict CRC in various populations [96,97,100,103,131,133,252,253,254,255,256]. A study published in 2017 identified a set of 22 genes associated with CRC [97]. Four of these genes, butyryl-CoA dehydrogenase from F. nucleatum, two transposases from Peptostreptococcus anaerobius, and RNA polymerase subunit (rpoB) from P. micra, were also present in Danish, French, and Austrian cohorts. Many recent studies have identified microbial markers associated with CRC. Huo et al. [257] identified 17 bacterial genera/families that could serve as potential biomarkers for CRC recurrence and patient prognosis. Avuthu and Guda [258] identified CRC-associated species including C. symbiosum, F. nucleatum, R. torque, G. morbillorum, S. moorei, P. micra, and Clostridium citroniae. Li et al. [259] identified six key genera that were consistently over-represented in tumor mucosa, including Fusobacterium, Gemella, Campylobacter, Peptostreptococcus, Alloprevotella, and Parvimonas.
F. nucleatum has the potential to serve as a biomarker for CRC [260,261]. Screening for F. nucleatum in fecal samples has shown to differentiate patients with colorectal adenomas from healthy subjects [260]. Studies have shown an inverse relationship between F. nucleatum abundance and CRC survival [146,153]. Serum antibodies against F. nucleatum can also serve as a biomarker for screening CRC [262]. Similarly, serological tests based on antibodies against S. gallolyticus have also been explored as potential biomarkers for CRC [263,264].
Microbial metabolites, such as SCFAs and bile acids, that have been associated with CRC progression may also serve as biomarkers for the early screening of CRC [265]. Many studies have reported differentiating levels of microbial metabolites in fecal samples of patients with CRC, including ursodeoxycholic acid, lower levels of butyrate, and higher levels of acetate [266]. Analysis of CRC metagenomes suggests an enrichment of protein and mucin catabolism genes and a depletion of carbohydrate degradation genes [102], which may be used as a marker for preliminary CRC diagnosis. Zhang et al. [267] identified biomarkers associated with CRC stage I (Peptostreptococcus and Parvimonas), stage II (Fusobacterium, Streptococcus, Parvimonas, Burkholderiales, Delftia, Caulobacteraceae, and Oxalobacteraceae), and stage III (Fusobacterium, Faecalibacterium Sutterella, Burkholderiales, Caulobacteraceae, and Oxalobacteraceae). Chang et al. [268] identified 37 CRC-enriched bacterial species, including Fusobacterium nucleatum, Parvimonas micra, Citrobacter portucalensis, Shigella sonnei, Gemella morbillorum, Alloprevotella sp., and Coriobacteriaceae. Shen et al. [269] identified five phage biomarkers, including Peptacetobacter hiranonis Phage, Fusobacterium nucleatum animalis 7_1 phage, Fusobacterium nucleatum polymorphum phage, Fusobacterium nucleatum animalis 4_8 phage, and Parvimonas micra phage. Liu et al. [270] reported that 25 species and 65 antibiotic resistance genes were significantly enriched in CRC patients. Of 65 antibiotic resistant genes, 12 were multidrug-resistant genes (MRGs), including acrB, AcrS, TolC, marA, H-NS, and Escherichia coli acrR mutation. Osman et al. [271] identified four bacterial markers that could distinguish CRC patients from control individuals. These biomarkers were Parvimonas micra, Fusobacterium nucleatum, Peptostreptococcus stomatis, and Akkermansia muciniphila. Löwenmark et al. [272] reported the use of Parvimonas micra as a potential non-invasive biomarker for CRC.
A study published in 2019 reported an increase in the abundance of genes involved in amino acid and sulphur metabolism and a relative decrease in the abundance of genes involved in methane metabolism in patients with preneoplastic polyps [103].
5. Microbial Mechanisms Involved in Colorectal Carcinogenesis
The key mechanisms by which gut microbiota induce colorectal carcinogenesis include genotoxins and virulence factors, gut microbial metabolites, inflammation pathways, oxidative stress, and anti-oxidative defense modulation.
5.1. Bacterial Genotoxins and Virulence Factors
During biological evolution, gut bacteria developed pathogenicity by acquiring various virulence factors that enabled them to penetrate the gut mucosal barrier and intestinal epithelial cells [172,273,274], which form a barrier between human tissues and microbiota. A breach in this barrier results in inflammation [275]. Virulence factors are responsible for pro-carcinogenic and disease-promoting effects [276]. CRC-associated E. coli strains have Afa and Eae adhesins, which allow them to adhere to and invade intestinal epithelial cells [277], and activate inflammation pathways [278]. F. nucleatum binds to E-cadherin via its FadA virulence factor, activating the β-catenin signaling pathway and promoting colorectal carcinogenesis [115,279,280].
Many pathogenic bacteria produce toxins that are associated with the development of CRC. ETBF produces B. fragilis toxin (BFT), which activates the NF-κB and Wnt/β-catenin pathways [186,188], leading to increased cell proliferation, DNA damage, and release of pro-inflammatory mediators [281,282,283]. BFT can also hydrolyze the extracellular domain of E-cadherin [117,284]. Many gut microbes produce genotoxins that damage DNA. Cyclomodulins, such as colibactin, cytolethal distending toxins (CDT), cycle inhibiting factors, and cytotoxic necrotizing factors (CNFs), are genotoxins that induce DNA damage and interfere with the cell cycle [127,128,285,286,287,288]. CDTs and colibactin are considered true genotoxins as they directly mediate DNA damage by inducing double-stranded DNA breaks [128,285,289]. CDTs are well-characterized toxins produced by most Gram-negative bacteria associated with CRC, such as Escherichia and Campylobacter [290]. The CdtA and CdtC subunits allow interactions with host cells, and the CdtB subunit can translocate to the nucleus and damage host cell DNA [128,285,291,292]. CDTs also induce the production of IL-6, TNF-α, NF-κB, and cyclooxygenase 2 [67]. Colibactin induces DNA damage, ROS formation, and cell cycle arrest [68,128]. Targeting colibactin production has been shown to reduce tumors in a mouse model [293]. Although toxin-producing bacteria represent a small proportion of the gut microbiota, an analysis of CRC tissue samples suggests a high expression of these toxins [294]. Therefore, targeting these toxins may have therapeutic implications in CRC.
5.2. Gut Microbial Metabolites and Products
Some metabolites produced by gut microbes are linked to CRC [295,296]. The microbial products that affect CRC development are secondary bile acids, acetaldehyde, trimethylamine-N-oxide (TMAO), and glucuronidase.
5.2.1. Secondary Bile Acids
Bile acids are a type of steroid acid found in bile. They are involved in the emulsification and absorption of fats and the elimination of cholesterol. Cholic acid (CA) and chenodeoxycholic acid (CDCA) are primary bile acids synthesized in the liver. Secondary bile acids, such as deoxycholic acid (DCA) and lithocholic acid (LCA), are produced from primary bile acids by the action of anaerobic microorganisms in the colon [297], which use bile acids as a source of energy [298]. It has been found that people with high-fat diets are susceptible to CRC [299,300], probably because a high-fat diet increases the secretion of primary bile acids, which are converted by gut microbes to secondary bile acids [299,301,302]. Feeding mice with secondary bile acids increases inflammation and induces CRC [301].
DCA can modulate intracellular signaling and gene expression [303]. It can also induce the expression of orphan nuclear receptor Nur77 [304] and downregulate the expression of miR-199a-5p [305]. Nur77 promotes tumorigenesis by upregulating anti-apoptotic BRE (brain and reproductive-organ-expressed protein) and angiogenic VEGF (vascular endothelial growth factor) [304]. miR-199a-5p can target CAC1 (CDK2-associated cullin domain 1), a novel cell cycle regulator widely expressed in CRC, for degradation and, therefore, functions as a tumor suppressor. DCA can directly induce oxidative DNA damage and tumor formation [301,306]. DCA induces epithelial-mesenchymal transition and activates vascular endothelial growth factor receptor 2, leading to intestinal carcinogenesis [307]. LCA is known to induce the expression of urokinase-type plasminogen activator receptor (uPAR), which may activate the MAPK signaling pathway and contribute to cancer progression and metastasis [308,309].
Secondary bile acids can activate G-protein-coupled bile acid receptor 1 (GPBAR1), inducing epithelial cell proliferation [310]. Secondary bile acids can promote DNA damage (by producing ROS and RNS) [311,312], regulate gene expression and membrane permeability [313], and activate epidermal growth factor receptor (EGFR) pathway signaling [303,314,315] and the protein kinase C pathway [316]. In addition, bile acids have strong antimicrobial properties and cause changes in the gut microbiome by selectively killing microbes. This leads to an increase in the population of CRC-associated Bacteroidetes and Gamma-proteobacteria [93].
A bile acid, ursodeoxycholic acid (UDCA), which is produced by Ruminococcus gnavus [317], suppresses colon carcinogenesis [302,318]. It inhibits COX-2 expression [319] and DCA-induced apoptosis through modulation of EGFR/Raf-1/ERK signaling in colon cancer cells [320].
5.2.2. Acetaldehyde
Acetaldehyde is produced from ethanol by the activity of aerobic and facultative anaerobic bacteria in the gut. Excessive consumption of ethanol is considered a risk factor for various cancers, including CRC [321]. Acetaldehyde is highly toxic and pro-carcinogenic. It can damage DNA and impair DNA excision repair, promoting colorectal carcinogenesis [322].
5.2.3. Trimethylamine-N-oxide (TMAO)
Trimethylamine-N-oxide (TMAO) is produced by a reaction between flavin monooxygenase and trimethylamine (TMA), which is a microbial metabolite produced from red meat and fats. A diet rich in fats and red meat leads to the production of more TMAO because L-carnitine (a TMA) is processed by gut microbes to form TMAO [323]. TMAO has been linked to increased risk of cardiovascular diseases [323,324,325] and CRC development [326,327]. TMAO causes CRC probably by inducing DNA damage, inflammation, oxidative stress, and protein misfolding [328,329].
5.2.4. Glucuronidase
High fecal glucuronidase activity is found in patients with CRC [330]. The liver inactivates some carcinogens by glucuronic-acid-mediated conjugation, which are excreted through the digestive tract. This process may be reversed in the colon by bacterial glucuronidase, reactivating carcinogens. Inhibiting bacterial glucuronidase in a mouse model can reduce the number of tumors [331], indicating that bacterial glucuronidase is responsible for CRC progression. Moreover, bacterial glucuronidase also affects the activity of some anti-tumor drugs [332], influencing the treatment outcome.
5.3. Inflammation and Host Immunity
Inflammation is an adaptive response of the host immune system. Significant inflammation is not caused by healthy microbiota as the host immune system is programmed to recognize normal gut microorganisms. There is a constant cross-talk between host immune cells and gut microbes, selecting and tolerating commensal microbes and eliminating pathogenic ones [333]. Alterations in the gut microbiota play an important role in inflammation [334], which promotes CRC development [335]. The inflammatory events associated with CRC development include DNA damage by ROS and RNS produced by macrophages, neutrophils, and dendritic cells (DCs) and the production of cyclooxygenase-2 [336]. Moreover, the invasion of the intestinal mucosa by pathogens triggers the activation of immune cells and the release of cytokines and growth factors [337], which drive the inflammation process. Persistent inflammation results in epithelial cell proliferation, angiogenesis, and inhibition of apoptosis, leading to cancer [338,339]. The pro-inflammatory cytokines secreted by macrophages and T cells, such as IL-6 and TNF, trigger the differentiation of pro-inflammatory Th17 cells. The prolonged presence of Th17 cells and elevated levels of associated cytokines, such as IL-17 and IL-22, are associated with poor survival in CRC [337]. Studies have found that limiting Th17 cells reduces the risk of carcinogenesis [127,340]. IL-6 is an important cytokine required for angiogenesis and the activation of STAT3 [341]. Increased serum IL-6 and TNF levels have been used as prognostic markers for poor survival of CRC patients [342,343]. Inflammation-associated factors can also activate oncogenes [344] or inactivate tumor suppressor genes [345].
Inflammation is associated with the development of inflammatory bowel diseases (IBD), such as ulcerative colitis and Crohn’s disease. IBDs are associated with increased risk of developing CRC, called colitis-associated cancer [346,347,348], as 20% of patients with ulcerative colitis develop CRC [349]. Individuals with pancolitis are at a higher risk of developing CRC than patients with limited colitis [350]. Meta-analyses have indicated a risk of 18.4% for patients with ulcerative colitis [351] and 8.3% for patients with Crohn’s disease [352] to develop CRC. Colitis stimulates carcinogenesis by inducing the expansion of genotoxic bacteria [68]. The gut microbiota of patients with IBDs shows increased abundance of Proteobacteria, particularly Enterobacteriaceae, such as E. coli [353,354]. The role of colibactin-producing E. coli in inducing inflammation and intestinal tumors was elucidated in a study carried out in IL10-deficient mice treated with the genotoxic agent azoxymethane. In such a mouse model lacking pks+ E. coli, fewer intestinal tumors developed than in similarly treated mice with pks+ E. coli [68]. ETBF promotes inflammation by activating STAT3 and NF-κB signaling in colonic epithelial cells [33,355]. The NF-κB pathway is an important regulator of the genes encoding TNF and COX-2, which are usually overexpressed in IBDs and CRC [356]. B. fragilis also induces the expression of spermine oxidase in colonocytes, which induces ROS production and DNA damage [283].
Other bacteria, such as Citrobacter rodentium and Mycobacterium, can also promote inflammation, inducing IBDs [357,358,359]. Patients with IBDs are more likely to be affected by CRC due to changes in the gut microflora homeostasis [360]. IBDs are also associated with dysbiosis with lower Firmicutes and Bacteriodetes as compared to healthy subjects [361]. Long-term use of NSAIDs is known to reduce the risk of CRC, suggesting a link between inflammation and CRC development [362].
Pattern-recognition receptors (PRRs), such as toll-like receptors (TLRs) and nucleotide-binding oligomerization (NOD)-like receptors (NLRs), play an important role in recognizing the specific molecular patterns of pathogenic microorganisms called microorganism-associated molecular patterns (MAMPs) [363]. PRRs recognize microbial surface molecules, such as peptidoglycan, flagellin, lipoproteins, lipoteichoic acid, and lipopolysaccharides. Lipoteichoic acid specifically binds to CD14 or TLR-2, inducing the secretion of pro-inflammatory factors [364,365]. TLRs are a major class of PPR expressed in macrophages and dendritic cells. They recognize microbes and activate an immune response if the mucosal barrier is disrupted. TLR signaling initiates immune defense by producing pro-inflammatory cytokines and also enhances barrier function, preventing microbial invasion [366]. There are two important TLR pathways: myeloid differentiation factor 88 (MyD88) adaptor-protein-dependent and TRIF-dependent [367,368]. In the MyD88-dependent pathway, the downstream activators are NF-κB and MAPK [368]. Studies have suggested the role of MyD88 in CRC induction [369]. In AOM-treated APCmin/+ mice, the inactivation of MyD88 resulted in a decrease in tumor number [370].
TLR-2 expression is important in maintaining the gut microbiota composition and suppressing inflammation [371,372]. It has been found that the number of tumors increases in TLR2-deficient mice compared to control mice [373], suggesting that TLR2 is important for maintaining gut homeostasis [374]. An increase in TLR4 activates NF-κB, which induces COX-2 expression and an increased risk of CRC [375]. High levels of TLR4 and MyD88 in CRC patients increase the risk of liver metastasis and also affect survival [376,377]. Inhibition of TLR4 expression protects against CRC [378]. A study also found that chronic activation of TLR9 may induce hyperproliferation and CRC development [379].
NLRs are located in the cytoplasm of immune and non-immune cells. NLR activation triggers the production of pro-inflammatory cytokines and autophagy [380]. A study found a significant difference in NLR signaling between the tumor and non-tumor tissues of patients with CRC [381]. NOD1- or NOD2-deficient APCmin/+ and AOM/DSS-treated mice show asignificant increase in CRC numbers [382]. NOD2 mutations are also associated with Crohn’s disease and increased risk of CRC [383,384,385,386].
5.4. Oxidative Stress
Oxidative stress is caused by imbalance between oxidative molecules, such as ROS and RNS, and anti-oxidative defenses [387]. Oxidative stress affects biomolecules, damages cell membranes, and induces DNA breaks and mutations [387,388]. Oxidative stress induces NF-κB and up-regulates the expression of pro-inflammatory cytokines and anti-apoptotic signaling [338]. The production of ROS has been directly linked with CRC induction. ROS are produced by the gut microbiota or host immune cells, such as macrophages and neutrophils, in response to inflammation induced by pathogenic bacteria or other external environmental factors [389,390,391]. RNS are produced by some bacterial species, such as Lactobacillus and Bifidobacterium [392,393,394]. Species, such as E. faecalis, can produce hydroxyl radicals [195,395] that contribute to CRC development by inducing point mutations and chromosomal breaks [388,396]. H. pylori also induces oxidative stress, resulting in gastric carcinogenesis [397].
Various anti-oxidative defense mechanisms, such as DNA repair, balance oxidative stress [398,399]. Only the base-excision repair system accounts for more than 10,000 repairs in the colon cells per day [400]). Anti-oxidative defense mechanisms are found to be altered in CRC [401,402]. Studies have found downregulation of the MMR system by some enteropathogenic E. coli strains [277,403] as well as in a colitis-induced CRC mouse model [404]. Using APCmin/+ MMR-deficient mice, Belcheva et al. [405] found that gut microbes could induce CRC in MMR-deficient epithelial cells.
6. Diet and the Risk of CRC
Diet plays an important role in the development and progression of many cancers, including CRC [406] (Table 3 and Figure 2). Diet is the major determinant of gut microbiota [407], and changes in diet are accompanied by changes in the fecal microbiota within a few days of diet change [28,408]. High intake of red meat, processed meat, and fats increases the risk of CRC [326,327,409,410], while a high intake of dietary fiber can decrease this risk [411]. Processed meat and red meat are classified as Class 1 and Class 2A carcinogens, respectively, by the WHO. The risk of CRC increases linearly with the intake of processed and red meat [412]. The heme iron in red meat is converted into carcinogenic N-nitroso compounds, which contribute to CRC development [413]. In addition, heme iron also increases mucin-degrading bacteria, such as Akkermansia muciniphila, leading to gut barrier function impairment and CRC [414]. Heme causes persistent intestinal dysbiosis, with an increase in Proteobacteria, resulting in inflammation and hyperproliferation of the intestinal epithelium [415].

Table 3.
Dietary components and their metabolic products in CRC tumorigenesis.
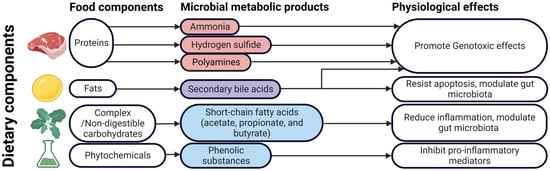
Figure 2.
Food components, microbial metabolites, and their physiological effects that play putative roles in colorectal carcinogensis. Figure created with BioRender.com.
A diet rich in meat was found to increase Bacteroidetes and decrease Firmicutes [408]. Zimmer et al. [416] reported that fecal samples from vegans showed a significant reduction in Enterobacteriaceae compared to omnivorous control subjects. Vegetarians, therefore, have a 20% lower risk of developing CRC than non-vegetarians [417].
A diet rich in complex carbohydrates increases the abundance of probiotic bacteria, such as Bifidobacterium longum, Bifidobacterium breve, and Bifidobacterium thetaiotaomicron [418] and reduces the growth of opportunistic bacteria, such as Enterobacteriaceae [419]. On the other hand, excessive consumption of refined sugars results in the proliferation of pathogenic bacteria, such as Clostridioides difficile and Clostridium perfringens [420]. A diet rich in resistant starch increases the abundance of Firmicutes, such as Ruminococcus bromii, while a diet rich in wheat increases the abundance of butyrate-producing Lachnospiraceae [419]. A low carbohydrate diet is associated with a decrease in butyrate-producing Firmicutes and Actinobacteria [419,421].
Dietary fibers are important factors that affect gut microbial composition and diversity [422]. It has been found that patients with colorectal adenomas have a low intake of dietary fibers [423]. Dietary rice bran intake has been shown to modify gut microbiota and increases the anti-cancer metabolites, myristoylcarnitine and palmitoylcarnitine, in a mouse model of CRC [424]. Cellulose intake has been found to decrease inflammation and tumor formation and increases survival rate in an AOM/DSS mouse model of CRC [425].
Non-digestible carbohydrates are processed by gut microbes to produce short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate [295]. The composition of the gut microbiota affects the production of these SCFAs and thus the risk of CRC development [426,427,428]. SCFAs interact with G-protein-coupled receptors (GPCRs), such as GPR41 (FFA3), GPR43 (FFA2), and GPR109A, expressed in human colon epithelial cells and activate them [429]. GPR43 recognizes acetate, propionate, and butyrate [430], while GPR109A interacts with only butyrate [431]. Butyrate activates GPR109A [432] and promotes the differentiation of regulatory T cells (Tregs) and also activates macrophages and CD+ T cells, reducing inflammation and exerting anti-carcinogenic effects [433,434,435]. It can modulate proliferation and promote apoptosis of colon cancer cells [405,436,437]. Butyrate causes the autophagy-mediated degradation of β-catenin, limiting CRC cell proliferation [438].
Butyrate and propionate can alter chromatin state (by inhibiting histone deacetylases) [439], downregulate pro-inflammatory cytokines, such as IL-6 and IL-12 [440,441], and induce apoptosis [442]. Butyrate and propionate can activate the AP-1 signaling pathway, which controls cell proliferation and apoptosis [443]. Faecalibacterium prausnitzii is an important butyrate-producing bacterium found in the intestine [444], which has drawn much attention in recent years. A decrease in F. prausnitzii has been reported in patients with IBDs [445] and CRC [81,153]. Propionate suppresses CRC by promoting the degradation of euchromatic histone-lysine N-methyltransferase 2 [446]. SCFAs show anti-inflammatory effects and regulate colonic regulatory T cells [435]. Although there is much supporting evidence that dietary fiber decreases the risk of CRC, several cohort studies have failed to find a link between high fibre intake and lower risk of CRC [447,448].
High-protein and low-carbohydrate diets increase the production of toxic metabolites, such as amines, ammonia, phenolic compounds, indoles, and N-nitroso compounds, due to bacterial fermentation [449,450,451]. Many of these toxic metabolites may cause mutations, increasing the risk of carcinogenesis [35,452]. N-nitroso compounds promote DNA alkylation [11]. Ammonia is a potent carcinogen that promotes mucosal damage and adenocarcinoma in rat models [453]. Protein-rich diets are also associated with high fecal glucuronidase activity [454]. The fermentation of sulfur-containing amino acids by sulfate-reducing bacteria, such as Desulfovibrio, Desulfobacter, and Desulfobulbus, results in the generation of sulfides. Desulfovibrio is a gut bacterium that uses lactate to generate hydrogen sulfide [455], which is genotoxic [456] and damages DNA, probably by generating ROS [452]. It also inhibits butyric acid oxidation [457] and increases cell proliferation in vitro [458]. High-sulfur microbial diets are associated with increased risk of distal colon and rectal cancers [459].
The association between fat intake and CRC was first established by Drasar and Irving in 1973 [460]. Several studies have confirmed that high fat intake is a risk factor for CRC. A high-fat diet increases secondary bile acid formation, affecting the composition of the gut microbiota. Increased levels of deoxycholic acid increase resistance to apoptosis [306], induce ROS formation, damage DNA, and activate NF-κB [313]. Therefore, a high-fat diet increases the risk of CRC [461]. Contrary to these findings, studies on low-fat or high-fibre diets have failed to demonstrate a decreased risk for CRC [462,463,464,465,466]. In a landmark epidemiological study involving 61,463 Swedish women, Terry et al. [463] concluded that fat intake is not associated with CRC. A study by Taira et al. [467] in rats found that switching from a low-fat diet to a high-fat diet resulted in an increase in Firmicutes and a decrease in Bacteriodetes. In a similar study, Higashimura et al. [468] found that a high-fat diet decreased Lactobacillales and increased Clostridium. A recent study found that a high fat diet induced gut microbial dysbiosis and gut barrier dysfunction in mice, driving colorectal tumorigenesis [469]. Another recent study found that a high-fat diet promoted gut barrier dysfunction and inflammation in the colorectum and liver, contributing to CRC tumorigenesis and metastasis [470].
Urolithins are microbial metabolites of fruits and nuts rich in ellagic acid that inhibit Wnt signaling and have anti-carcinogenic effects [471,472]. Glucosinolates are plant secondary metabolites that have a protective role against CRC [473]. Kaempferol, a polyphenol found in fruits and vegetables, reduces tumor burden and restores damaged intestinal barrier in Apcmin/+ mice [474]. Many dietary phytochemicals are modified by gut microbiota to produce phenolic substances that are known to inhibit pro-inflammatory mediators, such as NF-κB and TNF. Studies on mouse models have shown that ketogenic diets inhibit glycolysis and cancer cell proliferation [475,476]. The effect of ketogenic diet on gut microbiota wasstudied in a mouse model of autism, where ketogenic diet significantly increased the Firmicutes/Bacteroidetes ratio [477]. A Mediterranean diet modulates the gut microbiota by enriching anti-inflammatory-environment-promoting bacteria, thus preventing CRC [478]. Since diet affects the composition of gut microbiota, it may influence susceptibility to CRC.
7. Gut Microbiota in CRC Treatment
Studies have shown that the gut microbiota can influence the therapeutic effects of cancer therapies by modulating the response, efficacy, and toxicity of chemotherapy, radiotherapy, and immunotherapy [479,480]. Dietary interventions through probiotics and prebiotics have been shown to influence the outcome of most cancer therapies. Many clinical trials are being conducted to know the efficacy of probiotics on CRC treatment (Table 4).

Table 4.
Ongoing and completed clinical trials on the effects of probiotics and prebiotics on CRC.
7.1. Probiotics
Probiotics are live microorganisms that provide multiple health benefits when administered in adequate amounts [481]. Probiotics modify the gut microflora composition by replacing pathogenic microbes with beneficial microbes [482]. Probiotics provide many health benefits, including regulation of the immune system, reduction in colitis and blood cholesterol, inhibition of pathogenic bacteria, and prevention of CRC [483,484,485,486]. Studies have found that probiotics prevent the colonization of pathogenic microbes by competing for nutrients [487] or adhering to epithelial cells or mucus [488] and, thus, help prevent intestinal infections [489,490]. Additionally, probiotics can produce certain metabolites that inhibit pathogen growth [491,492]. By lowering the risks of intestinal infections and inflammation, probiotics may prevent CRC development.
Since CRC is linked to gut microflora dysbiosis, restoring normal gut microflora through probiotics is one of the new approaches for IBD and CRC treatment. Probiotics inhibit CRC by reducing inflammation and carcinogenic microbial metabolites [484,493], downregulating inflammation pathways [494], producing short-chain fatty acids, antioxidants, and anti-cancer compounds [495,496], reducing the expression of cyclooxygenase-2 [497] and cell proliferation [62,498], inducing cancer cell apoptosis [499], and stimulating the expression of tumor suppressor genes [500]. Many studies have confirmed the positive effects of probiotics in the treatment of IBDs, CRC, and other cancers [486,501,502]. The key probiotics are Bifidobacterium and Lactobacillus (Figure 3). The administration of Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bl-04 affects the gut microbial profile. Probiotics have been shown to increase butyrate-producing bacteria, such as Faecalibacterium, and decrease CRC-associated bacteria, such as Fusobacterium [503]. Kuugbee et al. [504] reported that a probiotic cocktail containing Lactobacillus acidophilus, Bifidobacterium bifidum, and Bifidobacterium infantum, with oligofructose and maltodextrin, modulates the gut microbiota and reduces colon cancer development by suppressing apoptosis and inflammation. A combination of Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 with inulin reduces cell proliferation and improves epithelial barrier function [505]. Benito et al. [506] reported that a combination of Bifidobacterium bifidum and Lactobacillus gasseri along with quercetin inhibited CRC development in Apcmin/+ mice. Clostridium butyricum can decrease the incidence of tumors in mice by decreasing the number of Th2 and Th17 cells and reducing the secretion of factors associated with inflammation, such as IL-22 and NF-κB [507]. C. butyricum is also known to inhibit high-fat diet-induced CRC development in Apcmin/+ mice [508]as well as change gut microbial composition, and decrease the incidence and size of CRC [509]. Apart from these genera, Streptococcus thermophilus has been found to reduce tumor formation in mice, through β-galactosidase-dependent production of galactose that activates oxidative phosphorylation and downregulates the Hippo pathway kinase [510].
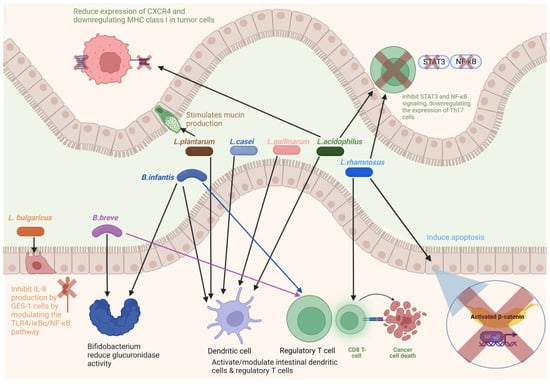
Figure 3.
Diagram showing bacteria as potential probiotic for colorectal cancer. Figure created with BioRender.com.
7.1.1. Bifidobacterium
Bifidobacterium is a Gram-positive, non-motile, anaerobic bacterium found in the human gut. The ratio of Bifidobacterium to E. coli has been used as an indicator of gut microflora. A decrease in Bifidobacterium and an increase in E. coli have been observed in CRC [511]. It has been found that an oral administration of Bifidobacterium alone can influence the immune response against CRC [512]. Bifidobacterium may also enhance chemotherapeutic efficacy by reducing glucuronidase activity [513]. An oral administration of B. breve significantly improves ulcerative colitis [514]. B. breve reduces tumor growth in MC38 colon carcinoma-bearing mice and boosted the efficacy of cancer therapeutics [515]. The strains of B. infantis and B. breve interact with toll-like receptors (TLRs) and can activate intestinal dendritic cells, Foxp3+ regulatory T cells, and IL-10-producing Tr1 cells [516,517]. Smoking reduces the abundance of butyrate-producing Bifidobacterium [518].
7.1.2. Lactobacillus
Lactobacillus is a Gram-positive, facultative anaerobic bacterium found in most probiotics. Lactobacillus can reduce the incidence of CRC by inducing apoptosis, reducing the expression of β-catenin and NF-κB [519], and modulating cytokine-producing dendritic cells [520]. It also regulates the expression of toll-like receptors and enhances intestinal epithelial barrier function [504,521,522]. L. rhamnosus GG and L. acidophilus inhibit STAT3 and NF-κB signaling, downregulating the expression of Th17 cells [523,524]. L. rhamnosus GG decreases tumor burden in a mouse model of CRC by increasing colonic CD8 T-cell responses [525].
Studies have confirmed that oral administration of L. casei strain significantly improves ulcerative colitis [526] and decreases the incidence of CRC in high-risk patients [527]. L. casei produces a metabolite, ferrichrome, which can trigger apoptosis in tumor cells through the JNK pathway [499]. Administration of L. salivarius Ren [528] and L. paracasei [529] can suppress CRC development in 1, 2-dimethylhydrazine-induced rat models. L. paracasei subsp. paracasei NTU 101 in combination with 5-fluorouracil (5-FU) was effective in reducing CRC cell viability [530]. It has been found that L. acidophilus NCFM suppresses tumor growth in mouse model by reducing the expression of CXCR4 and downregulating MHC class I in tumor cells [62]. L. rhamnosus and L. plantarum have been shown to stimulate mucin production [531,532]. It has been found that L. acidophilus and L. bulgaricus inhibit H. pylori adherence to GES-1 cells. L. bulgaricus, in particular, was found to inhibit IL-8 production by GES-1 cells by modulating the TLR4/IκBα/NF-κB pathway [533]. L. bulgaricus decreases intestinal inflammation by decreasing the levels of IL-6, TNF-α, IL-17, IL-23, and IL-1β and, thus, has a potential chemopreventive effect on colitis associated colon cancer [534]. L. reuteri strain ATCC PTA 6475 and ATCC 53608 were found to inhibit enteropathogenic E. coli [535]. L. reuteri restricts colon tumor growth and increases tumor reactive oxygen species. L. reuteri and its metabolite, reuterin, are downregulated in mouse and human CRC [536]. A study found that L. plantarum and L. salivarius could augment IL-18 production in a rat model of CRC [537]. A recent study found that administration of L. gallinarum could inhibit colorectal tumorigenesis in Apcmin/+ mice and in AOM/DSS-treated mice [538]. Another recent study found that L. coryniformis MXJ32 enhanced the expression of tight junction proteins and alleviated intestinal inflammation by downregulating the expression of inflammatory cytokines, decreasing the number of tumors and the average tumor diameter [539].
7.2. Prebiotics
Prebiotics are food components that provide health benefits by maintaining a healthy gut microbiota [540]. Many dietary components act as prebiotics. Clinical trials have found that prebiotic administration increases the abundance of probiotic strains, such as Ruminococcus, Faecalibacterium, Rosebura, and Akkermansia [541,542,543]. Prebiotic oligosaccharides can inhibit pathogen colonization by interacting with bacterial receptors and preventing pathogens from attaching to epithelial cells [544]. A study on polydextrose found its beneficial effects on maintaining healthy gut microbiota [545]. Fructans and galacto-oligosaccharides increase the abundance of beneficial bacteria, such as Bifidobacterium and Lactobacillus, and increase fecal butyrate concentration [546]. Inulin, a polysaccharide found in artichokes, bananas, asparagus, and wheat, decreases the formation of precancerous lesions by inhibiting the activity of glucuronidase and decreasing pH and concentration of indole, phenol, and p-cresol in the colon [547]. Inulin intake has also been shown to increase the abundance of Bifidobacterium [548]. Agro-oligosaccharides alter the production of SCFAs and secondary bile acids and, thus, control CRC development [468]. Polysaccharides from Lachnum sp. alters gut microbiota and reduces inflammation and tumor incidence [549].
Avenanthramide-C, found in oats, is metabolized by gut bacteria into bioactive compounds that show anti-tumor effects [550]. Nutmeg can prevent colon cancer by modulating gut microbiota and inflammation [551]. Eicosapentaenoic acid (EPA) is a type of omega-3 fatty acid that inhibits inflammation and colitis-associated CRC [552]. Investigations on the effects of fructo-oligosaccharides, xylo-oligosaccharides, polydextrose, and resistant dextrin on the gut microbiota of perioperative patients with CRC found an increase in the abundance of Bifidobacterium and Enterococcus and a reduction in Bacteroides [553]. Recent studies have found that berberine, found in Berberis, affects the proliferation, migration, and invasion of CRC cells and induces their apoptosis [554], decreases inflammatory modulators [555], and the expression of NF-κB [556] and increases fecal butyrate, acetate and propionate levels [557]. Ginsenoside compound K, produced from ginseng saponins, suppresses tumor growth and increases the abundance of A. muciniphila in an AOM/DSS-induced colitis-associated CRC Balb/c mouse model [558].
7.3. Chemotherapy
Many studies have shown that the gut microbiota influences the activity of various chemotherapeutic drugs and, thus, the efficacy of cancer treatment [559,560]. The gut microbiota modulates the host response by the ‘TIMER’ mechanistic framework: translocation, immunomodulation, metabolism, enzymatic degradation, and reduced diversity and ecological variation [479]. Gut microbial metabolites can have an enhanced killing effect on CRC [561]. Cyclophosphamide (CTX), a chemotherapeutic drug, can alter the gut microbiota and promote the translocation of Gram-positive bacteria into secondary lymphoid organs, promoting the generation of Th17 cells [560,562]. The mouse model, where Gram-positive bacteria were killed by antibiotics, was resistant to the anti-tumor effects of CTX [560]. Restoring the microbiota can improve the efficiency of CTX treatment. Studies carried out on mouse models have shown that the anti-tumor effect of CTX is enhanced by bacteria, such as Enterococcus hirae, Lactobacillus johnsonii, and Barnesiella intestinihominis [560,563].
Gut microbiota can influence the anti-tumor activity of oxaliplatin by affecting the production of ROS by immune cells [559]. A recent study explored the effect of gut microbial metabolites on the efficacy of oxaliplatin in combination with fluorouracil and leucovorin [564]. The study found that butyrate stimulated the anti-tumor cytotoxic CD8+ T cell response, and patients that responded to oxaliplatin had higher serum butyrate levels than non-responding patients. Gut microbes, particularly F. nucleatum, promote resistance to chemotherapy by modulating autophagy and inducing selective loss of miR-18a and miR-4802 [565]. A study showed that the effect of chemotherapy could be enhanced by administering irinotecan-loaded dextran nanoparticles that were covalently linked to azide-modified phages against F. nucleatum [566].
Bacterial genotoxins are also being targeted for CRC treatment. As described previously, colibactin produced by enterogenic E. coli plays an important role in CRC progression. The synthesis of colibactin requires a serine enzyme, ClbP [567,568], which can be inhibited by boronic acid, thus suppressing the genotoxicity of colibactin-producing E. coli [293]. The use of boronic acid is effective in preventing cell proliferation in a CRC mouse model. In vitro studies have found that healthy gut microbiota can increase the response to capecitabine or TAS-102 [569]. Gammaproteobacteria were found to metabolize the chemotherapeutic drug gemcitabine in a colon cancer mouse model [570]. Therefore, targeting these bacteria through antibiotics may promote the efficacy of gemcitabine treatment. A recent study found that the antibiotic vancomycin, which targets Gram-positive bacteria, potentiated the radiotherapy-induced antitumor immune response and tumor growth inhibition [571]. A recent study found that the patients that responded to radiochemotherapy showed an enrichment of butyrate-producing bacteria in fecal microbiota and had significantly higher levels of SCFAs, such as acetate, butyrate, and isobutyrate [572].
7.4. Immunotherapy
Immunotherapy has been used for the treatment of various cancers. Immunotherapy for CRC was approved by the FDA as a second-line therapy for tumors positive for deficient mismatch repair/microsatellite-high (dMMR/MSI-H) due to increase in overall survival [573]. Immune checkpoint inhibitors (ICIs) are widely used in immunotherapy. ICIs activate T cells to enable them to mount anti-tumor responses [574]. ICIs are usually monoclonal antibodies that prevent programmed cell death protein 1 (PD-1) from interacting with its ligand, PD-L1, or enable cytotoxic lymphocyte-mediated attack on tumor cells by targeting cytotoxic T-lymphocyte antigen 4 (CTLA-4) [575]. Compelling evidence suggests that gut microbiota can modulate cancer immunotherapy [512,559,560,576,577], and several studies have found that certain bacteria are positively correlated with immunotherapeutic response, including Bifidobacterium [512], Faecalibacterium [578], and Akkermansia [579] although not necessarily in the CRC context. Tanoue et al. [577] found 11 bacterial strains that could enhance the therapeutic effects of ICI. The oral administration of Bifidobacterium alone can influence the immune response against CRC, by maturing dendritic cell and promoting their function, upregulating cytokine secretion, and activating tumor-specific T cells [577]. The gut microbiota is also associated with ICI-induced colitis [580]. Administration of probiotics that include Bacteriodales and Burkholderiales, as well as fecal microbiota transplantation (FMT), improves ICI-induced colitis [581].
7.5. Fecal Microbiota Transplantation
Fecal microbiota transplantation (FMT) is an emerging biotherapeutic procedure that aims to restore normal gut microbial ecology to ameliorate various gastrointestinal disorders, including IBDs [582,583]. FMT involves the transplantation of a microbial population from a donor to a recipient. The prospects of using FMT to enhance the treatment of CRC are largely unexplored, and only a few studies have been carried out in this direction. A study carried out by Rosshart et al. [584] found that fecal microbiota transplantations from wild mice to laboratory mice could provide resistance against DSS/AOM-induced colorectal tumorigenesis. However, the logistics, safety, and potentially limited efficacy of FMT have precluded its wider use. Some patients that received FMT developed adverse effects, such as diarrhea, constipation, and abdominal distension [585]. Moreover, one possible risk with FMT is the transmission of multi-drug-resistant bacteria, potentially leading to life-threatening infections such as Escherichia coli bacteremia [586,587]. Stringent protocols for donor screening can prevent the occurrence of such infections. Another risk associated with FMT is the transmission of microbiome-associated chronic diseases, such as gastrointestinal, cardiometabolic, and autoimmune disorders [588]. In a study, it was found that transplanting human feces from obese individuals to germ-free mice induced obesity [589]. Similar results were found in humans where a woman developed obesity after receiving an FMT from an overweight donor [590]). Gregory et al. [591] reported the transmission of atherosclerosis after FMT.
8. Conclusions and Future Perspectives
A growing body of evidence suggests that the gut microbiota is strongly associated with the development and progression of CRC. The study of changes in the gut microbiome with CRC progression is critical to obtain a link between the gut microbiota and CRC. The presence of certain bacterial species may serve as a biomarker to assess the risk of developing CRC and the patient’s response to chemotherapy and immunotherapy. To determine the likelihood of developing CRC, intestinal microflora profiles may be used in conjunction with other factors, such as age, diet, family history, body mass index, and geographic location. The indiscriminate use of antibiotics should be stopped to prevent any disharmony in the intestinal microecology, which has been linked to IBDs and CRC. Dietary interventions can reshape the gut microbiota and may help in preventing and treating CRC. Gut-microbiota-based diagnostic tests can provide reliable and accurate identification of risk factors for developing CRC. This can also help in developing patient-specific probiotic therapy for CRC treatment.
Author Contributions
Conceptualization, H.P., S.H.W. and D.L.; resources, H.P., D.W.T.T., S.H.W. and D.L.; diagrams, D.W.T.T.; writing—original draft preparation, H.P., D.W.T.T., S.H.W. and D.L.; writing—review and editing, H.P., D.W.T.T., S.H.W. and D.L.; supervision, D.L. and S.H.W.; funding acquisition, H.P. and S.H.W. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to acknowledge the NTU Start Up Grant (021337-00001) (S.W.) and the Wang Lee Wah Memorial Fund for support of this work. The authors would also like to acknowledge Redcliffe Lifetech Inc. for paying article processing charge.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020, GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, W.F.; Bailey, C.J.; Bodmer, J.; Bussey, H.J.; Ellis, A.; Gorman, P.; Lucibello, F.C.; Murday, V.A.; Rider, S.H.; Scambler, P.; et al. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature 1987, 328, 614–616. [Google Scholar] [CrossRef]
- Carethers, J.M.; Jung, B.H. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 2015, 149, 1177–1190. [Google Scholar] [CrossRef]
- Czene, K.; Lichtenstein, P.; Hemminki, K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-cancer database. Int. J. Cancer 2002, 99, 260–266. [Google Scholar] [CrossRef]
- Blanpain, C. Tracing the cellular origin of cancer. Nat. Cell Biol. 2013, 15, 126–134. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Rabeneck, L.; Davila, J.A.; El-Serag, H.B. Is there a true ‘shift’ to the right colon in the incidence of colorectal cancer? Am. J. Gastroenterol. 2003, 98, 1400–1409. [Google Scholar] [CrossRef]
- Gill, C.I.R.; Rowland, I.R. Diet and cancer: Assessing the risk. Br. J. Nutr. 2002, 88, S73–S87. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Baquero, F.; Nombela, C. The microbiome as a human organ. Clin. Microbiol. Infect. 2012, 18, 2–4. [Google Scholar] [CrossRef]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Wells, J.M.; Rossi, O.; Meijerink, M.; van Baarlen, P. Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. USA 2011, 108, 4607–4614. [Google Scholar] [CrossRef]
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86, 13–15. [Google Scholar] [CrossRef]
- Collado, M.C.; Delgado, S.; Maldonado, A.; Rodríguez, J.M. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 2009, 48, 523–528. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, D.; Keijser, B.; Huse, S.; Rossen, J.; Veenhoven, R.; van Gils, E.; Bruin, J.; Montijn, R.; Bonten, M.; Sanders, E. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS ONE 2011, 6, e17035. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Eckburg, P.B.; Bik, E.M.; Relman, D.A. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 2006, 21, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Neish, A.S. Microbes in gastrointestinal health and disease. Gastroenterology 2009, 136, 65–80. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R89. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Frutos Rde, L.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef]
- Boleij, A.; Tjalsma, H. Gut bacteria in health and disease: A survey on the interface between intestinal microbiology and colorectal cancer. Biol. Rev. Camb. Philos. Soc. 2012, 87, 701–730. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Leung, A.; Tsoi, H.; Yu, J. Fusobacterium and Escherichia: Models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 651–657. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. Do microbiotas warm their hosts? Gut Microbes 2016, 7, 283–285. [Google Scholar] [CrossRef]
- Weinstein, P.D.; Cebra, J.J. The preference for switching to IgA expression by Peyer’s patch germinal center B cells is likely due to the intrinsic influence of their microenvironment. J. Immunol. 1991, 147, 4126–4135. [Google Scholar] [CrossRef]
- Cebra, J.J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999, 69, 1046S–1051S. [Google Scholar] [CrossRef]
- Shanahan, F. The host-microbe interface within the gut. Best Pract. Res. Clin. Gastroenterol. 2002, 16, 915–931. [Google Scholar] [CrossRef]
- Chung, H.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Nunez, G. Gut microbiotainduced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Spiljar, M.; Merkler, D.; Trajkovski, M. The immune system bridges the gut microbiota with systemic energy homeostasis: Focus on TLRs, mucosal barrier, and SCFAs. Front. Immunol. 2017, 8, 1353. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108, 4586–4591. [Google Scholar] [CrossRef]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Marchesi, J.R. Human distal gut microbiome. Environ. Microbiol. 2011, 13, 3088–3102. [Google Scholar] [CrossRef]
- Hopkins, M.J.; Sharp, R.; Macfarlane, G.T. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 2001, 48, 198–205. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Akkermans, A.D.L.; Akkermans-van Vilet, W.M.; de Visser, J.A.G.M.; de Vos, W.M. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb. Ecol. Health Dis. 2001, 13, 129–134. [Google Scholar] [CrossRef]
- Tap, J.; Mondot, S.; Levenez, F.; Pelletier, E.; Caron, C.; Furet, J.P.; Ugarte, E.; Muñoz-Tamayo, R.; Paslier, D.L.; Nalin, R.; et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009, 11, 2574–2584. [Google Scholar] [CrossRef]
- Petersonm, D.A.; Frank, D.N.; Pace, N.R.; Gordon, J.I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 2008, 3, 417–427. [Google Scholar] [CrossRef]
- Walter, J.; Ley, R. The human gut microbiome: Ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011, 65, 411–429. [Google Scholar] [CrossRef]
- Corr, S.C.; Hill, C.; Gahan, C.G. Understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv. Food Nutr. Res. 2009, 56, 1–15. [Google Scholar] [CrossRef]
- Sun, J.; Kato, I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef]
- Podolsky, D.K. The current future understanding of inflammatory bowel disease. Best Pract. Res. Clin. Gastroenterol. 2002, 16, 933–943. [Google Scholar] [CrossRef]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef]
- Sartor, R.B. Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology 2010, 139, 1816–1819. [Google Scholar] [CrossRef]
- Zur Hausen, H. The search for infectious causes of human cancers: Where and why. Virology 2009, 392, 1–10. [Google Scholar] [CrossRef]
- Collins, D.; Hogan, A.M.; Winter, D.C. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 2011, 12, 504–512. [Google Scholar] [CrossRef]
- Laqueur, G.L.; McDaniel, E.G.; Matsumoto, H. Tumor induction in germfree rats with methylazoxymethanol (MAM) and synthetic MAM acetate. J. Natl. Cancer Inst. 1967, 39, 355–371. [Google Scholar]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Tran Van Nhieu, J.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Lin, W.C.; Kong, M.S.; Shi, H.N.; Walker, W.A.; Lin, C.Y.; Huang, C.T.; Lin, Y.C.; Jung, S.M.; Lin, T.Y. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br. J. Nutr. 2012, 107, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Randall, T.A.; Galanko, J.; Benson, A.; Sandler, R.S.; Rawls, J.F.; Abdo, Z.; Fodor, A.A.; Keku, T.O. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012, 6, 1858–1868. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Wu, N.; Yang, X.; Zhang, R.; Li, J.; Xiao, X.; Hu, Y.; Chen, Y.; Yang, F.; Lu, N.; Wang, Z.; et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013, 66, 462–470. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Zhan, Y.; Chen, P.J.; Sadler, W.D.; Wang, F.; Poe, S.; Núñez, G.; Eaton, K.A.; Chen, G.Y. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 2013, 73, 7199–7210. [Google Scholar] [CrossRef]
- Bonnet, M.; Buc, E.; Sauvanet, P.; Darcha, C.; Dubois, D.; Pereira, B.; Déchelotte, P.; Bonnet, R.; Pezet, D.; Darfeuille-Michaud, A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 2014, 20, 859–867. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Brown, K.; Godovannyi, A.; Ma, C.; Zhang, Y.; Ahmadi-Vand, Z.; Dai, C.; Gorzelak, M.A.; Chan, Y.; Chan, J.M.; Lochner, A.; et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016, 10, 321–332. [Google Scholar] [CrossRef]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 2018, 175, 947–961. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Ma, Y.; Zhang, F.; Zhao, C.; Nie, Y. Insights into the role of gut microbiota in obesity: Pathogenesis, mechanisms, and therapeutic perspectives. Protein Cell 2018, 9, 397–403. [Google Scholar] [CrossRef]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L.; et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11, eaav1892. [Google Scholar] [CrossRef]
- Maini Rekdal, V.; Bess, E.N.; Bisanz, J.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019, 364, eaau6323. [Google Scholar] [CrossRef]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The intestinal microbiota in colorectal cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell 2019, 176, 998–1013. [Google Scholar] [CrossRef]
- Raskov, H.; Pommergaard, H.C.; Burcharth, J.; Rosenberg, J. Colorectal carcinogenesis—Update and perspectives. World J. Gastroenterol. 2014, 20, 18151–18164. [Google Scholar] [CrossRef] [PubMed]
- Borges-Canha, M.; Portela-Cidade, J.P.; Dinis-Ribeiro, M.; Leite-Moreira, A.F.; Pimentel-Nunes, P. Role of colonic microbiota in colorectal carcinogenesis: A systematic review. Rev. Esp. Enferm. Dig. 2015, 107, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Ahn, J.; Sampson, J.N.; Shi, J.; Yu, G.; Xiong, X.; Hayes, R.B.; Goedert, J.J. Fecal microbiota, fecal metabolome, and colorectal cancer interrelations. PLoS ONE 2016, 11, e0152126. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Collins, S.M.; Bercik, P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology 2009, 136, 2003–2014. [Google Scholar] [CrossRef]
- Cao, Y.; Wu, K.; Mehta, R.; Drew, D.A.; Song, M.; Lochhead, P.; Nguyen, L.H.; Izard, J.; Fuchs, C.S.; Garrett, W.S.; et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut 2018, 67, 672–678. [Google Scholar] [CrossRef]
- Bai, X.; Wei, H.; Liu, W.; Coker, O.O.; Gou, H.; Liu, C.; Zhao, L.; Li, C.; Zhou, Y.; Wang, G.; et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut 2022, 71, 2439–2450. [Google Scholar] [CrossRef]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 2017, 153, 1621–1633. [Google Scholar] [CrossRef]
- Vannucci, L.; Stepankova, R.; Kozakova, H.; Fiserova, A.; Rossmann, P.; Tlaskalova-Hogenova, H. Colorectal carcinogenesis in germ-free and conventionally reared rats: Different intestinal environments affect the systemic immunity. Int. J. Oncol. 2008, 32, 609–617. [Google Scholar] [CrossRef]
- Li, Y.; Kundu, P.; Seow, S.W.; de Matos, C.T.; Aronsson, L.; Chin, K.C.; Kärre, K.; Pettersson, S.; Greicius, G. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APC Min/+ mice. Carcinogenesis 2012, 33, 1231–1238. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Pierluigi Di Simone, M.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018, 9, 131–142. [Google Scholar] [CrossRef]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Q.; Liao, Q.; Zhao, Y. Pancreatic cancer, gut microbiota, and therapeutic efficacy. J. Cancer 2020, 11, 2749–2758. [Google Scholar] [CrossRef]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef]
- Vogtmann, E.; Goedert, J.J. Epidemiologic studies of the human microbiome and cancer. Br. J. Cancer 2016, 114, 237–242. [Google Scholar] [CrossRef]
- Touchefeu, Y.; Duchalais, E.; Bruley des Varannes, S.; Alameddine, J.; Mirallie, E.; Matysiak-Budnik, T.; Le Bastard, Q; Javaudin, F.; Rimbert, M.; Jotereau, F.; et al. Concomitant decrease of double-positive lymphocyte population CD4CD8αα and Faecalibacterium prausnitzii in patients with colorectal cancer. Eur. J. Gastroenterol. Hepatol. 2021, 32, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Baxter, N.T.; Ruffin, M.T., 4th; Rogers, M.A.; Schloss, P.D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016, 8, 37. [Google Scholar] [CrossRef]
- Yazici, C.; Wolf, P.G.; Kim, H.; Cross, T.L.; Vermillion, K.; Carroll, T.; Augustus, G.J.; Mutlu, E.; Tussing-Humphreys, L.; Braunschweig, C.; et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017, 66, 1983–1994. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-Analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomics analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Geng, J.; Tang, X.; Fan, H.; Xu, J.; Wen, X.; Ma, Z.S.; Shi, P. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. ISME J. 2014, 8, 881–893. [Google Scholar] [CrossRef]
- Allali, I.; Delgado, S.; Marron, P.I.; Astudillo, A.; Yeh, J.J.; Ghazal, H.; Amzazi, S.; Keku, T.; Azcarate-Peril, M.A. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 2015, 6, 161–172. [Google Scholar] [CrossRef]
- Nakatsu, G.; Li, X.; Zhou, H.; Sheng, J.; Wong, S.H.; Wu, W.K.; Ng, S.C.; Tsoi, H.; Dong, Y.; Zhang, N.; et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015, 6, 8727. [Google Scholar] [CrossRef]
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017, 3, 34. [Google Scholar] [CrossRef]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-Cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef]
- Shah, M.S.; DeSantis, T.Z.; Weinmaier, T.; McMurdie, P.J.; Cope, J.L.; Altrichter, A.; Yamal, J.M.; Hollister, E.B. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 2018, 67, 882–891. [Google Scholar] [CrossRef]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Goedert, J.J.; Shi, J.; Bork, P.; Sinha, R. Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef]
- Zackular, J.P.; Baxter, N.T.; Chen, G.Y.; Schloss, P.D. Manipulation of the gut microbiota reveals role in colon tumorigenesis. mSphere 2016, 1, e00001-15. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, R.; Shu, R.; Yu, J.; Li, H.; Long, H.; Jin, S.; Li, S.; Hu, Q.; Yao, F.; et al. Study of the relationship between microbiome and colorectal cancer susceptibility using 16S rRNA sequencing. Biomed. Res. Int. 2020, 2020, 7828392. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Xiang, J.; Xiang, S.; Zhao, Y.; Xiao, M.; Du, F.; Ji, H.; Kaboli, P.J.; Wu, X.; et al. Metagenome analysis of intestinal bacteria in healthy people, patients with inflammatory bowel disease and colorectal cancer. Front. Cell. Infect. Microbiol. 2021, 11, 599734. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Martin, H.M.; Campbell, B.J.; Hart, C.A.; Mpofu, C.; Nayar, M.; Singh, R.; Englyst, H.; Williams, H.F.; Rhodes, J.M. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004, 127, 80–93. [Google Scholar] [CrossRef]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef]
- Toprak, N.U.; Yagci, A.; Gulluoglu, B.M.; Akin, M.L.; Demirkalem, P.; Celenk, T.; Soyletir, G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 2006, 12, 782–786. [Google Scholar] [CrossRef]
- Housseau, F.; Sears, C.L. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/-) mice: A human commensal-based murine model of colon carcinogenesis. Cell Cycle 2010, 9, 3–5. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Bakar, F.A. The association of Streptococcus bovis/gallolyticus with colorectal tumors: The nature and the underlying mechanisms of its etiological role. J. Exp. Clin. Cancer Res. 2011, 30, 11. [Google Scholar] [CrossRef]
- Hermsen, J.L.; Schurr, M.J.; Kudsk, K.A.; Faucher, L.D. Phenotyping Clostridium septicum infection: A surgeon’s infectious disease. J. Surg. Res. 2008, 148, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Mirza, N.N.; McCloud, J.M.; Cheetham, M.J. Clostridium septicum sepsis and colorectal cancer—A reminder. World J. Surg. Oncol. 2009, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, R.; Rajendiran, E.; George, S.; Samuel, G.V.; Ramakrishna, B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 2008, 23, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Y.; Moore, D.R.; Nimmo, S.L.; Lightfoot, S.A.; Huycke, M.M. 4-hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology 2012, 142, 543–547. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y; Yu, J. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 2017, 152, 1419–1433. [Google Scholar] [CrossRef]
- Long, X.; Wong, C.C.; Tong, L.; Chu, E.S.H.; Ho Szeto, C.; Go, M.Y.Y.; Coker, O.O.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 2019, 4, 2319–2330. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrède, J.P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Sheu, L.F.; Meng, C.L.; Lee, W.H.; Lin, J.C. Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction. Gut 1995, 37, 87–90. [Google Scholar] [CrossRef]
- Bender, C.; Zipeto, D.; Bidoia, C.; Costantini, S.; Zamò, A.; Menestrina, F.; Bertazzoni, U. Analysis of colorectal cancers for human cytomegalovirus presence. Infect. Agent Cancer 2009, 4, 6. [Google Scholar] [CrossRef]
- Nakatsu, G.; Zhou, H.; Wu, W.K.K.; Wong, S.H.; Coker, O.O.; Dai, Z.; Li, X.; Szeto, C.H.; Sugimura, N.; Lam, T.Y.; et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 2018, 155, 529–541. [Google Scholar] [CrossRef]
- Gao, R.; Kong, C.; Li, H.; Huang, L.; Qu, X.; Qin, N.; Qin, H. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2457–2468. [Google Scholar] [CrossRef]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, Y.; Huang, Y.; Yu, X.; Yang, Y.; Wang, D.; Shi, L.; Tao, K.; Wang, G.; Wu, K. Fungal dysbiosis of the gut microbiota is associated with colorectal cancer in Chinese patients. Am. J. Transl. Res. 2021, 13, 11287–11301. [Google Scholar]
- Lin, Y.; Lau, H.C.; Liu, Y.; Kang, X.; Wang, Y.; Ting, N.L.; Kwong, T.N.; Han, J.; Liu, W.; Liu, C.; et al. Altered mycobiota signatures and enriched pathogenic Aspergillus rambellii are associated with colorectal cancer based on multi-cohort fecal metagenomic analyses. Gastroenterology 2022, 163, 908–921. [Google Scholar] [CrossRef]
- Gao, R.; Xia, K.; Wu, M.; Zhong, H.; Sun, J.; Zhu, Y.; Huang, L.; Wu, X.; Yin, L.; Yang, R.; et al. Alterations of gut mycobiota profiles in adenoma and colorectal cancer. Front. Cell. Infect. Microbiol. 2022, 12, 839435. [Google Scholar] [CrossRef]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.Y.; Yu, J. Altered gut archaea composition and interaction with bacteria are associated with colorectal cancer. Gastroenterology 2020, 159, 1459–1470. [Google Scholar] [CrossRef]
- Liu, N.N.; Jiao, N.; Tan, J.C.; Wang, Z.; Wu, D.; Wang, A.J.; Chen, J.; Tao, L.; Zhou, C.; Fang, W.; et al. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 2022, 7, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef]
- Russo, E.; Bacci, G.; Chiellini, C.; Fagorzi, C.; Niccolai, E.; Taddei, A.; Ricci, F.; Ringressi, M.N.; Borrelli, R.; Melli, F.; et al. Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: A pilot study. Front. Microbiol. 2017, 8, 2699. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Song, Q.; Tang, X.; Liang, X.; Fan, H.; Peng, H.; Guo, Q.; Zhang, Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Keku, T.O.; McCoy, A.N.; Azcarate-Peril, A.M. Fusobacterium spp. and colorectal cancer: Cause or consequence? Trends Microbiol. 2013, 21, 506–508. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Shaw, S.; Berry, S.; Thomson, J.; Murray, G.I.; El-Omar, E.; Hold, G.L. Gut mucosal microbiome signatures of colorectal cancer differ according to BMI status. Front. Med. 2022, 8, 800566. [Google Scholar] [CrossRef]
- Tahara, T.; Yamamoto, E.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Jelinek, J.; Yamano, H.O.; Sugai, T.; An, B.; et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014, 74, 1311–1318. [Google Scholar] [CrossRef]
- McCoy, A.N.; Araújo-Pérez, F.; Azcárate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium is associated with colorectal adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef]
- Repass, J.; Maherali, N.; Owen, K. Reproducibility Project: Cancer Biology Registered report: Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. eLife 2016, 5, e10012. [Google Scholar] [CrossRef]
- Viljoen, K.S.; Dakshinamurthy, A.; Goldberg, P.; Blackburn, J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE 2015, 10, e0119462. [Google Scholar] [CrossRef]
- Yan, X.; Liu, L.; Li, H.; Qin, H.; Sun, Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. OncoTargets Ther. 2017, 10, 5031–5046. [Google Scholar] [CrossRef]
- Wei, Z.; Cao, S.; Liu, S.; Yao, Z.; Sun, T.; Li, Y.; Li, J.; Zhang, D.; Zhou, Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget 2016, 7, 46158–46172. [Google Scholar] [CrossRef]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef]
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, N.; Tommasino, M.; et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1381–1390. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Zheng, S.; Li, M.; Xu, C.; Jia, D.; Qi, Y.; Hou, T.; Wang, L.; Wang, B.; et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut Microbes 2022, 14, 2038852. [Google Scholar] [CrossRef]
- Yin, H.; Miao, Z.; Wang, L.; Su, B.; Liu, C.; Jin, Y.; Wu, B.; Han, H.; Yuan, X. Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging 2022, 14, 1941–1958. [Google Scholar] [CrossRef]
- Salvucci, M.; Crawford, N.; Stott, K.; Bullman, S.; Longley, D.B.; Prehn, J.H.M. Patients with mesenchymal tumours and high Fusobacteriales prevalence have worse prognosis in colorectal cancer (CRC). Gut 2022, 71, 1600–1612. [Google Scholar] [CrossRef]
- Fardini, Y.; Wang, X.; Témoin, S.; Nithianantham, S.; Lee, D.; Shoham, M.; Han, Y.W. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol. Microbiol. 2011, 82, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappab, and up-regulating expression of microRNA-21. Gastroenterology 2017, 152, 851–866.e24. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, J.; Chen, T.; Li, Q.; Peng, W.; Li, H.; Tang, X.; Fu, X. Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll-like receptor 4/p21-activated kinase 1 cascade. Dig. Dis. Sci. 2018, 63, 1210–1218. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Li, M.; Zhang, Y.; Sun, M.; Wang, L.; Lin, J.; Cui, Y.; Chen, Q.; Jin, C.; et al. Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis. Nat. Commun. 2022, 13, 1248. [Google Scholar] [CrossRef]
- Ternes, D.; Tsenkova, M.; Pozdeev, V.I.; Meyers, M.; Koncina, E.; Atatri, S.; Schmitz, M.; Karta, J.; Schmoetten, M.; Heinken, A.; et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat. Metab. 2022, 4, 458–475. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Arthur, J.C.; Jobin, C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes 2013, 4, 253–258. [Google Scholar] [CrossRef]
- Arthur, J.C.; Gharaibeh, R.Z.; Mühlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724. [Google Scholar] [CrossRef]
- Darfeuille-Michaud, A.; Neut, C.; Barnich, N.; Lederman, E.; Di Martino, P.; Desreumaux, P.; Gambiez, L.; Joly, B.; Cortot, A.; Colombel, J.F. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 1998, 115, 1405–1413. [Google Scholar] [CrossRef]
- Darfeuille-Michaud, A.; Boudeau, J.; Bulois, P.; Neut, C.; Glasser, A.L.; Barnich, N.; Bringer, M.A.; Swidsinski, A.; Beaugerie, L.; Colombel, J.F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 2004, 127, 412–421. [Google Scholar] [CrossRef]
- Martins, S.A.; Prazeres, D.M.; Cabral, J.M.; Monteiro, G.A. Comparison of real-time polymerase chain reaction and hybridization assays for the detection of Escherichia coli genomic DNA in process samples and pharmaceutical-grade plasmid DNA products. Anal. Biochem. 2003, 322, 127–129. [Google Scholar] [CrossRef]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef]
- Nougayrède, J.P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Taieb, F.; Petit, C.; Nougayrede, J.P.; Oswald, E. The enterobacterial genotoxins: Cytolethal distending toxin and colibactin. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Wilson, M.R.; Jiang, Y.; Villalta, P.W.; Stornetta, A.; Boudreau, P.D.; Carrá, A.; Brennan, C.A.; Chun, E.; Ngo, L.; Samson, L.D.; et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 2019, 363, eaar7785. [Google Scholar] [CrossRef]
- Cougnoux, A.; Dalmasso, G.; Martinez, R.; Buc, E.; Delmas, J.; Gibold, L.; Sauvanet, P.; Darcha, C.; Déchelotte, P.; Bonnet, M.; et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014, 63, 1932–1942. [Google Scholar] [CrossRef]
- Ge, Z.; Schauer, D.B.; Fox, J. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell. Microbiol. 2008, 10, 1599–1607. [Google Scholar] [CrossRef]
- Ge, Z.; Feng, Y.; Whary, M.T.; Nambiar, P.R.; Xu, S.; Ng, V.; Taylor, N.S.; Fox, J.G. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect. Immun. 2005, 73, 3559–3567. [Google Scholar] [CrossRef]
- Ge, Z.; Rogers, A.B.; Feng, Y.; Lee, A.; Xu, S.; Taylor, N.S.; Fox, J.G. Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell. Microbiol. 2007, 9, 2070–2080. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Salesse, L.; Lucas, C.; Hoang, M.H.T.; Sauvanet, P.; Rezard, A.; Rosenstiel, P.; Damon-Soubeyrand, C.; Barnich, N.; Godfraind, C.; Dalmasso, G.; et al. Colibactin-producing Escherichia coli induce the formation of invasive carcinomas in a chronic inflammation-associated mouse model. Cancers 2021, 13, 2060. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Woodmansey, E.J.; Macfarlane, G.T. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol. 2005, 71, 7483–7492. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Lee, S.M.; Mazmanian, S.K. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 2011, 17, 137–141. [Google Scholar] [CrossRef]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C.; et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef]
- Sears, C.L. Enterotoxigenic Bacteroides fragilis: A rogue among symbiotes. Clin. Microbiol. Rev. 2009, 22, 349–369. [Google Scholar] [CrossRef]
- Sears, C.L.; Geis, A.L.; Housseau, F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J. Clin. Investig. 2014, 124, 4166–4172. [Google Scholar] [CrossRef]
- Rhee, K.J.; Wu, S.; Wu, X.; Huso, D.L.; Karim, B.; Franco, A.A.; Rabizadeh, S.; Golub, J.E.; Mathews, L.E.; Shin, J.; et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 2009, 77, 1708–1718. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Geis, A.L.; Fan, H.; Wu, X.; Wu, S.; Huso, D.L.; Wolfe, J.L.; Sears, C.L.; Pardoll, D.M.; Housseau, F. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17- dependent colon carcinogenesis. Cancer Discov. 2015, 5, 1098–1109. [Google Scholar] [CrossRef]
- Thiele Orberg, E.; Fan, H.; Tam, A.J.; Dejea, C.M.; Destefano Shields, C.E.; Wu, S.; Chung, L.; Finard, B.B.; Wu, X.; Fathi, P.; et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017, 10, 421–433. [Google Scholar] [CrossRef]
- Kordahi, M.C.; Stanaway, I.B.; Avril, M.; Chac, D.; Blanc, M.P.; Ross, B.; Diener, C.; Jain, S.; McCleary, P.; Parker, A.; et al. Genomic and functional characterization of a mucosal symbiont involved in early-stage colorectal cancer. Cell Host Microbe 2021, 29, 1589–1598.e6. [Google Scholar] [CrossRef]
- De Almeida, C.V.; Taddei, A.; Amedei, A. The controversial role of Enterococcus faecalis in colorectal cancer. Therap. Adv. Gastroenterol. 2018, 11, 1756284818783606. [Google Scholar] [CrossRef]
- Huycke, M.M.; Moore, D.R. In vivo production of hydroxyl radical by Enterococcus faecalis colonizing the intestinal tract using aromatic hydroxylation. Free Radic. Biol. Med. 2002, 33, 818–826. [Google Scholar] [CrossRef]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef]
- Fearon, E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 2011, 6, 479–507. [Google Scholar] [CrossRef]
- Steck, N.; Hoffmann, M.; Sava, I.G.; Kim, S.C.; Hahne, H.; Tonkonogy, S.L.; Mair, K.; Krueger, D.; Pruteanu, M.; Shanahan, F.; et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 2011, 141, 959–971. [Google Scholar] [CrossRef]
- Gong, J.; Bai, T.; Zhang, L.; Qian, W.; Song, J.; Hou, X. Inhibition effect of Bifidobacterium longum, Lactobacillus acidophilus, Streptococcus thermophilus and Enterococcus faecalis and their related products on human colonic smooth muscle in vitro. PLoS ONE 2017, 12, e0189257. [Google Scholar] [CrossRef]
- Grootaert, C.; Van de Wiele, T.; Van Roosbroeck, I.; Possemiers, S.; Vercoutter-Edouart, A.S.; Verstraete, W.; Bracke, M.; Vanhoecke, B. Bacterial monocultures, propionate, butyrate and H2O2 modulate the expression, secretion and structure of the fasting-induced adipose factor in gut epithelial cell lines. Environ. Microbiol. 2011, 13, 1778–1789. [Google Scholar] [CrossRef]
- Shen, F.; Feng, J.; Wang, X.; Qi, Z.; Shi, X.; An, Y.; Zhang, Q.; Wang, C.; Liu, M.; Liu, B. Vinegar treatment prevents the development of murine experimental colitis via inhibition of inflammation and apoptosis. J. Agric. Food Chem. 2016, 64, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Komiya, M.; Fujii, G.; Hamoya, T.; Nakanishi, R.; Fujimoto, K.; Tamura, S.; Kurokawa, Y.; Takahashi, M.; Ijichi, T.; et al. Preventive effects of heat-killed Enterococcus faecalis strain EC-12 on mouse intestinal tumor development. Int. J. Mol. Sci. 2017, 18, 826. [Google Scholar] [CrossRef] [PubMed]
- Corredoira, J.; Alonso, M.P.; Coira, A.; Casariego, E.; Arias, C.; Alonso, D.; Pita, J.; Rodriguez, A.; López, M.J.; Varela, J. Characteristics of Streptococcus bovis endocarditis and its differences with Streptococcus viridans endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 285–291. [Google Scholar] [CrossRef] [PubMed]
- McCoy, W.C.; Mason, J.M., 3rd. Enterococcal endocarditis associated with carcinoma of the sigmoid; Report of a case. J. Med. Assoc. State Ala. 1951, 21, 162–166. [Google Scholar] [PubMed]
- Hoppes, W.L.; Lerner, P.I. Nonenterococcal group-D streptococcal endocarditis caused by Streptococcus bovis. Ann. Intern. Med. 1974, 81, 588–593. [Google Scholar] [CrossRef]
- Klein, R.S.; Catalano, M.T.; Edberg, S.C.; Casey, J.I.; Steigbigel, N.H. Streptococcus bovis septicemia and carcinoma of the colon. Ann. Intern. Med. 1979, 91, 560–562. [Google Scholar] [CrossRef]
- Gupta, A.; Madani, R.; Mukhtar, H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Color. Dis. 2010, 12, 164–171. [Google Scholar] [CrossRef]
- Boleij, A.; van Gelder, M.M.; Swinkels, D.W.; Tjalsma, H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: Systematic review and meta-analysis. Clin. Infect. Dis. 2011, 53, 870–878. [Google Scholar] [CrossRef]
- Kwong, T.N.Y.; Wang, X.; Nakatsu, G.; Chow, T.C.; Tipoe, T.; Dai, R.Z.W.; Tsoi, K.K.K.; Wong, M.C.S; Tse, G.; Chan, M.T.V.; et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology 2018, 155, 383–390.e8. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Bakar, F.A. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: Inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer 2010, 9, 249. [Google Scholar] [CrossRef]
- Pasquereau-Kotula, E.; Martins, M.; Aymeric, L.; Dramsi, S. Significance of Streptococcus gallolyticus subsp. gallolyticus association with colorectal cancer. Front. Microbiol. 2018, 9, 614. [Google Scholar] [CrossRef]
- Martins, M.; Aymeric, L.; du Merle, L.; Danne, C.; Robbe-Masselot, C.; Trieu-Cuot, P.; Sansonetti, P.; Dramsi, S. Streptococcus gallolyticus Pil3 pilus is required for adhesion to colonic mucus and for colonization of mouse distal colon. J. Infect. Dis. 2015, 212, 1646–1655. [Google Scholar] [CrossRef]
- Martins, M.; du Merle, L.; Trieu-Cuot, P.; Dramsi, S. Heterogeneous expression of Pil3 pilus is critical for Streptococcus gallolyticus translocation across polarized colonic epithelial monolayers. Microbes Infect. 2020, 22, 55–59. [Google Scholar] [CrossRef]
- Taylor, J.C.; Gao, X.; Xu, J.; Holder, M.; Petrosino, J.; Kumar, R.; Liu, W.; Höök, M.; Mackenzie, C.; Hillhouse, A.; et al. A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog. 2021, 17, e1009182. [Google Scholar] [CrossRef]
- Kumar, R.; Herold, J.L.; Schady, D.; Davis, J.; Kopetz, S.; Martinez-Moczygemba, M.; Murray, B.E.; Han, F.; Li, Y.; Callaway, E.; et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 2017, 13, e1006440. [Google Scholar] [CrossRef]
- Zhang, Y.; Weng, Y.; Gan, H.; Zhao, X.; Zhi, F. Streptococcus gallolyticus conspires myeloid cells to promote tumorigenesis of inflammatory bowel disease. Biochem. Biophys. Res. Commun. 2018, 506, 907–911. [Google Scholar] [CrossRef]
- Watari, J.; Chen, N.; Amenta, P.S.; Fukui, H.; Oshima, T.; Tomita, T.; Miwa, H.; Lim, K.J.; Das, K.M. Helicobacter pylori associated chronic gastritis, clinical syndromes, precancerous lesions, and pathogenesis of gastric cancer development. World J. Gastroenterol. 2014, 20, 5461–5473. [Google Scholar] [CrossRef]
- Doorakkers, E.; Lagergren, J.; Engstrand, L.; Brusselaers, N. Helicobacter pylori eradication treatment and the risk of gastric adenocarcinoma in a Western population. Gut 2018, 67, 2092–2096. [Google Scholar] [CrossRef]
- Alfarouk, K.O.; Bashir, A.H.H.; Aljarbou, A.N.; Ramadan, A.M.; Muddathir, A.K.; AlHoufie, S.T.S.; Hifny, A.; Elhassan, G.O.; Ibrahim, M.E.; Alqahtani, S.S.; et al. The possible role of Helicobacter pylori in gastric cancer and its management. Front. Oncol. 2019, 9, 75. [Google Scholar] [CrossRef]
- Doorakkers, E.; Lagergren, J.; Engstrand, L.; Brusselaers, N. Eradication of Helicobacter pylori and gastric cancer: A systematic review and meta-analysis of cohort studies. J. Natl. Cancer Inst. 2016, 108, djw132. [Google Scholar] [CrossRef]
- Polk, D.B.; Peek, R.M. Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef]
- Lv, Y.P.; Cheng, P.; Zhang, J.Y.; Mao, F.Y.; Teng, Y.S.; Liu, Y.G.; Kong, H.; Wu, X.L.; Hao, C.J.; Han, B.; et al. Helicobacter pylori-induced matrix metallopeptidase-10 promotes gastric bacterial colonization and gastritis. Sci. Adv. 2019, 5, eaau6547. [Google Scholar] [CrossRef] [PubMed]
- Zumkeller, N.; Brenner, H.; Zwahlen, M.; Rothenbacher, D. Helicobacter pylori infection and colorectal cancer risk: A metaanalysis. Helicobacter 2006, 11, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, H.Y. Association between Helicobacter pylori infection and colorectal neoplasm risk: A meta-analysis based on East Asian population. J. Cancer Res. Ther. 2014, 10, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Jing, Z.; Bie, M.; Xu, C.; Hao, X.; Wang, B. Association between Helicobacter pylori infection and the risk of colorectal cancer: A systematic review and meta-analysis. Medicine 2020, 99, e21832. [Google Scholar] [CrossRef]
- Jones, M.; Helliwell, P.; Pritchard, C.; Tharakan, J.; Mathew, J. Helicobacter pylori in colorectal neoplasms: Is there an aetiological relationship? World J. Surg. Oncol. 2007, 5, 51. [Google Scholar] [CrossRef]
- Cover, T.L. Helicobacter pylori diversity and gastric cancer risk. mBio 2016, 7, e01869-15. [Google Scholar] [CrossRef]
- Higashi, H.; Tsutsumi, R.; Fujita, A.; Yamazaki, S.; Asaka, M.; Azuma, T.; Hatakeyama, M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA 2002, 99, 14428–14433. [Google Scholar] [CrossRef]
- Odenbreit, S.; Puls, J.; Sedlmaier, B.; Gerland, E.; Fischer, W.; Haas, R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 2000, 287, 1497–1500. [Google Scholar] [CrossRef]
- Kwok, T.; Zabler, D.; Urman, S.; Rohde, M.; Hartig, R.; Wessler, S.; Misselwitz, R.; Berger, J.; Sewald, N.; König, W.; et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 2007, 449, 862–866. [Google Scholar] [CrossRef]
- Shmuely, H.; Passaro, D.; Figer, A.; Niv, Y.; Pitlik, S.; Samra, Z.; Koren, R.; Yahav, J. Relationship between Helicobacter pylori CagA status and colorectal cancer. Am. J. Gastroenterol. 2001, 96, 3406–3410. [Google Scholar] [CrossRef]
- Epplein, M.; Pawlita, M.; Michel, A.; Peek, R.M., Jr.; Cai, Q.; Blot, W.J. Helicobacter pylori protein-specific antibodies and risk of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1964–1974. [Google Scholar] [CrossRef]
- Butt, J.; Varga, M.G.; Blot, W.J.; Teras, L.; Visvanathan, K.; Le Marchand, L.; Haiman, C.; Chen, Y.; Bao, Y.; Sesso, H.D. Serologic response to Helicobacter pylori proteins associated with risk of colorectal cancer among diverse populations in the United States. Gastroenterology 2019, 156, 175–186. [Google Scholar] [CrossRef]
- Moyat, M.; Velin, D. Immune responses to Helicobacter pylori infection. World J. Gastroenterol. 2014, 20, 5583–5593. [Google Scholar] [CrossRef]
- Yong, X.; Tang, B.; Li, B.S.; Xie, R.; Hu, C.J.; Luo, G.; Qin, Y.; Dong, H.; Yang, S.M. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun. Signal. 2015, 13, 30. [Google Scholar] [CrossRef]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef]
- Mashima, H.; Suzuki, J.; Hirayama, T.; Yoshikumi, Y.; Ohno, H.; Ohnishi, H.; Yasuda, H.; Fujita, T.; Omata, M. Involvement of vesicle-associated membrane protein 7 in human gastric epithelial cell vacuolation induced by Helicobacter pylori-produced VacA. Infect. Immun. 2008, 76, 2296–2303. [Google Scholar] [CrossRef]
- Ki, M.R.; Lee, H.R.; Goo, M.J.; Hong, I.H.; Do, S.H.; Jeong, D.H.; Yang, H.J.; Yuan, D.W.; Park, J.K.; Jeong, K.S. Differential regulation of ERK1/2 and p38 MAP kinases in VacA-induced apoptosis of gastric epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G635–G647. [Google Scholar] [CrossRef]
- Song, X.; Xin, N.; Wang, W.; Zhao, C. Wnt/b-catenin, an oncogenic pathway targeted by H. pylori in gastric carcinogenesis. Oncotarget 2015, 6, 35579–35588. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, N.; Chai, N.; Liu, X.; Jiang, H.; Wu, Q.; Li, Q. Helicobacter pylori promotes angiogenesis depending on Wnt/beta-catenin-mediated vascular endothelial growth factor via the cyclooxygenase-2 pathway in gastric cancer. BMC Cancer 2016, 16, 321. [Google Scholar] [CrossRef]
- Sato, F.; Meltzer, S.J. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer 2006, 106, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Contreras, A.; Goldfarb, K.C.; Godoy-Vitorino, F.; Karaoz, U.; Contreras, M.; Blaser, M.J.; Brodie, E.L.; Dominguez-Bello, M.G. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011, 5, 574–579. [Google Scholar] [CrossRef]
- Iizasa, H.; Ishihara, S.; Richardo, T.; Kanehiro, Y.; Yoshiyama, H. Dysbiotic infection in the stomach. World J. Gastroenterol. 2015, 21, 11450–11457. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.F.; Lindberg, M.; Jakobsson, H.; Bäckhed, F.; Nyrén, P.; Engstrand, L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 2008, 3, e2836. [Google Scholar] [CrossRef]
- Li, T.H.; Qin, Y.; Sham, P.C.; Lau, K.S.; Chu, K.M.; Leung, W.K. Alterations in gastric microbiota after H. pylori eradication and in different histological stages of gastric carcinogenesis. Sci. Rep. 2017, 7, 44935. [Google Scholar] [CrossRef] [PubMed]
- Koeth, L.M.; Good, C.E.; Appelbaum, P.C.; Goldstein, E.J.; Rodloff, A.C.; Claros, M.; Dubreuil, L.J. Surveillance of susceptibility patterns in 1297 European and US anaerobic and capnophilic isolates to co-amoxiclav and five other antimicrobial agents. J. Antimicrob. Chemother. 2004, 53, 1039–1044. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; Zhou, Y.; Fu, K.; Lau, H.C.; Chun, T.W.; Cheung, A.H.; Coker, O.O.; Wei, H.; Wu, W.K.; et al. Parvimonas micra promotes colorectal tumorigenesis and is associated with prognosis of colorectal cancer patients. Oncogene 2022, 41, 4200–4210. [Google Scholar] [CrossRef]
- Lauby-Secretan, B.; Vilahur, N.; Bianchini, F.; Guha, N.; Straif, K. International Agency for Research on Cancer Handbook Working Group, The IARC perspective on colorectal cancer screening. N. Engl. J. Med. 2018, 378, 1734–1740. [Google Scholar] [CrossRef]
- Lee, J.K.; Liles, E.G.; Bent, S.; Levin, T.R.; Corley, D.A. Accuracy of fecal immunochemical tests for colorectal cancer: Systematic review and meta-analysis. Ann. Intern. Med. 2014, 160, 171. [Google Scholar] [CrossRef]
- Monahan, K.J.; Davies, M.M.; Abulafi, M.; Banerjea, A.; Nicholson, B.D.; Arasaradnam, R.; Barker, N.; Benton, S.; Booth, R.; Burling, D.; et al. Faecal immunochemical testing (FIT) in patients with signs or symptoms of suspected colorectal cancer (CRC): A joint guideline from the Association of Coloproctology of Great Britain and Ireland (ACPGBI) and the British Society of Gastroenterology (BSG). Gut 2022, 71, 1939–1962. [Google Scholar] [CrossRef]
- Hundt, S.; Haug, U.; Brenner, H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann. Intern. Med. 2009, 150, 162–169. [Google Scholar] [CrossRef]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef]
- Liang, Q.; Chiu, J.; Chen, Y.; Huang, Y.; Higashimori, A.; Fang, J.; Brim, H.; Ashktorab, H.; Ng, S.C.; Ng, S.S.M.; et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin. Cancer Res. 2017, 23, 2061–2070. [Google Scholar] [CrossRef]
- Suehiro, Y.; Zhang, Y.; Hashimoto, S.; Takami, T.; Higaki, S.; Shindo, Y.; Suzuki, N.; Hazama, S.; Oka, M.; Nagano, H.; et al. Highly sensitive faecal DNA testing of TWIST1 methylation in combination with faecal immunochemical test for haemoglobin is a promising marker for detection of colorectal neoplasia. Ann. Clin. Biochem. 2018, 55, 59–68. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef]
- Huo, R.X.; Wang, Y.J.; Hou, S.B.; Wang, W.; Zhang, C.Z.; Wan, X.H. Gut mucosal microbiota profiles linked to colorectal cancer recurrence. World J. Gastroenterol. 2022, 28, 1946–1964. [Google Scholar] [CrossRef]
- Avuthu, N.; Guda, C. Meta-analysis of altered gut microbiota reveals microbial and metabolic biomarkers for colorectal cancer. Microbiol. Spectr. 2022, 29, e0001322. [Google Scholar] [CrossRef]
- Li, Y.; Cao, H.; Fei, B.; Gao, Q.; Yi, W.; Han, W.; Bao, C.; Xu, J.; Zhao, W.; Zhang, F. Gut microbiota signatures in tumor, para-cancerous, normal mucosa, and feces in colorectal cancer patients. Front. Cell Dev. Biol. 2022, 10, 916961. [Google Scholar] [CrossRef]
- Wong, S.H.; Kwong, T.N.Y.; Chow, T.C.; Luk, A.K.C.; Dai, R.Z.W.; Nakatsu, G.; Lam, T.Y.T.; Zhang, L.; Wu, J.C.Y; Chan, F.K.L.; et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017, 66, 1441–1448. [Google Scholar] [CrossRef]
- Guo, S.; Li, L.; Xu, B.; Li, M.; Zeng, Q.; Xiao, H.; Xue, Y.; Wu, Y.; Wang, Y.; Liu, W.; et al. A simple and novel fecal biomarker for colorectal cancer: Ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin. Chem. 2018, 64, 1327–1337. [Google Scholar] [CrossRef]
- Wang, H.F.; Li, L.F.; Guo, S.H.; Zeng, Q.Y.; Ning, F.; Liu, W.L.; Zhang, G. Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci. Rep. 2016, 6, 33440. [Google Scholar] [CrossRef]
- Butt, J.; Blot, W.J.; Teras, L.R.; Visvanathan, K.; Le Marchand, L.; Haiman, C.A.; Chen, Y.; Bao, Y.; Sesso, H.D.; Wassertheil-Smoller, S.; et al. Antibody responses to Streptococcus gallolyticus subspecies gallolyticus proteins in a large prospective colorectal cancer cohort consortium. Cancer Epidemiol. Biomark. Prev. 2018, 27, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Butt, J.; Jenab, M.; Willhauck-Fleckenstein, M.; Michel, A.; Pawlita, M.; Kyrø, C.; Tjønneland, A.; Boutron-Ruault, M.C.; Carbonnel, F.; Severi, G.; et al. Prospective evaluation of antibody response to Streptococcus gallolyticus and risk of colorectal cancer. Int. J. Cancer 2018, 143, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, A.H.; Taghinezhad, S.S.; Fu, X. Gut microbiota-derived metabolites and colorectal cancer: New insights and updates. Microb. Pathog. 2020, 149, 104569. [Google Scholar] [CrossRef] [PubMed]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef]
- Zhang, M.; Lv, Y.; Hou, S.; Liu, Y.; Wang, Y.; Wan, X. Differential mucosal microbiome profiles across stages of human colorectal cancer. Life 2021, 11, 831. [Google Scholar] [CrossRef]
- Chang, H.; Mishra, R.; Cen, C.; Tang, Y.; Ma, C.; Wasti, S.; Wang, Y.; Ou, Q.; Chen, K.; Zhang, J. Metagenomic analyses expand bacterial and functional profiling biomarkers for colorectal cancer in a Hainan cohort, China. Curr. Microbiol. 2021, 78, 705–712. [Google Scholar] [CrossRef]
- Shen, S.; Huo, D.; Ma, C.; Jiang, S.; Zhang, J. Expanding the colorectal cancer biomarkers based on the human gut phageome. Microbiol. Spectr. 2021, 9, e0009021. [Google Scholar] [CrossRef]
- Liu, C.; Li, Z.; Ding, J.; Zhen, H.; Fang, M.; Nie, C. Species-level analysis of the human gut microbiome shows antibiotic resistance genes associated with colorectal cancer. Front. Microbiol. 2021, 12, 765291. [Google Scholar] [CrossRef]
- Osman, M.A.; Neoh, H.M.; Ab Mutalib, N.S.; Chin, S.F.; Mazlan, L.; Raja Ali, R.A.; Zakaria, A.D.; Ngiu, C.S.; Ang, M.Y.; Jamal, R. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 2021, 11, 2925. [Google Scholar] [CrossRef]
- Löwenmark, T.; Löfgren-Burström, A.; Zingmark, C.; Eklöf, V.; Dahlberg, M.; Wai, S.N.; Larsson, P.; Ljuslinder, I.; Edin, S.; Palmqvist, R. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci. Rep. 2020, 10, 15250. [Google Scholar] [CrossRef]
- Escobar-Páramo, P.; Grenet, K.; Le Menach, A.; Rode, L.; Salgado, E.; Amorin, C.; Gouriou, S.; Picard, B.; Rahimy, M.C.; Andremont, A.; et al. Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 2004, 70, 5698–5700. [Google Scholar] [CrossRef]
- Le Gall, T.; Clermont, O.; Gouriou, S.; Picard, B.; Nassif, X.; Denamur, E.; Tenaillon, O. Extraintestinal virulence is a coincidental byproduct of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 2007, 24, 2373–2384. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Maddocks, O.D.; Short, A.J.; Donnenberg, M.S.; Bader, S.; Harrison, D.J. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS ONE 2009, 4, e5517. [Google Scholar] [CrossRef]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2014, 63, 761–770. [Google Scholar] [CrossRef]
- Han, Y.W.; Ikegami, A.; Rajanna, C.; Kawsar, H.I.; Zhou, Y.; Li, M.; Sojar, H.T.; Genco, R.J.; Kuramitsu, H.K.; Deng, C.X. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 2005, 187, 5330–5340. [Google Scholar] [CrossRef]
- Ma, C.T.; Luo, H.S.; Gao, F.; Tang, Q.C.; Chen, W. Fusobacterium nucleatum promotes the progression of colorectal cancer by interacting with E-cadherin. Oncol. Lett. 2018, 16, 2606–2612. [Google Scholar] [CrossRef]
- Wu, S.; Powell, J.; Mathioudakis, N.; Kane, S.; Fernandez, E.; Sears, C.L. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappaB pathway. Infect. Immun. 2004, 72, 5832–5839. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Shin, J.; Zhang, G.; Cohen, M.; Franco, A.; Sears, C.L. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect. Immun. 2006, 74, 5382–5390. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [PubMed]
- Holton, J. Enterotoxigenic Bacteroides fragilis. Curr. Infect. Dis. Rep. 2008, 10, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Nesić, D.; Hsu, Y.; Stebbins, C.E. Assembly and function of a bacterial genotoxin. Nature 2004, 429, 429–433. [Google Scholar] [CrossRef]
- Nougayrède, J.P.; Taieb, F.; De Rycke, J.; Oswald, E. Cyclomodulins: Bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005, 13, 103–110. [Google Scholar] [CrossRef]
- Oswald, E.; Nougayrède, J.P.; Taieb, F.; Sugai, M. Bacterial toxins that modulate host cell-cycle progression. Curr. Opin. Microbiol. 2005, 8, 83–91. [Google Scholar] [CrossRef]
- Travaglione, S.; Fabbri, A.; Fiorentini, C. The Rho-activating CNF1 toxin from pathogenic E. coli: A risk factor for human cancer development? Infect. Agent Cancer 2008, 3, 4. [Google Scholar] [CrossRef]
- Tomkovich, S.; Yang, Y.; Winglee, K.; Gauthier, J.; Mühlbauer, M.; Sun, X.; Mohamadzadeh, M.; Liu, X.; Martin, P.; Wang, G.P.; et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017, 77, 2620–2632. [Google Scholar] [CrossRef]
- Smith, J.L.; Bayles, D.O. The contribution of cytolethal distending toxin to bacterial pathogenesis. Crit. Rev. Microbiol. 2006, 32, 227–248. [Google Scholar] [CrossRef]
- Bezine, E.; Vignard, J.; Mirey, G. The cytolethal distending toxin effects on mammalian cells: A DNA damage perspective. Cells 2014, 3, 592–615. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Cougnoux, A.; Delmas, J.; Gibold, L.; Faïs, T.; Romagnoli, C.; Robin, F.; Cuevas-Ramos, G.; Oswald, E.; Darfeuille-Michaud, A.; Prati, F.; et al. Small-molecule inhibitors prevent the genotoxic and protumoural effects induced by colibactin-producing bacteria. Gut 2016, 65, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Dutilh, B.E.; Backus, L.; van Hijum, S.A.; Tjalsma, H. Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Belcheva, A.; Irrazabal, T.; Martin, A. Gut microbial metabolism and colon cancer: Can manipulations of the microbiota be useful in the management of gastrointestinal health? Bioessays 2015, 37, 403–412. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef]
- Philipp, B. Bacterial degradation of bile salts. Appl. Microbiol. Biotechnol. 2011, 89, 903–915. [Google Scholar] [CrossRef]
- Ou, J.; DeLany, J.P.; Zhang, M.; Sharma, S.; O’Keefe, S.J. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr. Cancer 2012, 64, 34–40. [Google Scholar] [CrossRef]
- O’Keefe, S.J.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329–3340. [Google Scholar] [CrossRef]
- Barrasa, J.I.; Olmo, N.; Lizarbe, M.A.; Turnay, J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol. In Vitro 2013, 27, 964–977. [Google Scholar] [CrossRef]
- Farhana, L.; Nangia-Makker, P.; Arbit, E.; Shango, K.; Sarkar, S.; Mahmud, H.; Hadden, T.; Yu, Y.; Majumdar, A.P. Bile acid: A potential inducer of colon cancer stem cells. Stem Cell Res. Ther. 2016, 7, 181. [Google Scholar] [CrossRef]
- Wu, H.; Lin, Y.; Li, W.; Sun, Z.; Gao, W.; Zhang, H.; Xie, L.; Jiang, F.; Qin, B.; Yan, T.; et al. Regulation of Nur77 expression by β-catenin and its mitogenic effect in colon cancer cells. FASEB J. 2011, 25, 192–205. [Google Scholar] [CrossRef]
- Kong, Y.; Bai, P.S.; Sun, H.; Nan, K.J.; Chen, N.Z.; Qi, X.G. The deoxycholic acid targets miRNA-dependent CAC1 gene expression in multidrug resistance of human colorectal cancer. Int. J. Biochem. Cell Biol. 2012, 44, 2321–2332. [Google Scholar] [CrossRef]
- Bernstein, C.; Holubec, H.; Bhattacharyya, A.K.; Nguyen, H.; Payne, C.M.; Zaitlin, B.; Bernstein, H. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 2011, 85, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; An, Y.; Chen, D.; Zhang, W.; Wu, X.; Li, C.; Wang, S.; Dong, W.; Wang, B.; Liu, T.; et al. Microbial metabolite deoxycholic acid promotes vasculogenic mimicry formation in intestinal carcinogenesis. Cancer Sci. 2022, 113, 459–477. [Google Scholar] [CrossRef]
- Yang, J.L.; Seetoo, D.q.; Wang, Y.; Ranson, M.; Berney, C.R.; Ham, J.M.; Russell, P.J.; Crowe, P.J. Urokinase-type plasminogen activator and its receptor in colorectal cancer: Independent prognostic factors of metastasis and cancer-specific survival and potential therapeutic targets. Int. J. Cancer 2000, 89, 431–439. [Google Scholar] [CrossRef]
- Baek, M.K.; Park, J.S.; Park, J.H.; Kim, M.H.; Kim, H.D.; Bae, W.K.; Chung, I.J.; Shin, B.A.; Jung, Y.D. Lithocholic acid upregulates uPAR and cell invasiveness via MAPK and AP-1 signaling in colon cancer cells. Cancer Lett. 2010, 290, 123–128. [Google Scholar] [CrossRef]
- Carino, A.; Graziosi, L.; D’Amore, C.; Cipriani, S.; Marchianò, S.; Marino, E.; Zampella, A.; Rende, M.; Mosci, P.; Distrutti, E.; et al. The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget 2016, 7, 61021–61035. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorakova, K.; Garewal, H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 2005, 589, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, K.; Payne, C.M.; Chavarria, M.; Ramsey, L.; Dvorakova, B.; Bernstein, H.; Holubec, H.; Sampliner, R.E.; Guy, N.; Condon, A.; et al. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: Relevance to the pathogenesis of Barrett’s oesophagus. Gut 2007, 56, 763–771. [Google Scholar] [CrossRef]
- Ajouz, H.; Mukherji, D.; Shamseddine, A. Secondary bile acids: An underrecognized cause of colon cancer. World J. Surg. Oncol. 2014, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Crawley, S.; Hokari, R.; Kwon, S.; Kim, Y.S. Bile acid regulates MUC2 transcription in colon cancer cells via positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and p38/ MSK1/CREB pathways and negative JNK/c-Jun/AP-1 pathway. Int. J. Oncol. 2010, 36, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Centuori, S.M.; Gomes, C.J.; Trujillo, J.; Borg, J.; Brownlee, J.; Putnam, C.W.; Martinez, J.D. Deoxycholic acid mediates non-canonical EGFR-MAPK activation through the induction of calcium signaling in colon cancer cells. Biochim. Biophys. Acta 2016, 1861, 663–670. [Google Scholar] [CrossRef]
- Byrne, A.M.; Foran, E.; Sharma, R.; Davies, A.; Mahon, C.; O’Sullivan, J.; O’Donoghue, D.; Kelleher, D.; Long, A. Bile acids modulate the Golgi membrane fission process via a protein kinase Ceta and protein kinase D-dependent pathway in colonic epithelial cells. Carcinogenesis 2010, 31, 737–744. [Google Scholar] [CrossRef]
- Lee, J.Y.; Arai, H.; Nakamura, Y.; Fukiya, S.; Wada, M.; Yokota, A. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 2013, 54, 3062–3069. [Google Scholar] [CrossRef]
- Centuori, S.M.; Martinez, J.D. Differential regulation of EGFR-MAPK signaling by deoxycholic acid (DCA) and ursodeoxycholic acid (UDCA) in colon cancer. Dig. Dis. Sci. 2014, 59, 2367–2380. [Google Scholar] [CrossRef]
- Khare, S.; Mustafi, R.; Cerda, S.; Yuan, W.; Jagadeeswaran, S.; Dougherty, U.; Tretiakova, M.; Samarel, A.; Cohen, G.; Wang, J.; et al. Ursodeoxycholic acid suppresses Cox-2 expression in colon cancer: Roles of Ras, p38, and CCAAT/enhancer-binding protein. Nutr. Cancer 2008, 60, 389–400. [Google Scholar] [CrossRef]
- Im, E.; Martinez, J.D. Ursodeoxycholic acid (UDCA) can inhibit deoxycholic acid (DCA)-induced apoptosis via modulation of EGFR/Raf-1/ERK signaling in human colon cancer cells. J. Nutr. 2004, 134, 483–486. [Google Scholar] [CrossRef]
- Seitz, H.K.; Stickel, F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 2007, 7, 599–612. [Google Scholar] [CrossRef]
- Homann, N. Alcohol and upper gastrointestinal tract cancer: The role of local acetaldehyde production. Addict. Biol. 2001, 6, 309–323. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef]
- Bae, S.; Ulrich, C.M.; Neuhouser, M.L.; Malysheva, O.; Bailey, L.B.; Xiao, L.; Brown, E.C.; Cushing-Haugen, K.L.; Zheng, Y.; Cheng, T.Y.; et al. Plasma choline metabolites and colorectal cancer risk in the Women’s Health Initiative Observational Study. Cancer Res. 2014, 74, 7442–7452. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Q.; Li, L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genom. 2015, 16, S4. [Google Scholar] [CrossRef]
- Chan, C.W.H.; Law, B.M.H.; Waye, M.M.Y.; Chan, J.Y.W.; So, W.K.W.; Chow, K.M. Trimethylamine-N-oxide as one hypothetical link for the relationship between intestinal microbiota and cancer—Where we are and where shall we go? J. Cancer 2019, 10, 5874–5882. [Google Scholar] [CrossRef]
- Jalandra, R.; Dalal, N.; Yadav, A.K.; Verma, D.; Sharma, M.; Singh, R.; Khosla, A.; Kumar, A.; Solanki, P.R. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl. Microbiol. Biotechnol. 2021, 105, 7651–7660. [Google Scholar] [CrossRef]
- Kim, D.H.; Jin, Y.H. Intestinal bacterial betaglucuronidase activity of patients with colon cancer. Arch. Pharm. Res. 2001, 24, 564–567. [Google Scholar] [CrossRef]
- Takada, H.; Hirooka, T.; Hiramatsu, Y.; Yamamoto, M. Effect of beta-glucuronidase inhibitor on azoxymethane-induced colonic carcinogenesis in rats. Cancer Res. 1982, 42, 331–334. [Google Scholar] [PubMed]
- Haiser, H.J.; Turnbaugh, P.J. Is it time for a metagenomic basis of therapeutics? Science 2012, 336, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Tlaskalová-Hogenová, H.; Stepánková, R.; Hudcovic, T.; Tucková, L.; Cukrowska, B.; Lodinová-Zádníková, R.; Kozáková, H.; Rossmann, P.; Bártová, J.; Sokol, D.; et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 2004, 93, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Todoric, J.; Antonucci, L.; Karin, M. Targeting inflammation in cancer prevention and therapy. Cancer Prev. Res. 2016, 9, 895–905. [Google Scholar] [CrossRef]
- Savari, S.; Vinnakota, K.; Zhang, Y.; Sjölander, A. Cysteinyl leukotriene and their receptors: Bridging inflammation and colorectal cancer. World J. Gastroenterol. 2014, 20, 968–977. [Google Scholar] [CrossRef]
- Mager, L.F.; Wasmer, M.H.; Rau, T.T.; Krebs, P. Cytokine-induced modulation of colorectal cancer. Front. Oncol. 2016, 6, 96. [Google Scholar] [CrossRef]
- Klampfer, L. Cytokines, inflammation and colon cancer. Curr. Cancer Drug Targets 2011, 11, 451–464. [Google Scholar] [CrossRef]
- Bultman, S.J. Emerging roles of the microbiome in cancer. Carcinogenesis 2014, 35, 249–255. [Google Scholar] [CrossRef]
- Drewes, J.L.; Housseau, F.; Sears, C.L. Sporadic colorectal cancer: Microbial contributors to disease prevention, development and therapy. Br. J. Cancer 2016, 115, 273–280. [Google Scholar] [CrossRef]
- Morikawa, T.; Baba, Y.; Yamauchi, M.; Kuchiba, A.; Nosho, K.; Shima, K.; Tanaka, N.; Huttenhower, C.; Frank, D.A.; Fuchs, C.S.; et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin. Cancer Res. 2011, 17, 1452–1462. [Google Scholar] [CrossRef]
- Belluco, C.; Nitti, D.; Frantz, M.; Toppan, P.; Basso, D.; Plebani, M.; Lise, M.; Jessup, J.M. Interleukin-6 blood level is associated with circulating carcinoembryonic antigen and prognosis in patients with colorectal cancer. Ann. Surg. Oncol. 2000, 7, 133–138. [Google Scholar] [CrossRef]
- Stanilov, N.; Miteva, L.; Dobreva, Z.; Stanilova, S. Colorectal cancer severity and survival in correlation with tumour necrosis factor-alpha. Biotechnol. Biotechnol. Equip. 2014, 28, 911–917. [Google Scholar] [CrossRef]
- Raponi, M.; Winkler, H.; Dracopoli, N.C. KRAS mutations predict response to EGFR inhibitors. Curr. Opin. Pharmacol. 2008, 8, 413–418. [Google Scholar] [CrossRef]
- Hussain, S.P.; Amstad, P.; Raja, K.; Ambs, S.; Nagashima, M.; Bennett, W.P.; Shields, P.G.; Ham, A.J.; Swenberg, J.A.; Marrogi, A.J.; et al. Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: A cancer-prone chronic inflammatory disease. Cancer Res. 2000, 60, 3333–3337. [Google Scholar]
- Danese, S.; Malesci, A.; Vetrano, S. Colitis-associated cancer: The dark side of inflammatory bowel disease. Gut 2011, 60, 1609–1610. [Google Scholar] [CrossRef]
- Beaugerie, L.; Itzkowitz, S.H. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 2015, 373, 195. [Google Scholar] [CrossRef]
- Loddo, I.; Romano, C. Inflammatory bowel disease: Genetics, epigenetics, and pathogenesis. Front. Immunol. 2015, 6, 551. [Google Scholar] [CrossRef]
- Yamamoto, M.; Matsumoto, S. Gut microbiota and colorectal cancer. Genes Environ. 2016, 38, 11. [Google Scholar] [CrossRef]
- Tomasello, G.; Tralongo, P.; Damiani, P.; Sinagra, E.; Di Trapani, B.; Zeenny, M.N.; Hussein, I.H.; Jurjus, A.; Leone, A. Dismicrobism in inflammatory bowel disease and colorectal cancer: Changes in response of colocytes. World J. Gastroenterol. 2014, 20, 18121–18130. [Google Scholar] [CrossRef]
- Eaden, J.A.; Abrams, K.R.; Mayberry, J.F. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut 2001, 48, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Canavan, C.; Abrams, K.R.; Mayberry, J. Metaanalysis: Colorectal and small bowel cancer risk in patients with Crohn’s disease. Aliment. Pharmacol. Ther. 2006, 23, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Barnich, N.; Darfeuille-Michaud, A. Adherent-invasive Escherichia coli and Crohn’s disease. Curr. Opin. Gastroenterol. 2007, 23, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD—What role do Proteobacteria play? Nature Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Rabizadeh, S.; Rhee, K.J.; Wu, S.; Huso, D.; Gan, C.M.; Golub, J.E.; Wu, X.; Zhang, M.; Sears, C. Enterotoxigenic Bacteroides fragilis: A potential instigator of colitis. Inflamm. Bowel Dis. 2007, 13, 1475–1483. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene 2010, 29, 781–788. [Google Scholar] [CrossRef]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; Devinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Ryz, N.R.; Patterson, S.J.; Zhang, Y.; Ma, C.; Huang, T.; Bhinder, G.; Wu, X.; Chan, J.; Glesby, A.; Sham, H.P.; et al. Active vitamin D (1,25- dihydroxyvitamin D3) increases host susceptibility to Citrobacter rodentium by suppressing mucosal Th17 responses. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1299–G1311. [Google Scholar] [CrossRef]
- Nazareth, N.; Magro, F.; Machado, E.; Ribeiro, T.G.; Martinho, A.; Rodrigues, P.; Alves, R.; Macedo, G.N.; Gracio, D.; Coelho, R.; et al. Prevalence of Mycobacterium avium subsp. paratuberculosis and Escherichia coli in blood samples from patients with inflammatory bowel disease. Med. Microbiol. Immunol. 2015, 204, 681–692. [Google Scholar] [CrossRef]
- Bellaguarda, E.; Chang, E.B. IBD and the gut microbiota from bench to personalized medicine. Curr. Gastroenterol. Rep. 2015, 17, 15. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Hawk, E.T.; Levin, B. Colorectal cancer prevention. J. Clin. Oncol. 2005, 23, 378–391. [Google Scholar] [CrossRef]
- Lavelle, E.C.; Murphy, C.; O’Neill, L.A.; Creagh, E.M. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 2010, 3, 17–28. [Google Scholar] [CrossRef]
- Ginsburg, I. Role of lipoteichoic acid in infection and inflammation. Lancet Infect. Dis. 2002, 2, 171–179. [Google Scholar] [CrossRef]
- Hermann, C.; Spreitzer, I.; Schröder, N.W.; Morath, S.; Lehner, M.D.; Fischer, W.; Schütt, C.; Schumann, R.R.; Hartung, T. Cytokine induction by purified lipoteichoic acids from various bacterial species–role of LBP, sCD14, CD14 and failure to induce IL-12 and subsequent IFN-gamma release. Eur. J. Immunol. 2002, 32, 541–551. [Google Scholar] [CrossRef]
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability—A new target for disease prevention and therapy. BMC Gastroenterol. 2014, 14, 189. [Google Scholar] [CrossRef]
- Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010, 10, 131–144. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Medzhitov, R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science 2007, 317, 124–127. [Google Scholar] [CrossRef]
- Salcedo, R.; Worschech, A.; Cardone, M.; Jones, Y.; Gyulai, Z.; Dai, R.M.; Wang, E.; Ma, W.; Haines, D.; O’hUigin, C.; et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: Role of interleukin 18. J. Exp. Med. 2010, 207, 1625–1636. [Google Scholar] [CrossRef]
- Asquith, M.; Powrie, F. An innately dangerous balancing act: Intestinal homeostasis, inflammation, and colitis-associated cancer. J. Exp. Med. 2010, 207, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Abreu, M.T. Microflora in colorectal cancer: A friend to fear. Nat. Med. 2010, 16, 639–641. [Google Scholar] [CrossRef] [PubMed]
- Lowe, E.L.; Crother, T.R.; Rabizadeh, S.; Hu, B.; Wang, H.; Chen, S.; Shimada, K.; Wong, M.H.; Michelsen, K.S.; Arditi, M. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS ONE 2010, 5, e13027. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Hao, L.; Medzhitov, R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity 2006, 25, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Yesudhas, D.; Gosu, V.; Anwar, M.A.; Choi, S. Multiple roles of toll-like receptor 4 in colorectal cancer. Front. Immunol. 2014, 5, 334. [Google Scholar] [CrossRef]
- Wang, A.C.; Su, Q.B.; Wu, F.X.; Zhang, X.L.; Liu, P.S. Role of TLR4 for paclitaxel chemotherapy in human epithelial ovarian cancer cells. Eur. J. Clin. Investig. 2009, 39, 157–164. [Google Scholar] [CrossRef]
- Wang, E.L.; Qian, Z.R.; Nakasono, M.; Tanahashi, T.; Yoshimoto, K.; Bando, Y.; Kudo, E.; Shimada, M.; Sano, T. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br. J. Cancer 2010, 102, 908–915. [Google Scholar] [CrossRef]
- Beutler, B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 2004, 430, 257–263. [Google Scholar] [CrossRef]
- Lin, L.; Zhang, J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 2. [Google Scholar] [CrossRef]
- Gao, R.; Gao, Z.; Huang, L.; Qin, H. Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 757–769. [Google Scholar] [CrossRef]
- Lascorz, J.; Hemminki, K.; Forsti, A. Systematic enrichment analysis of gene expression profiling studies identifies consensus pathways implicated in colorectal cancer development. J. Carcinog. 2011, 10, 7. [Google Scholar] [CrossRef]
- Couturier-Maillard, A.; Secher, T.; Rehman, A.; Normand, S.; De Arcangelis, A.; Haesler, R.; Huot, L.; Grandjean, T.; Bressenot, A.; Delanoye-Crespin, A.; et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Investig. 2013, 123, 700–711. [Google Scholar] [CrossRef]
- Tian, Y.; Li, Y.; Hu, Z.; Wang, D.; Sun, X.; Ren, C. Differential effects of NOD2 polymorphisms on colorectal cancer risk: A meta-analysis. Int. J. Color. Dis. 2010, 25, 161–168. [Google Scholar] [CrossRef]
- Caruso, R.; Warner, N.; Inohara, N.; Nunez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD proteins: Regulators of inflammation in health and disease. Nat. Rev. Immunol. 2014, 14, 9–23. [Google Scholar] [CrossRef]
- Nabatov, A.A. The vesicle-associated function of NOD2 as a link between Crohn’s disease and mycobacterial infection. Gut Pathog. 2015, 7, 1. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Kamendulis, L.M.; Hocevar, B.A. Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 2010, 38, 96–109. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Bartsch, H.; Nair, J. Potential role of lipid peroxidation derived DNA damage in human colon carcinogenesis: Studies on exocyclic base adducts as stable oxidative stress markers. Cancer Detect. Prev. 2002, 26, 308–312. [Google Scholar] [CrossRef]
- Roessner, A.; Kuester, D.; Malfertheiner, P.; Schneider-Stock, R. Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol. Res. Pract. 2008, 204, 511–524. [Google Scholar] [CrossRef]
- Irrazábal, T.; Belcheva, A.; Girardin, S.E.; Martin, A.; Philpott, D.J. The multifaceted role of the intestinal microbiota in colon cancer. Mol. Cell 2014, 54, 309–320. [Google Scholar] [CrossRef]
- Lundberg, J.O.; Weitzberg, E.; Cole, J.A.; Benjamin, N. Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2004, 2, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Sobko, T.; Huang, L.; Midtvedt, T.; Norin, E.; Gustafsson, L.E.; Norman, M.; Jansson, E.A.; Lundberg, J.O. Generation of NO by probiotic bacteria in the gastrointestinal tract. Free Radic. Biol. Med. 2006, 41, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Sobko, T.; Reinders, C.I.; Jansson, E.; Norin, E.; Midtvedt, T.; Lundberg, J.O. Gastrointestinal bacteria generate nitric oxide from nitrate and nitrite. Nitric Oxide 2005, 13, 272–278. [Google Scholar] [CrossRef]
- Huycke, M.M.; Moore, D.; Joyce, W.; Wise, P.; Shepard, L.; Kotake, Y.; Gilmore, M.S. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol. Microbiol. 2001, 42, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA damage and disease: Induction, repair and significance. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef]
- Handa, O.; Naito, Y.; Yoshikawa, T. Helicobacter pylori: A ROS-inducing bacterial species in the stomach. Inflamm. Res. 2010, 59, 997–1003. [Google Scholar] [CrossRef]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Hazane-Puch, F.; Bonnet, M.; Valenti, K.; Schnebert, S.; Kurfurst, R.; Favier, A.; Sauvaigo, S.l. Study of fibroblast gene expression in response to oxidative stress induced by hydrogen peroxide or UVA with skin aging. Eur. J. Dermatol. 2010, 20, 308–320. [Google Scholar] [CrossRef]
- Lang, T.; Maitra, M.; Starcevic, D.; Li, S.X.; Sweasy, J.B. A DNA polymerase beta mutant from colon cancer cells induces mutations. Proc. Natl. Acad. Sci. USA 2004, 101, 6074–6079. [Google Scholar] [CrossRef]
- Peltomäki, P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003, 21, 1174–1179. [Google Scholar] [CrossRef]
- Moreno, V.; Gemignani, F.; Landi, S.; Gioia-Patricola, L.; Chabrier, A.; Blanco, I.; González, S.; Guino, E.; Capellà, G.; Canzian, F. Polymorphisms in genes of nucleotide and base excision repair: Risk and prognosis of colorectal cancer. Clin. Cancer Res. 2006, 12, 2101–2108. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Scanlon, K.M.; Donnenberg, M.S. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio 2013, 4, e00152-13. [Google Scholar] [CrossRef]
- Mangerich, A.; Knutson, C.G.; Parry, N.M.; Muthupalani, S.; Ye, W.; Prestwich, E.; Cui, L.; McFaline, J.L.; Mobley, M.; Ge, Z.; et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E1820–E1829. [Google Scholar] [CrossRef]
- Belcheva, A.; Irrazabal, T.; Robertson, S.J.; Streutker, C.; Maughan, H.; Rubino, S.; Moriyama, E.H.; Copeland, J.K.; Surendra, A.; Kumar, S.; et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 2014, 158, 288–299. [Google Scholar] [CrossRef]
- Doll, R.; Peto, R. The causes of cancer: Quantitative estimates of avoidable risks of cancer in the United States today. J. Natl. Cancer Inst. 1981, 66, 1191–1308. [Google Scholar] [CrossRef]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M., Jr.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- O’Keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Zhang, F.F.; Cudhea, F.; Shan, Z.; Michaud, D.S.; Imamura, F.; Eom, H.; Ruan, M.; Rehm, C.D.; Liu, J.; Du, M.; et al. Preventable cancer burden associated with poor diet in the United States. JNCI Cancer Spectr. 2019, 3, pkz034. [Google Scholar] [CrossRef]
- Bernstein, A.M.; Song, M.; Zhang, X.; Pan, A.; Wang, M.; Fuchs, C.S.; Le, N.; Chan, A.T.; Willett, W.C.; Ogino, S.; et al. Processed and unprocessed red meat and risk of colorectal cancer: Analysis by tumor location and modification by time. PLoS ONE 2015, 10, e0135959. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.S.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and processed meat and colorectal cancer incidence: Meta-analysis of prospective studies. PLoS ONE 2011, 6, e20456. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.J.; Ferrucci, L.M.; Risch, A.; Graubard, B.I.; Ward, M.H.; Park, Y.; Hollenbeck, A.R.; Schatzkin, A.; Sinha, R. A large prospective study of meat consumption and colorectal cancer risk: An investigation of potential mechanisms underlying this association. Cancer Res. 2010, 70, 2406–2414. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Chan, A.T. Environmental factors, gut microbiota, and colorectal cancer prevention. Clin. Gastroenterol. Hepatol. 2019, 17, 275–289. [Google Scholar] [CrossRef]
- Seiwert, N.; Adam, J.; Steinberg, P.; Wirtz, S.; Schwerdtle, T.; Adams-Quack, P.; Hövelmeyer, N.; Kaina, B.; Foersch, S.; Fahrer, J. Chronic intestinal inflammation drives colorectal tumor formation triggered by dietary heme iron in vivo. Arch. Toxicol. 2021, 95, 2507–2522. [Google Scholar] [CrossRef]
- Zimmer, J.; Lange, B.; Frick, J.S.; Sauer, H.; Zimmermann, K.; Schwiertz, A.; Rusch, K.; Klosterhalfen, S.; Enck, P. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur. J. Clin. Nutr. 2012, 66, 53–60. [Google Scholar] [CrossRef]
- Orlich, M.J.; Singh, P.N.; Sabaté, J.; Fan, J.; Sveen, L.; Bennett, H.; Knutsen, S.F.; Beeson, W.L.; Jaceldo-Siegl, K.; Butler, T.L.; et al. Vegetarian dietary patterns and the risk of colorectal cancers. JAMA Intern. Med. 2015, 175, 767–776. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Fitzgerald, G.F.; Van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef]
- Berg, A.M.; Kelly, C.P.; Farraye, F.A. Clostridium difficile infection in the inflammatory bowel disease patient. Inflamm. Bowel Dis. 2013, 19, 194–204. [Google Scholar] [CrossRef]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Backhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef]
- Chen, H.M.; Yu, Y.N.; Wang, J.L.; Lin, Y.W.; Kong, X.; Yang, C.Q.; Yang, L.; Liu, Z.J.; Yuan, Y.Z.; Liu, F.; et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013, 97, 1044–1052. [Google Scholar] [CrossRef]
- Parker, K.D.; Maurya, A.K.; Ibrahim, H.; Rao, S.; Hove, P.R.; Kumar, D.; Kant, R.; Raina, B.; Agarwal, R.; Kuhn, K.A.; et al. Dietary rice bran-modified human gut microbial consortia confers protection against colon carcinogenesis following fecal transfaunation. Biomedicines 2021, 9, 144. [Google Scholar] [CrossRef]
- Li, Q.; Chen, C.; Liu, C.; Sun, W.; Liu, X.; Ci, Y.; Song, Y. The effects of cellulose on AOM/DSS-treated C57BL/6 colorectal cancer mice by changing intestinal flora composition and inflammatory factors. Nutr. Cancer 2021, 73, 502–513. [Google Scholar] [CrossRef]
- Russell, W.R.; Hoyles, L.; Flint, H.J.; Dumas, M.E. Colonic bacterial metabolites and human health. Curr. Opin. Microbiol. 2013, 16, 246–254. [Google Scholar] [CrossRef]
- Bultman, S.J.; Jobin, C. Microbial-derived butyrate: An oncometabolite or tumor-suppressive metabolite? Cell Host Microbe 2014, 16, 143–145. [Google Scholar] [CrossRef]
- Hester, C.M.; Jala, V.R.; Langille, M.G.; Umar, S.; Greiner, K.A.; Haribabu, B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J. Gastroenterol. 2015, 21, 2759–2769. [Google Scholar] [CrossRef]
- Ang, Z.; Ding, J.L. GPR41 and GPR43 in obesity and inflammation—Protective or causative? Front. Immunol. 2016, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Augenlicht, L.H.; Mariadason, J.M.; Wilson, A.; Arango, D.; Yang, W.; Heerdt, B.G.; Velcich, A. Short chain fatty acids and colon cancer. J. Nutr. 2002, 132, 3804S–3808S. [Google Scholar] [CrossRef]
- Fung, K.Y.; Brierley, G.V.; Henderson, S.; Hoffmann, P.; McColl, S.R.; Lockett, T.; Head, R.; Cosgrove, L. Butyrate-induced apoptosis in HCT116 colorectal cancer cells includes induction of a cell stress response. J. Proteome Res. 2011, 10, 1860–1869. [Google Scholar] [CrossRef]
- Garavaglia, B.; Vallino, L.; Ferraresi, A.; Esposito, A.; Salwa, A.; Vidoni, C.; Gentilli, S.; Isidoro, C. Butyrate inhibits colorectal cancer cell proliferation through autophagy degradation of β-catenin regardless of apc and β-catenin mutational status. Biomedicines 2022, 10, 1131. [Google Scholar] [CrossRef]
- Hinnebusch, B.F.; Meng, S.; Wu, J.T.; Archer, S.Y.; Hodin, R.A. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J. Nutr. 2002, 132, 1012–1017. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Kreznar, J.H.; Romano, K.A.; Vivas, E.I.; Barrett-Wilt, G.A.; Rabaglia, M.E.; Keller, M.P.; Attie, A.D.; Rey, F.E.; Denu, J.M. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 2016, 64, 982–992. [Google Scholar] [CrossRef]
- Buda, A.; Qualtrough, D.; Jepson, M.A.; Martines, D.; Paraskeva, C.; Pignatelli, M. Butyrate downregulates alpha2beta1 integrin: A possible role in the induction of apoptosis in colorectal cancer cell lines. Gut 2003, 52, 729–734. [Google Scholar] [CrossRef]
- Nepelska, M.; Cultrone, A.; Béguet-Crespel, F.; Le Roux, K.; Doré, J.; Arulampalam, V.; Blottière, H.M. Butyrate produced by commensal bacteria potentiates phorbol esters induced AP-1 response in human intestinal epithelial cells. PLoS ONE 2012, 7, e52869. [Google Scholar] [CrossRef]
- Duncan, S.H.; Hold, G.L.; Harmsen, H.J.; Stewart, C.S.; Flint, H.J. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2141–2146. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Martinez-Medina, M.; Abellà, C.; Busquets, D.; Sabat-Mir, M.; Duncan, S.H.; Aldeguer, X.; Flint, H.J.; Garcia-Gil, L.J. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl. Environ. Microbiol. 2015, 81, 7582–7592. [Google Scholar] [CrossRef]
- Ryu, T.Y.; Kim, K.; Han, T.S.; Lee, M.O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.J.; An, D.M.; Jung, C.R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022, 16, 1205–1221. [Google Scholar] [CrossRef]
- Butler, L.M.; Wang, R.; Koh, W.P.; Yu, M.C. Prospective study of dietary patterns and colorectal cancer among Singapore Chinese. Br. J. Cancer 2008, 99, 1511–1516. [Google Scholar] [CrossRef]
- Kabat, G.C.; Shikany, J.M.; Beresford, S.A.; Caan, B.; Neuhouser, M.L.; Tinker, L.F.; Rohan, T.E. Dietary carbohydrate, glycemic index, and glycemic load in relation to colorectal cancer risk in the Women’s Health Initiative. Cancer Causes Control 2008, 19, 1291–1298. [Google Scholar] [CrossRef]
- Rowland, I.R. The role of the gastrointestinal microbiota in colorectal cancer. Curr. Pharm. Des. 2009, 15, 1524–1527. [Google Scholar] [CrossRef]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.E.; Wallace, R.J.; et al. High-protein, reduced carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Attene-Ramos, M.S.; Wagner, E.D.; Gaskins, H.R.; Plewa, M.J. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 2007, 5, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Windey, K.; de Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef]
- McIntosh, F.M.; Maison, N.; Holtrop, G.; Young, P.; Stevens, V.J.; Ince, J.; Johnstone, A.M.; Lobley, G.E.; Flint, H.J.; Louis, P. Phylogenetic distribution of genes encoding β-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ. Microbiol. 2012, 14, 1876–1887. [Google Scholar] [CrossRef] [PubMed]
- Marquet, P.; Duncan, S.H.; Chassard, C.; Bernalier-Donadille, A.; Flint, H.J. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol. Lett. 2009, 299, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Attene-Ramos, M.S.; Nava, G.M.; Muellner, M.G.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 int cells. Environ. Mol. Mutag. 2010, 51, 304–314. [Google Scholar] [CrossRef]
- Hulin, S.J.; Singh, S.; Chapman, M.A.S.; Allan, A.; Langman, M.J.S.; Eggo, M.C. Sulphide-induced energy deficiency in colonic cells is prevented by glucose but not by butyrate. Aliment. Pharm. Ther. 2002, 16, 325–331. [Google Scholar] [CrossRef]
- Christl, S.U.; Eisner, H.D.; Dusel, G.; Kasper, H.; Scheppach, W. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: A potential role for these agents in the pathogenesis of ulcerative colitis. Dig. Dis. Sci. 1996, 41, 2477–2481. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Ma, W.; Wang, D.D.; Cao, Y.; Mallick, H.; Gerbaba, T.K.; Lloyd-Price, J.; Abu-Ali, G.; Hall, A.B.; Sikavi, D.; et al. Association between sulfur-metabolizing bacterial communities in stool and risk of distal colorectal cancer in men. Gastroenterology 2020, 158, 1313–1325. [Google Scholar] [CrossRef]
- Drasar, B.S.; Irving, D. Environmental factors and cancer of the colon and breast. Br. J. Cancer 1973, 27, 167–172. [Google Scholar] [CrossRef]
- Cao, H.; Luo, S.; Xu, M.; Zhang, Y.; Song, S.; Wang, S.; Kong, X.; He, N.; Cao, X.; Yan, F.; et al. The secondary bile acid, deoxycholate accelerates intestinal adenoma-adenocarcinoma sequence in Apcmin/+ mice through enhancing Wnt signaling. Fam. Cancer 2014, 13, 563–571. [Google Scholar] [CrossRef]
- Schatzkin, A.; Lanza, E.; Corle, D.; Lance, P.; Iber, F.; Caan, B.; Shike, M.; Weissfeld, J.; Burt, R.; Cooper, M.R.; et al. Lack of effect of a low-fat, high-fiber diet on the recurrence of colorectal adenomas. Polyp Prevention Trial Study Group. N. Engl. J. Med. 2000, 342, 1149–1155. [Google Scholar] [CrossRef]
- Terry, P.; Bergkvist, L.; Holmberg, L.; Wolk, A. No association between fat and fatty acids intake and risk of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2001, 10, 913–914. [Google Scholar]
- Lanza, E.; Yu, B.; Murphy, G.; Albert, P.S.; Caan, B.; Marshall, J.R.; Lance, P.; Paskett, E.D.; Weissfeld, J.; Slattery, M.; et al. The Polyp Prevention trial continued follow-up study: No effect of a low-fat, high-fiber, high-fruit, and -vegetable diet on adenoma recurrence eight years after randomization. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1745–1752. [Google Scholar] [CrossRef]
- Weijenberg, M.P.; Luchtenborg, M.; de Goeij, A.F.; Brink, M.; van Muijen, G.N.; de Bruine, A.P.; Goldbohm, R.A.; van den Brandt, P.A. Dietary fat and risk of colon and rectal cancer with aberrant MLH1 expression, APC or KRAS genes. Cancer Causes Control 2007, 18, 865–879. [Google Scholar] [CrossRef]
- Thomson, C.A.; Van Horn, L.; Caan, B.J.; Aragaki, A.K.; Chlebowski, R.T.; Manson, J.E.; Rohan, T.E.; Tinker, L.F.; Kuller, L.H.; Hou, L.; et al. Cancer incidence and mortality during the intervention and postintervention periods of the Women’s Health Initiative dietary modification trial. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2924–2935. [Google Scholar] [CrossRef]
- Taira, T.; Yamaguchi, S.; Takahashi, A.; Okazaki, Y.; Yamaguchi, A.; Sakaguchi, H.; Chiji, H. Dietary polyphenols increase fecal mucin and immunoglobulin A and ameliorate the disturbance in gut microbiota caused by a high fat diet. J. Clin. Biochem. Nutr. 2015, 57, 212–216. [Google Scholar] [CrossRef]
- Higashimura, Y.; Naito, Y.; Takagi, T.; Uchiyama, K.; Mizushima, K.; Ushiroda, C.; Ohnogi, H.; Kudo, Y.; Yasui, M.; Inui, S.; et al. Protective effect of agaro-oligosaccharides on gut dysbiosis and colon tumorigenesis in high-fat diet-fed mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G367–G375. [Google Scholar] [CrossRef]
- Yang, J.; Wei, H.; Zhou, Y.; Szeto, C.H.; Li, C.; Lin, Y.; Coker, O.O.; Lau, H.C.H.; Chan, A.W.H.; Sung, J.J.Y.; et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology 2022, 162, 135–149.e2. [Google Scholar] [CrossRef]
- Yu, Y.; Cai, Y.; Yang, B.; Xie, S.; Shen, W.; Wu, Y.; Sui, Z.; Cai, J.; Ni, C.; Ye, J. High-fat diet enhances the liver metastasis potential of colorectal cancer through microbiota dysbiosis. Cancers 2022, 14, 2573. [Google Scholar] [CrossRef]
- Sharma, M.; Li, L.; Celver, J.; Killian, C.; Kovoor, A.; Seeram, N.P. Effects of fruit ellagitannin extracts, ellagic acid, and their colonic metabolite, urolithin A., on Wnt signaling. J. Agric. Food Chem. 2010, 58, 3965–3969. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Bastida, J.A.; Ávila-Gálvez, M.Á.; Espín, J.C.; González-Sarrías, A. The gut microbiota metabolite urolithin A, but not other relevant urolithins, induces p53-dependent cellular senescence in human colon cancer cells. Food Chem. Toxicol. 2020, 139, 111260. [Google Scholar] [CrossRef] [PubMed]
- Herr, I.; Büchler, M.W. Dietary constituents of broccoli and other cruciferous vegetables: Implications for prevention and therapy of cancer. Cancer Treat. Rev. 2010, 36, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Khan, I.; Huang, G.; Lu, Y.; Wang, L.; Liu, Y.; Lu, L.; Hsiao, W.L.W.; Liu, Z. Kaempferol acts on bile acid signaling and gut microbiota to attenuate the tumor burden in Apcmin/+ mice. Eur. J. Pharmacol. 2022, 918, 174773. [Google Scholar] [CrossRef] [PubMed]
- Klement, R.J. Beneficial effects of ketogenic diets for cancer patients: A realist review with focus on evidence and confirmation. Med. Oncol. 2017, 34, 132. [Google Scholar] [CrossRef]
- Weber, D.D.; Aminazdeh-Gohari, S.; Kofler, B. Ketogenic diet in cancer therapy. Aging 2018, 10, 164–165. [Google Scholar] [CrossRef]
- Newell, C.; Bomhof, M.R.; Reimer, R.A.; Hittel, D.S.; Rho, J.M.; Shearer, J. Ketogenic diet modifies the gut microbiota in a murine model of autism spectrum disorder. Mol. Autism 2016, 7, 37. [Google Scholar] [CrossRef]
- Illescas, O.; Rodríguez-Sosa, M.; Gariboldi, M. Mediterranean diet to prevent the development of colon diseases: A meta-analysis of gut microbiota studies. Nutrients 2021, 13, 2234. [Google Scholar] [CrossRef]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Yi, M.; Qin, S.; Chu, Q.; Wu, K. The role of gut microbiota inimmune checkpoint inhibitor therapy. Hepatobiliary Surg. Nutr. 2018, 7, 481–483. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Mackowiak, P.A. Recycling Metchnikoff: Probiotics, the intestinal microbiome and the quest for long life. Front. Public Health 2013, 1, 52. [Google Scholar] [CrossRef]
- Zhu, Y.; Michelle Luo, T.; Jobin, C.; Young, H.A. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011, 309, 119–127. [Google Scholar] [CrossRef]
- Raman, M.; Ambalam, P.; Kondepudi, K.K.; Pithva, S.; Kothari, C.; Patel, A.T.; Purama, R.K.; Dave, J.M.; Vyas, B.R. Potential of probiotics, prebiotics and synbiotics for management of colorectal cancer. Gut Microbes 2013, 4, 181–192. [Google Scholar] [CrossRef]
- Tojo, R.; Suárez, A.; Clemente, M.G.; de los Reyes-Gavilán, C.G.; Margolles, A.; Gueimonde, M.; Ruas-Madiedo, P. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World J. Gastroenterol. 2014, 20, 15163–15176. [Google Scholar] [CrossRef]
- Ambalam, P.; Raman, M.; Purama, R.K.; Doble, M. Probiotics, prebiotics and colorectal cancer prevention. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 119–131. [Google Scholar] [CrossRef]
- Kamada, N.; Kim, Y.G.; Sham, H.P.; Vallance, B.A.; Puente, J.L.; Martens, E.C.; Núñez, G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 2012, 336, 1325–1329. [Google Scholar] [CrossRef]
- Tuomola, E.M.; Ouwehand, A.C.; Salminen, S.J. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol. Med. Microbiol. 1999, 26, 137–142. [Google Scholar] [CrossRef]
- Mills, J.P.; Rao, K.; Young, V.B. Probiotics for prevention of Clostridium difficile infection. Curr. Opin. Gastroenterol. 2018, 34, 3–10. [Google Scholar] [CrossRef]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef]
- Fayol-Messaoudi, D.; Berger, C.N.; Coconnier-Polter, M.H.; Lievin-Le Moal, V.; Servin, A.L. pH-, Lactic acid-, and non-lactic acid-dependent activities of probiotic Lactobacilli against Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2005, 71, 6008–6013. [Google Scholar] [CrossRef] [PubMed]
- Gillor, O.; Etzion, A.; Riley, M.A. The dual role of bacteriocins as anti- and probiotics. Appl. Microbiol. Biotechnol. 2008, 81, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Khazaie, K.; Zadeh, M.; Khan, M.W.; Bere, P.; Gounari, F.; Dennis, K.; Blatner, N.R.; Owen, J.L.; Klaenhammer, T.R.; Mohamadzadeh, M. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. USA 2012, 109, 10462–10467. [Google Scholar] [CrossRef] [PubMed]
- Saber, R.; Zadeh, M.; Pakanati, K.C.; Bere, P.; Klaenhammer, T.; Mohamadzadeh, M. Lipoteichoic acid-deficient Lactobacillus acidophilus regulates downstream signals. Immunotherapy 2011, 3, 337–347. [Google Scholar] [CrossRef]
- Chong, E.S. A potential role of probiotics in colorectal cancer prevention: Review of possible mechanisms of action. World J. Microbiol. Biotechnol. 2014, 30, 351–374. [Google Scholar] [CrossRef]
- Sharma, M.; Shukla, G. Metabiotics: One step ahead of probiotics; an insight into mechanisms involved in anticancerous effect in colorectal cancer. Front. Microbiol. 2016, 7, 1940. [Google Scholar] [CrossRef]
- Walia, S.; Kamal, R.; Kanwar, S.S.; Dhawan, D.K. Cyclooxygenase as a target in chemoprevention by probiotics during 1,2-dimethylhydrazine induced colon carcinogenesis in rats. Nutr. Cancer 2015, 67, 603–611. [Google Scholar] [CrossRef]
- Thirabunyanon, M.; Boonprasom, P.; Niamsup, P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol. Lett. 2009, 31, 571–576. [Google Scholar] [CrossRef]
- Konishi, H.; Fujiya, M.; Tanaka, H.; Ueno, N.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Akutsu, H.; Tanabe, H.; Kohgo, Y. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat. Commun. 2016, 7, 12365. [Google Scholar] [CrossRef]
- Lightfoot, Y.L.; Yang, T.; Sahay, B.; Mohamadzadeh, M. Targeting aberrant colon cancer-specific DNA methylation with lipoteichoic acid-deficient Lactobacillus acidophilus. Gut Microbes 2013, 4, 84–88. [Google Scholar] [CrossRef]
- Theodoropoulos, G.E.; Memos, N.A.; Peitsidou, K.; Karantanos, T.; Spyropoulos, B.G.; Zografos, G. Synbiotics and gastrointestinal function-related quality of life after elective colorectal cancer resection. Ann. Gastroenterol. 2016, 29, 56–62. [Google Scholar]
- Flesch, A.T.; Tonial, S.T.; Contu, P.C.; Damin, D.C. Perioperative synbiotics administration decreases postoperative infections in patients with colorectal cancer: A randomized, double-blind clinical trial. Rev. Col. Bras. Cir. 2017, 44, 567–573. [Google Scholar] [CrossRef]
- Hibberd, A.A.; Lyra, A.; Ouwehand, A.C.; Rolny, P.; Lindegren, H.; Cedgård, L.; Wettergren, Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017, 4, e000145. [Google Scholar] [CrossRef]
- Kuugbee, E.D.; Shang, X.; Gamallat, Y.; Bamba, D.; Awadasseid, A.; Suliman, M.A.; Zang, S.; Ma, Y.; Chiwala, G.; Xin, Y.; et al. Structural change in microbiota by a probiotic cocktail enhances the gut barrier and reduces cancer via TLR2 signaling in a rat model of colon cancer. Dig. Dis. Sci. 2016, 61, 2908–2920. [Google Scholar] [CrossRef]
- Rafter, J.; Bennett, M.; Caderni, G.; Clune, Y.; Hughes, R.; Karlsson, P.C.; Klinder, A.; O’Riordan, M.; O’Sullivan, G.C.; Pool-Zobel, B.; et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 2007, 85, 488–496. [Google Scholar] [CrossRef]
- Benito, I.; Encío, I.J.; Milagro, F.I.; Alfaro, M.; Martínez-Peñuela, A.; Barajas, M.; Marzo, F. Microencapsulated Bifidobacterium bifidum and Lactobacillus gasseri in combination with quercetin inhibit colorectal cancer development in Apcmin/+ mice. Int. J. Mol. Sci. 2021, 22, 4906. [Google Scholar] [CrossRef]
- Chen, Z.F.; Ai, L.Y.; Wang, J.L.; Ren, L.L.; Yu, Y.N.; Xu, J.; Chen, H.Y.; Yu, J.; Li, M.; Qin, W.X.; et al. Probiotics Clostridium butyricum and Bacillus subtilis ameliorate intestinal tumorigenesis. Future Microbiol. 2015, 10, 1433–1445. [Google Scholar] [CrossRef]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020, 469, 456–467. [Google Scholar] [CrossRef]
- Liu, M.; Xie, W.; Wan, X.; Deng, T. Clostridium butyricum modulates gut microbiota and reduces colitis associated colon cancer in mice. Int. Immunopharmacol. 2020, 88, 106862. [Google Scholar] [CrossRef]
- Li, Q.; Hu, W.; Liu, W.X.; Zhao, L.Y.; Huang, D.; Liu, X.D.; Chan, H.; Zhang, Y.; Zeng, J.D.; Coker, O.O.; et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting β-galactosidase. Gastroenterology 2021, 160, 1179–1193.e14. [Google Scholar] [CrossRef]
- Liu, D.; Jiang, X.Y.; Zhou, L.S.; Song, J.H.; Zhang, X. Effects of probiotics on intestinal mucosa barrier in patients with colorectal cancer after operation: Meta-analysis of randomized controlled trials. Medicine 2016, 95, e3342. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, D.; Kim, D.; Cho, J.; Yang, J.; Chung, M.; Kim, K.; Ha, N. Inhibition of proliferation in colon cancer cell lines and harmful enzyme activity of colon bacteria by Bifidobacterium adolescentis SPM0212. Arch. Pharm. Res. 2008, 31, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Matsumoto, S.; Ohashi, Y.; Imaoka, A.; Setoyama, H.; Umesaki, Y.; Tanaka, R.; Otani, T. Beneficial effects of probiotic bifidobacterium and galacto-oligosaccharide in patients with ulcerative colitis: A randomized controlled study. Digestion 2011, 84, 128–133. [Google Scholar] [CrossRef]
- Yoon, Y.; Kim, G.; Jeon, B.N.; Fang, S.; Park, H. Bifidobacterium strain-specific enhances the efficacy of cancer therapeutics in tumor-bearing mice. Cancers 2021, 13, 957. [Google Scholar] [CrossRef]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Konieczna, P.; Groeger, D.; Ziegler, M.; Frei, R.; Ferstl, R.; Shanahan, F.; Quigley, E.M.; Kiely, B.; Akdis, C.A.; O’Mahony, L. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: Potential role for myeloid and plasmacytoid dendritic cells. Gut 2012, 61, 354–366. [Google Scholar] [CrossRef]
- Huang, C.; Shi, G. Smoking and microbiome in oral, airway, gut and some systemic diseases. J. Transl. Med. 2019, 17, 225. [Google Scholar] [CrossRef]
- Gamallat, Y.; Meyiah, A.; Kuugbee, E.D.; Hago, A.M.; Chiwala, G.; Awadasseid, A.; Bamba, D.; Zhang, X.; Shang, X.; Luo, F.; et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed. Pharmacother. 2016, 83, 536–541. [Google Scholar] [CrossRef]
- Evrard, B.; Coudeyras, S.; Dosgilbert, A.; Charbonnel, N.; Alamé, J.; Tridon, A.; Forestier, C. Dose-dependent immunomodulation of human dendritic cells by the probiotic Lactobacillus rhamnosus Lcr35. PLoS ONE 2011, 6, e18735. [Google Scholar] [CrossRef]
- Ciorba, M.A.; Riehl, T.E.; Rao, M.S.; Moon, C.; Ee, X.; Nava, G.M.; Walker, M.R.; Marinshaw, J.M.; Stappenbeck, T.S.; Stenson, W.F. Lactobacillus probiotic protects intestinal epithelium from radiation injury in a TLR-2/cyclo-oxygenase-2-dependent manner. Gut 2012, 61, 829–838. [Google Scholar] [CrossRef]
- Dong, L.; Li, J.; Liu, Y.; Yue, W.; Luo, X. Toll-like receptor 2 monoclonal antibody or/and Toll-like receptor 4 monoclonal antibody increase counts of Lactobacilli and Bifidobacteria in dextran sulfate sodium-induced colitis in mice. J. Gastroenterol. Hepatol. 2012, 27, 110–119. [Google Scholar] [CrossRef]
- Ghadimi, D.; Helwig, U.; Schrezenmeir, J.; Heller, K.J.; de Vrese, M. Epigenetic imprinting by commensal probiotics inhibits the IL-23/IL-17 axis in an in vitro model of the intestinal mucosal immune system. J. Leukoc. Biol. 2012, 92, 895–911. [Google Scholar] [CrossRef]
- Chen, L.; Zou, Y.; Peng, J.; Lu, F.; Yin, Y.; Li, F.; Yang, J. Lactobacillus acidophilus suppresses colitis-associated activation of the IL-23/Th17 axis. J. Immunol. Res. 2015, 2015, 909514. [Google Scholar] [CrossRef]
- Owens, J.A.; Saeedi, B.J.; Naudin, C.R.; Hunter-Chang, S.; Barbian, M.E.; Eboka, R.U.; Askew, L.; Darby, T.M.; Robinson, B.S.; Jones, R.M. Lactobacillus rhamnosus GG orchestrates an antitumor immune response. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Mitsuyama, K.; Matsumoto, S.; Yamasaki, H.; Masuda, J.; Kuwaki, K.; Takedatsu, H.; Nagaoka, M.; Andoh, A.; Tsuruta, O.; Sata, M. Beneficial effects of Lactobacillus casei in ulcerative colitis: A pilot study. J. Clin. Biochem. Nutr. 2008, 43, 78–81. [Google Scholar] [CrossRef]
- Ishikawa, H.; Akedo, I.; Otani, T.; Suzuki, T.; Nakamura, T.; Takeyama, I.; Ishiguro, S.; Miyaoka, E.; Sobue, T.; Kakizoe, T. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int. J. Cancer 2005, 116, 762–767. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, X.; Fang, B.; Zhu, C.; Zhu, J.; Ren, F. Effects of Lactobacillus salivarius Ren on cancer prevention and intestinal microbiota in 1, 2-dimethylhydrazine-induced rat model. J. Microbiol. 2015, 53, 398–405. [Google Scholar] [CrossRef]
- Mousavi Jam, S.A.; Morshedi, M.; Yari Khosroushahi, A.; Eftekharsadat, A.T.; Alipour, M.; Alipour, B. Preventive and tumor-suppressive effects of Lactobacillus paracasei X12 in rat model of colorectal cancer. Iran J. Pharm. Res. 2020, 19, 330–342. [Google Scholar] [CrossRef]
- Chang, C.Y.; Pan, T.M. Anticancer and antimigration effects of a combinatorial treatment of 5-fluorouracil and Lactobacillus paracasei subsp. paracasei NTU 101 fermented skim milk extracts on colorectal cancer cells. J. Agric. Food Chem. 2018, 66, 5549–5555. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.; Liu, L.; Wang, B.; Walker, W.A.; Acra, S.A.; Yan, F. Activation of epidermal growth factor receptor mediates mucin production stimulated by p40, a Lactobacillus rhamnosus GG-derived protein. J. Biol. Chem. 2014, 289, 20234–20244. [Google Scholar] [CrossRef]
- Martin, R.; Chamignon, C.; Mhedbi-Hajri, N.; Chain, F.; Derrien, M.; Escribano-Vazquez, U.; Garault, P.; Cotillard, A.; Pham, H.P.; Chervaux, C.; et al. The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci. Rep. 2019, 9, 5398. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhou, L.; Liu, D.; Ge, L.; Li, Y. Probiotic effect on Helicobacter pylori attachment and inhibition of inflammation in human gastric epithelial cells. Exp. Ther. Med. 2019, 18, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- Silveira, D.S.C.; Veronez, L.C.; Lopes-Júnior, L.C.; Anatriello, E.; Brunaldi, M.O.; Pereira-da-Silva, G. Lactobacillus bulgaricus inhibits colitis-associated cancer via a negative regulation of intestinal inflammation in azoxymethane/dextran sodium sulfate model. World J. Gastroenterol. 2020, 26, 6782–6794. [Google Scholar] [CrossRef] [PubMed]
- Walsham, A.D.; MacKenzie, D.A.; Cook, V.; Wemyss-Holden, S.; Hews, C.L.; Juge, N.; Schüller, S. Lactobacillus reuteri inhibition of enteropathogenic Escherichia coli adherence to human intestinal epithelium. Front. Microbiol. 2016, 7, 244. [Google Scholar] [CrossRef]
- Bell, H.N.; Rebernick, R.J.; Goyert, J.; Singhal, R.; Kuljanin, M.; Kerk, S.A.; Huang, W.; Das, N.K.; Andren, A.; Solanki, S.; et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 2022, 40, 185–200.e6. [Google Scholar] [CrossRef]
- Hradicka, P.; Beal, J.; Kassayova, M.; Foey, A.; Demeckova, V. A novel lactic acid bacteria mixture: Macrophage-targeted prophylactic intervention in colorectal cancer management. Microorganisms 2020, 8, 387. [Google Scholar] [CrossRef]
- Sugimura, N.; Li, Q.; Chu, E.S.H.; Lau, H.C.H.; Fong, W.; Liu, W.; Liang, C.; Nakatsu, G.; Su, A.C.Y.; Coker, O.O.; et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut 2021, 71, 2011–2021. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, L.; Wang, P.; Liu, Y.; Wang, G.; Shan, Y.; Yi, Y.; Zhou, Y.; Liu, B.; Wang, X.; et al. Lactobacillus coryniformis MXJ32 administration ameliorates azoxymethane/dextran sulfate sodium-induced colitis-associated colorectal cancer via reshaping intestinal microenvironment and alleviating inflammatory response. Eur. J. Nutr. 2022, 61, 85–99. [Google Scholar] [CrossRef]
- Pandey, K.R.; Naik, S.R.; Vakil, B.V. Probiotics, prebiotics and synbiotics—A review. J. Food Sci. Technol. 2015, 52, 7577–7587. [Google Scholar] [CrossRef]
- Dewulf, E.M.; Cani, P.D.; Claus, S.P.; Fuentes, S.; Puylaert, P.G.; Neyrinck, A.M.; Bindels, L.B.; de Vos, W.M.; Gibson, G.R.; Thissen, J.P.; et al. Insight into the prebiotic concept: Lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013, 62, 1112–1121. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Ritter, A.J.; Savaiano, D.; Monteagudo-Mera, A.; Anderson, C.; Magness, S.T.; Klaenhammer, T.R. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proc. Natl. Acad. Sci. USA 2017, 114, E367–E375. [Google Scholar] [CrossRef]
- Maier, T.V.; Lucio, M.; Lee, L.H.; VerBerkmoes, N.C.; Brislawn, C.J.; Bernhardt, J.; Lamendella, R.; McDermott, J.E.; Bergeron, N.; Heinzmann, S.S.; et al. Impact of dietary resistant starch on the human gut microbiome, metaproteome, and metabolome. mBio 2017, 8, e01343-17. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef]
- Costabile, A.; Fava, F.; Röytiö, H.; Forssten, S.D.; Olli, K.; Klievink, J.; Rowland, I.R.; Ouwehand, A.C.; Rastall, R.A.; Gibson, G.R.; et al. Impact of polydextrose on the faecal microbiota: A double-blind, crossover, placebo-controlled feeding study in healthy human subjects. Br. J. Nutr. 2012, 108, 471–481. [Google Scholar] [CrossRef]
- So, D.; Whelan, K.; Rossi, M.; Morrison, M.; Holtmann, G.; Kelly, J.T.; Shanahan, E.R.; Staudacher, H.M.; Campbell, K.L. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018, 107, 965–983. [Google Scholar] [CrossRef]
- Pattananandecha, T.; Sirilun, S.; Duangjitcharoen, Y.; Sivamaruthi, B.S.; Suwannalert, P.; Peerajan, S.; Chaiyasut, C. Hydrolysed inulin alleviates the azoxymethane-induced preneoplastic aberrant crypt foci by altering selected intestinal microbiota in Sprague-Dawley rats. Pharm. Biol. 2016, 54, 1596–1605. [Google Scholar] [CrossRef]
- Fehlbaum, S.; Prudence, K.; Kieboom, J.; Heerikhuisen, M.; van den Broek, T.; Schuren, F.H.J.; Steinert, R.E.; Raederstorff, D. In vitro fermentation of selected prebiotics and their effects on the composition and activity of the adult gut microbiota. Int. J. Mol. Sci. 2018, 19, 3097. [Google Scholar] [CrossRef]
- Zong, S.; Ye, H.; Ye, Z.; He, Y.; Zhang, X.; Ye, M. Polysaccharides from Lachnum sp. inhibited colitis-associated colon tumorigenesis in mice by modulating fecal microbiota and metabolites. Int. Immunopharmacol. 2022, 108, 108656. [Google Scholar] [CrossRef]
- Wang, P.; Chen, H.; Zhu, Y.; McBride, J.; Fu, J.; Sang, S. Oat avenanthramide-C (2c) is biotransformed by mice and the human microbiota into bioactive metabolites. J. Nutr. 2015, 145, 239–245. [Google Scholar] [CrossRef]
- Li, F.; Yang, X.W.; Krausz, K.W.; Nichols, R.G.; Xu, W.; Patterson, A.D.; Gonzalez, F.J. Modulation of colon cancer by nutmeg. J. Proteome Res. 2015, 14, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Piazzi, G.; D’Argenio, G.; Prossomariti, A.; Lembo, V.; Mazzone, G.; Candela, M.; Biagi, E.; Brigidi, P.; Vitaglione, P.; Fogliano, V.; et al. Eicosapentaenoic acid free fatty acid prevents and suppresses colonic neoplasia in colitis-associated colorectal cancer acting on Notch signaling and gut microbiota. Int. J. Cancer 2014, 135, 2004–2013. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; He, Y.; Li, H.; Yu, D.; Na, L.; Sun, T.; Zhang, D.; Shi, X.; Xia, Y.; Jiang, T.; et al. Effects of prebiotics on immunologic indicators and intestinal microbiota structure in perioperative colorectal cancer patients. Nutrition 2019, 61, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yang, H.; Liu, M.; Ren, S.; Zhao, H.; Ming, T.; Tang, S.; Tao, Q.; Chen, L.; Zeng, S.; et al. Berberine suppresses colorectal cancer by regulation of Hedgehog signaling pathway activity and gut microbiota. Phytomedicine 2022, 103, 154227. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Zhao, L.; Yuan, X.; Li, Y.; Shi, J.; Zhang, H.; Zhao, Y.; Han, L.; Wang, H.; Yan, Y.; et al. Pre-Administration of berberine exerts chemopreventive effects in AOM/DSS-induced colitis-associated carcinogenesis mice via modulating inflammation and intestinal microbiota. Nutrients 2022, 14, 726. [Google Scholar] [CrossRef]
- Chen, H.; Ye, C.; Cai, B.; Zhang, F.; Wang, X.; Zhang, J.; Zhang, Z.; Guo, Y.; Yao, Q. Berberine inhibits intestinal carcinogenesis by suppressing intestinal pro-inflammatory genes and oncogenic factors through modulating gut microbiota. BMC Cancer 2022, 22, 566. [Google Scholar] [CrossRef]
- Yan, S.; Chang, J.; Hao, X.; Liu, J.; Tan, X.; Geng, Z.; Wang, Z. Berberine regulates short-chain fatty acid metabolism and alleviates the colitis-associated colorectal tumorigenesis through remodeling intestinal flora. Phytomedicine 2022, 102, 154217. [Google Scholar] [CrossRef]
- Shao, L.; Guo, Y.P.; Wang, L.; Chen, M.Y.; Zhang, W.; Deng, S.; Huang, W.H. Effects of ginsenoside compound K on colitis-associated colorectal cancer and gut microbiota profiles in mice. Ann. Transl. Med. 2022, 10, 408. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Gonzalez-Sarrias, A.; Tome-Carneiro, J.; Bellesia, A.; Tomas-Barberan, F.A.; Espin, J.C. The ellagic acid-derived gut microbiota metabolite, urolithin A, potentiates the anticancer effects of 5-fluorouracil chemotherapy on human colon cancer cells. Food Funct. 2015, 6, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Flament, C.; Zoubir, M.; Pautier, P.; LeCesne, A.; Ribrag, V.; Soria, J.C.; Marty, V.; Vielh, P.; Robert, C.; et al. Cyclophosphamide induces differentiation of Th17 cells in cancer patients. Cancer Res. 2011, 71, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fu, L.; Li, Y.; Wang, W.; Gong, M.; Zhang, J.; Dong, X.; Huang, J.; Wang, Q.; Mackay, C.R.; et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 2021, 33, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563. [Google Scholar] [CrossRef]
- Zheng, D.W.; Dong, X.; Pan, P.; Chen, K.W.; Fan, J.X.; Cheng, S.X.; Zhang, X.Z. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019, 3, 717–728. [Google Scholar] [CrossRef]
- Dubois, D.; Baron, O.; Cougnoux, A.; Delmas, J.; Pradel, N.; Boury, M.; Bouchon, B.; Bringer, M.A.; Nougayrède, J.P.; Oswald, E.; et al. ClbP is a prototype of a peptidase subgroup involved in biosynthesis of nonribosomal peptides. J. Biol. Chem. 2011, 286, 35562–35570. [Google Scholar] [CrossRef]
- Cougnoux, A.; Gibold, L.; Robin, F.; Dubois, D.; Pradel, N.; Darfeuille-Michaud, A.; Dalmasso, G.; Delmas, J.; Bonnet, R. Analysis of structure-function relationships in the colibactin-maturating enzyme ClbP. J. Mol. Biol. 2012, 424, 203–214. [Google Scholar] [CrossRef]
- Aarnoutse, R.; de Vos-Geelen, J.M.P.G.M.; Penders, J.; Boerma, E.G.; Warmerdam, F.A.R.M.; Goorts, B.; Olde Damink, S.W.M.; Soons, Z.; Rensen, S.S.M.; Smidt, M.L. Study protocol on the role of intestinal microbiota in colorectal cancer treatment: A pathway to personalized medicine 2.0. Int. J. Colorectal Dis. 2017, 32, 1077–1084. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Uribe-Herranz, M.; Rafail, S.; Beghi, S.; Gil-de-Gómez, L.; Verginadis, I.; Bittinger, K.; Pustylnikov, S.; Pierini, S.; Perales-Linares, R.; Blair, I.A.; et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J. Clin. Investig. 2020, 130, 466–479. [Google Scholar] [CrossRef]
- Sánchez-Alcoholado, L.; Laborda-Illanes, A.; Otero, A.; Ordóñez, R.; González-González, A.; Plaza-Andrades, I.; Ramos-Molina, B.; Gómez-Millán, J.; Queipo-Ortuño, M.I. Relationships of gut microbiota composition, short-chain fatty acids and polyamines with the pathological response to neoadjuvant radiochemotherapy in colorectal cancer patients. Int. J. Mol. Sci. 2021, 22, 9549. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef]
- Pardoll, D. Cancer and the immune system: Basic concepts and targets for intervention. Semin. Oncol. 2015, 42, 523–538. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A defined commensal Consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef]
- Wang, Y.; Wiesnoski, D.H.; Helmink, B.A.; Gopalakrishnan, V.; Choi, K.; DuPont, H.L.; Jiang, Z.D.; Abu-Sbeih, H.; Sanchez, C.A.; Chang, C.C.; et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 2018, 24, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- Brandt, L.J.; Borody, T.J.; Campbell, J. Endoscopic fecal microbiota transplantation: “first-line” treatment for severe Clostridium difficile infection? J. Clin. Gastroenterol. 2011, 45, 655–657. [Google Scholar] [CrossRef]
- Smits, L.P.; Bouter, K.E.; de Vos, W.M.; Borody, T.J.; Nieuwdorp, M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology 2013, 145, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J.; et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 2017, 171, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-K.; Yang, Y.-S.; Chen, Y.; Yuan, J.; Sun, G.; Peng, L.-H. Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 14805–14820. [Google Scholar] [CrossRef]
- Quera, R.; Espinoza, R.; Estay, C.; Rivera, D. Bacteremia as an adverse event of fecal microbiota transplantation in a patient with Crohn’s disease and recurrent Clostridium difficile infection. J. Crohn’s Colitis 2014, 8, 252–253. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Bloom, P.P.; Soto, M.T.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.B.; Hohmann, E.L. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef]
- Alang, N.; Kelly, C.R. Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2015, 2, ofv004. [Google Scholar] [CrossRef]
- Gregory, J.C.; Buffa, J.A.; Org, E.; Wang, Z.; Levison, B.S.; Zhu, W.; Wagner, M.A.; Bennett, B.J.; Li, L.; DiDonato, J.A.; et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J. Biol. Chem. 2015, 290, 5647–5660. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).