Simple Summary
PARP inhibitors (PARPi) have been administered to treat BRCA1/2-mutated/deficient malignancies. Nevertheless, the resistance to PARPi is emerging in experimental and clinical interventions. Importantly, the resistance originated from diverse mechanisms, therefore requiring tremendous efforts to identify mechanistic aspects and develop combinational therapies to prevent the resistance and/or restore the efficiency of PARPi in cancer cells. Here, we review pre-existing and acquired resistance to PARPi and propose potential therapeutic solutions.
Abstract
The advanced development of synthetic lethality has opened the doors for specific anti-cancer medications of personalized medicine and efficient therapies against cancers. One of the most popular approaches being investigated is targeting DNA repair pathways as the implementation of the PARP inhibitor (PARPi) into individual or combinational therapeutic schemes. Such treatment has been effectively employed against homologous recombination-defective solid tumors as well as hematopoietic malignancies. However, the resistance to PARPi has been observed in both preclinical research and clinical treatment. Therefore, elucidating the mechanisms responsible for the resistance to PARPi is pivotal for the further success of this intervention. Apart from mechanisms of acquired resistance, the bone marrow microenvironment provides a pre-existing mechanism to induce the inefficiency of PARPi in leukemic cells. Here, we describe the pre-existing and acquired mechanisms of the resistance to PARPi-induced synthetic lethality. We also discuss the potential rationales for developing effective therapies to prevent/repress the PARPi resistance in cancer cells.
1. Introduction
Synthetic lethality is a biological process inducing cell death, which is based on the simultaneous inhibition of two pathways that act parallelly in a process required for cell survival. Meanwhile, the inhibition of only one pathway results in cell survival. The synthetic lethality strategy has been widely implemented in anti-cancer therapies. As one pathway may be inactivated in cancer cells due to transformation-related changes, targeting the other pathway triggers cell death while sparing healthy cells [1].
One of the critical features of cancer cells is genomic instability generated by the accumulation of DNA damage, including DNA double-strand breaks (DSBs), which are one of the most lethal DNA lesions in cells [2,3]. However, cancer cells are able to survive and proliferate by modulating their DNA repair pathways, which may differ from those in normal cells [4].
DSBs can be repaired by two major mechanisms: BRCA1/2-mediated homologous recombination (HR) and canonical DNA-PKcs-mediated non-homologous end joining (c-NHEJ) [5]. HR is the major DSB repair mechanism in the S cell cycle phase, whereas c-NHEJ repairs DSBs throughout the cell cycle [6,7]. When HR is inactivated due to deficiencies in BRCA1/2, the prevention and repair of DSBs highly depend on poly-ADP ribose polymerase 1 (PARP1)-mediated base excision repair (BER) and alternative-non-homologous end-joining (a-NHEJ) [8,9]. a-NHEJ is also called microhomology-mediated end-joining (MMEJ) [10], and a-NHEJ/MMEJ involving DNA polymerase theta (Polθ) is called Polθ-mediated end-joining (TMEJ) [11]. Therefore, the inhibition of PARP1 can lead to the induction of the synthetic lethality in proliferating cells harboring HR deficiency (HRD) due to mutations in BRCA1 and BRCA2—for example, [12,13,14,15,16]. Those studies led to the development and implementation of the synthetic lethality triggered by the PARP inhibitor (PARPi), which is currently one of the most effective agents against HR-deficient malignancies [17]. Concomitant c-NHEJ deficiencies enhance PARPi-mediated synthetic lethality in HR-deficient cells [18].
FDA-approved PARPi has been administered to patients with BRCA1/2-mutated cancers such as breast and ovarian carcinomas [19,20,21,22]. Although leukemia has not been recognized as a typical BRCA1/2-mutated cancer, our group and others have recently reported that certain types of leukemias and other hematopoietic malignancies display HR with/without concomitant c-NHEJ functional deficiency caused by leukemia-inducing mutations [18,23,24,25,26]. In addition, HR and/or c-NHEJ deficiency could be induced by the treatment of leukemia/solid tumors with the tyrosine kinase inhibitors (TKi) against the cancer-driven oncogenic tyrosine kinases (e.g., FLT3(ITD), JAK2(V617F), c-KIT(N822K), IGF-1R, EGFR). Therefore, oncogenic tyrosine kinase (OTK)-driven malignancies can effectively respond to PARPi after the inhibition of OTK [27,28,29,30,31].
Unfortunately, the resistance to PARPi has been reported in both preclinical research and clinical treatment. The acquired (time-dependent) resistant phenotype has been mediated by the functional recovery of HR, the abrogation of PARP1 expression, alterations in DSB end resection, the prevention of the replication fork from degradation, the loss of replication gaps and the arising of alternative factors (e.g., RAD52, Polθ) [29,32,33,34]. Moreover, recent studies have shown that the resistance to PARPi could be pre-existing (time-independent), induced by certain oncogenes triggering malignant transformation and by the tumor microenvironment [35]. In this review, we discuss the pre-existing and acquired mechanisms of PARPi resistance and possible therapeutic solutions.
2. Synthetic Lethality in the Context of DNA Repair
Genomic instability is one of the classical hallmarks of cancer [2]. Among other reasons, it occurs when the activation of oncogenes and/or the inactivation of tumor suppressor genes induce the production of endogenous reactive oxygen species (ROS). This leads to the accumulation of oxidative DNA damage in cancer cells, which is accompanied by secondary DNA mutations (e.g., derived from the initial treatment with chemotherapeutic agents) [36]. As a result, mutations in certain genes may lead to the inhibition of processes, which could counteract the cancer cells propagation such as apoptosis, senescence or DNA checkpoint pathways. To survive under the elevated DNA damage rate, cancer cells modulate DNA repair pathways [4]. This not only provides a pro-survival effect but also reveals an “Achilles heel” of cancer cells that can be therapeutically exploited by an anticancer concept called synthetic lethality. In this theory, cell death caused by synthetic lethality is based on mutations in two genes encoding two parallel proteins/pathways that perform functionality in a compensational process required for cell survival. In such case, the functional maintenance of one gene enables a compensatory response to prevent cell death when the other parallel protein/pathway is inactive due to gene mutations [37]. Historically, the first study describing the interplay between two parallel genes was conducted in Drosophila melanogaster in 1922 [38], and the term “synthetic lethality” was introduced in 1946 when the same result was found in Drosophila pseudoobscura [39]. Since then, the concept of synthetic lethality has been developed in the field of anti-cancer therapy.
In cancer cells, the accumulation of DNA damage can result in a mutation in one gene from the synthetically lethal pair, therefore putting the dependence of cell survival strictly on the second gene from this pair. In this instance, the mutation-free second gene becomes a weakness of cancer cells that should be therapeutically targeted to trigger synthetic lethality. This would eliminate malignant cells without an unwanted cytotoxic effect on normal cells. As DNA repair pathways are altered and/or enhanced to support the survival of cancers cell under high levels of spontaneous and/or drug-induced DNA damage, the synthetic lethality-based targeting of DNA repair pathways is a promising approach to develop a novel therapeutic strategy for anti-cancer treatment.
Among all types of DNA damage, DSBs are one of the most lethal types of DNA lesions in cells. Several exogenous (e.g., γ-irradiation, anti-cancer drugs) and endogenous metabolic factors (e.g., ROS) can cause DSBs as a consequence of direct damage or a stalled replication fork [40]. Once generated, DSBs should be effectively repaired; if not, they will result in cell death [3]. Altogether, redundant DSB repair mechanisms represent a perfect opportunity for the implementation of the concept of synthetic lethality in anti-cancer therapy.
In general, DSB repair consists of two major pathways: HR and NHEJ [5] (Figure 1). HR is considered an accurate DSB repair pathway because it depends on a sister chromatid of a cell in the S/G2 phases of a cell cycle as a template for DNA synthesis and the repair of a DSB [7]. Therefore, HR is capable of repairing DSBs, mostly in proliferating cells [6]. The HR repair pathway is comprehensively modulated by proteins encoded by two classical tumor suppressor genes, BRCA1 and BRCA2, followed by the recruitment of RAD51, its paralogs (including RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3) and RAD54 [6,41,42] to recognize a homologous DNA template and perform the strand invasion to repair DSBs. Mutations in BRCA1/2 have been found in a wide range of cancers, leading to the inactivation of the HR pathway. In such cases, cancer cells usually employ alternative DNA repair pathway(s) to survive.
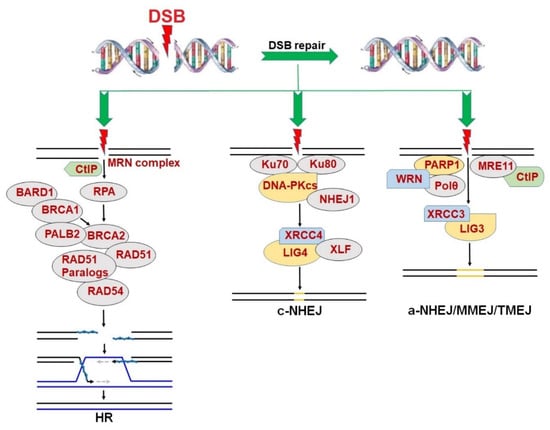
Figure 1.
DNA double strand break (DSB) repair pathways including homologous recombination (HR), canonical non-homologous end joining (c-NHEJ) and alternative non-homologous end joining (a-NHEJ)/microhomology-mediated end joining (MMEJ)/DNA polymerase theta (Polθ)-mediated end joining (TMEJ).
On the other hand, NHEJ is an error-prone DSB repair mechanism. NHEJ can be divided into two pathways including the c-NHEJ and a-NHEJ, also known as MMEJ or TMEJ [8,9,10,43]. Both pathways allow for DSB repair in cells throughout all phases of cell cycle [6]. Unlike HR, the DSBs in both quiescent and proliferating cells can be repaired by c-NHEJ, with joint participation of Ku70/80 protein, catalyzing kinase subunit of DNA-dependent protein kinase (DNA-PKcs), NHEJ1 and the complex of XRCC4/LIG4/XLF [44].
Although a-NHEJ also performs an error-prone DSB repair, it is more likely to generate more extensive alterations in DNA sequence than c-NHEJ, leading to an increased risk of chromosomal translocations [45]. The a-NHEJ pathway is critically regulated by PARP1, serving a backup role of both HR and c-NHEJ when one/both of the two DSB repair pathways is/are not fully functional [9]. In fact, PARP1 co-operates with MRE11, Polθ, and WRN helicase to promote DSB end re-section process, eventually leading to DNA ligation (which depends on catalytic activities of LIG1 and/or LIG3) [46,47].
In addition, PARP1 has been reported to mediate DNA single-strand break (SSB) repair by recruiting proteins of the base excision repair (BER) pathway [48]. In this pathway, PARP1 conducts a search throughout a single DNA strand until the enzyme recognizes an SSB and binds to the break. At this point, the elevated activity of a process named PARylation (catalyzed by the PARP enzymes family) occurs. This includes covalent bonds of long ADP-ribose to generate poly ADP-ribose (PAR) on PARP1 [8], whereas essential proteins comprising XRCC1 complex, DNA polymerase β and LIG3 are recruited to repair the SSB. Notably, if the SSB remains unrepaired during DNA replication, it will cause transcriptional arrest, leading to the formation of a lethal DSB [49]. Therefore, PARP1-dependent SSB repair is considered very important for the survival of HR-deficient proliferating cells because it prevents the conversion of an SSB to a DSB during the DNA replication. This establishes a rationale for the therapeutic application of PARPi-induced synthetic lethality in BRCA1/2-mutated/deficient cancer cells.
3. PARPi-Induced Synthetic Lethality in BRCA1/2-Mutated Cancers
The usage of PARPi, which predominantly blocks the activity of PARP1, PARP2 and PARP3, is a well-established example of synthetic lethality-based therapy in BRCA1/2-mutated cancers with a limited toxicity towards normal cells and tissues [12,13,14,15]. In fact, the effectiveness of PARPi in the BRCA1/2-mutated breast/ovarian tumors has initiated an era of personalized medicine with the utilization of PARPi [50,51,52]. Mechanistically, mutations in BRCA1/2 inactivate the HR pathway, and in order to survive, BRCA1/2-mutated cancer cells require the activity of PARP1 in BER and/or a-NHEJ, to prevent the formation of DSBs from unrepaired SSBs during DNA replication. Therefore, the inhibition of PARP1 by PARPi results in stalled replication forks and the accumulation of lethal DSBs, leading to cell death.
Recently, another mechanism has been proposed to regulate PARPi-triggered synthetic lethality in BRCA1/2-mutated cells: the single-strand DNA replication gaps [53,54]. Enhanced replication gaps in BRCA1/2-deficient cells were coupled with PARPi sensitivity. Besides working effectively in BRCA1/2-mutated cancers, PARPi-mediated synthetic lethality is capable of sensitizing c-NHEJ-deficient cancer cells. For example, the downregulation of LIG4 (involved in the c-NHEJ pathway to perform DNA ligation) induced the sensitivity of melanoma cells to PARPi (olaparib), without a cytotoxic effect on normal melanocytes [55].
Initially, the major mechanism of the efficiency of PARPi has been associated with the interference of the accessibility of NAD+ to the PARP1 catalytic domain, leading to the inactivation of the PARylation process and the inhibition of BER and/or a-NHEJ [56]. However, recent studies have shown that the inhibition of the catalytic activity of PARPs is not the only mechanism triggering synthetic lethality [57]. Additionally, PARPi can cause the trapping of PARP1 (and probably also PARP2), resulting in DNA replication, transcriptional arrest and the accumulation of DSBs. The magnitude of synthetic lethality triggered by PARPi corresponds to their capability of PARP1 entrapment [58]. PARPi talazoparib (also known as BMN673) has been reported to be approximately 20–200 times more efficient than previous versions of PARPi, such as olaparib [59]. The elevated efficacy of talazoparib results from its enhanced PARP1-trapping capacity, thus making talazoparib one of the best PARP-trapping agents among currently available PARPi [60].
4. PARPi in Clinical Trials of BRCA1/2-Mutated Cancers
Olaparib (commercial name—Lynparza®) is the first pharmacological PARPi that has been administered in clinical trials. Until now, olaparib is the most common PARPi used in BRCA1/2-deficient cancers. Historically, olaparib became the first PARPi approved by the FDA in December 2014, based on its significant efficacy in the treatment of relapsed ovarian cancer individuals with BRCA1/2 mutations [61]. In August 2017, olaparib obtained the second approval from the FDA as an extensive therapy for patients with recurrent fallopian tube, peritoneal or epithelial ovarian cancer who have achieved partial or complete remission after the systematic standard chemotherapy [62,63]. Additionally, the potential of olaparib in the anticancer therapy has been extended in January 2018, when the FDA licensed the PARPi as a therapeutic strategy for germline BRCA1/2-mutated metastatic breast cancer patients who previously received chemotherapy [64]. This marked olaparib as the first FDA-approved compound working effectively in individuals with hereditary breast cancer. Besides the trials in BRCA1/2-mutated breast and ovarian cancer, olaparib was also granted approval by the FDA in different solid tumors. This includes BRCA1/2-mutated metastatic pancreatic cancer in 2019 [65], fallopian and primary peritoneal carcinoma in a combinational intervention with bevacizumab [66] and HR-deficient metastatic castration-resistant prostate cancer in 2020 [67].
In addition, two other PARP inhibitors, rucaparib and niraparib, which also target polymerase enzymatic activity, have obtained approval for clinical trials. In detail, the FDA accepted the clinical trials of rucaparib for BRCA1/2-mutated advanced ovarian carcinomas undergoing multiple chemotherapy treatments in 2016 [68], reoccurring ovarian, fallopian and primary peritoneal carcinoma without BRCA1/2 mutational status in 2018 [69] and BRCA1/2-mutated metastatic castration-resistant prostate cancer in 2020 [70]. Meanwhile, niraparib achieved the approval of the FDA for reoccurring ovarian, fallopian and primary peritoneal carcinoma with complete or partial chemotherapeutic response in 2017 [71], HR-deficient reoccurring ovarian, fallopian and primary peritoneal carcinoma without chemotherapeutic response in 2019 [72] and advanced ovarian carcinomas with complete or partial chemotherapeutic response in 2020 [73].
On the other hand, based on the significant PARP1 trapping capacity, talazoparib has been clinically employed in breast cancer patients with germline mutations of BRCA1/2 and other types of cancer that contain impaired DNA damage responses [22,74]. For example, phase III clinical trials of talazoparib demonstrated the increased overall survival rate of metastatic breast cancer patients [75], and it has been approved by the FDA since 2018 [76]. Besides talazoparib, another orally available PARPi (veliparib) is currently undergoing clinical trials [77]. It shows the best selectivity against PARP1/2/3 catalysis, though this PARPi exhibits a limited efficacy of PARP1 trapping [78]. This demonstrated that PARPi, which exerts a more potent and selective inhibitory effect on the PARylation process, is also capable of entering clinical trials.
5. Therapeutic Potential of PARPi in Hematopoietic Malignancies
Many recent studies, including ours, have shown that even if BRCA1/2 mutations are rarely detected in leukemias, PARPi-induced synthetic lethality can be effectively exploited in BRCA1/2-deficient hematopoietic malignant cells. Using a comprehensive Gene Expression and Mutation Analysis strategy, we were able to identify acute myeloid leukemias/acute lymphoblastic leukemias (AMLs/ALLs) that displayed HR and/or c-NHEJ deficiency and were also sensitive to PARPi [18]. These DSB repair defects were detected by direct measurements of the expression of HR and c-NHEJ genes by mRNA microarrays, real-time PCR and/or flow cytometry. In addition, genetic alterations inducing hematopoietic malignancies, such as oncogenes driving myeloid and lymphoid malignancies, including AML1-ETO (also known as RUNX1-RUNX1T1), BCR-ABL1, PML-RARα, TCF3-HLF, IDH1/2mut and IGH-MYC, and loss-of-function mutations in tumor suppressor genes (e.g., TET2, WT1), can lead to the deregulation of HR and/or c-NHEJ activity, thus rendering cells susceptible to a synthetically lethal effect triggered by PARPi [23,24,25,79,80,81,82,83,84,85,86,87,88,89,90,91] (Figure 2A and Table 1). In addition, mutations in the core cohesion complex gene STAG2 (Stromal Antigen 2) induce DNA damage, stalled replication forks and a high genetic dependency on PARP1 in AML/myelodysplastic syndrome (MDS) cells. Therefore, those cells are sensitive to PARPi talazoparib both in vitro and in vivo; however, the mechanism remains unexplored [92].
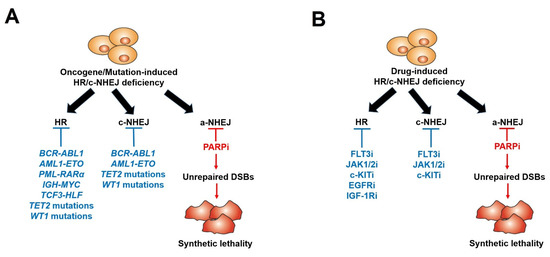
Figure 2.
Scheme of PARP inhibitors administered in hematopoietic malignancies and other tumors displaying HR/c-NHEJ deficiency induced by oncogenes/mutations (A) and tyrosine kinase inhibitors (B).

Table 1.
Oncogenes/Mutations inducing HR/c-NHEJ deficiency.
Furthermore, we and others described that those malignant hematopoietic cells expressing oncogenic tyrosine kinases (OTK) (e.g., BCR-ABL1, FLT3(ITD), JAK2(V617F)) spontaneously accumulate high levels of oxidative DNA damage and DSBs due to the increase in ROS production [93,94,95]. However, OTK-positive cells were capable of escaping from the cytotoxic effect of DSBs due to enhanced/modulated DSB repair. Remarkably, the inhibition of these OTKs by FDA-approved specific tyrosine kinase inhibitors (TKi) (JAK1/2 inhibitor ruxolitinib, FLT3 inhibitor quizartinib, ABL1 inhibitor imatinib) resulted in acute HR/c-NHEJ deficiency (due to the downregulation of BRCA1, BRCA2, RAD51 and/or LIG4) and the sensitivity to PARPi (Figure 2B) (Table 2). Therefore, the combination of TKi and PARPi was capable of eradicating both proliferating and quiescent malignant hematopoietic stem and progenitor cells [18,27,28,29,79]. All these promising results have made up a rationale for clinical trials with PARPi in patients with leukemias and other related hematopoietic malignancies [26].

Table 2.
Therapeutic drugs inducing HR/c-NHEJ deficiency.
As a result, the first trial (NCT01399840) of PARPi in hematopoietic malignancies began in 2014, when the efficacy of talazoparib was tested in 25 AML/MDS patients and 8 other individuals with chronic lymphocytic leukemia (CLL) and mantle cell lymphoma [96]. During 2017, there were three phase I clinical trials registered, including a combinational therapy of veliparib + temozolomide in 48 patients with relapsed/refractory AML (NCT01139970) [97], veliparib combination with topotecan and carboplatin in a clinical study of 99 patients with relapsed/refractory AML, chronic myelomonocytic leukemia or aggressive myeloproliferative neoplasms (NCT00588991) [98], and olaparib in 15 patients with relapsed CLL, T-prolymphocytic leukemia or mantle cell lymphoma [99]. In 2021, the results of a clinical trial (NCT04326023) examining the efficiency of PARP inhibitors, including olaparib, rucaparib, niraparib, talazoparib and veliparib, in 178 patients with MDS and AML were reported [100]. Although 104 in 178 MDS/AML participants were recorded with positive outcomes, PARPi increased the risk of MDS/AML in adults over 18 [100]. Additionally, a phase I clinical trial of the DNA methyltransferase inhibitor decitabine and talazoparib has been demonstrated in 25 patients with relapsed/refractory AML [101].
6. Acquired Resistance to PARPi-Mediated Synthetic Lethality
Despite the strong potency of PARPi in DSB repair-defective cancer cells, the resistance to PARPi-induced synthetic lethality has been reported in BRCA1/2-deficient and HRD tumor cells (Table 3). The most common mechanism responsible for the development of resistance to PARPi is the restoration of HR repair activity in BRCA1/2-mutated cancer cells [102]. In detail, the secondary mutations in BRCA1/2 were associated with the abrogation of the chain terminator/frameshift resulting from original mutations, leading to the restoration of the full-length BRCA1/2 open reading frame. Therefore, the active BRCA1/BRCA2 protein expression is recovered, thus restarting the fully functional HR pathway [34,103,104]. Besides the resistance mediated by the additional mutations in BRCA1/2, the reduction/loss of methylation of BRCA1 promoter has been suggested, which restores the expression of BRCA1, thereby leading to resistance to PARPi. Indeed, PARPi-sensitive primary breast cancer cells exhibited elevated BRCA1 promoter methylation, which was associated with impaired BRCA1 expression. Meanwhile, decreased promoter methylation and proficient BRCA1 expression were observed in individuals who did not respond to PARPi treatment [105]. Moreover, in a subset of BRCA1-deficient triple-negative breast cancer cells showing refractory against PARPi, a reduced expression of EMI1 impairs EMI-dependent RAD51 degradation, restoring HR repair activity [106]. Additionally, EMSY is a negative regulator of type I interferon response and also counteracts the HR repair pathway, and KEAP1 targets EMSY for ubiquitin-mediated degradation. Therefore, the overexpression of KEAP1 destabilizes EMSY, leading to HR restoration and resistance to PARPi. On the contrary, the inactivation of KEAP1 due to gene mutations causes non-small cell lung cancer to become sensitive to PARPi thanks to the stabilization of EMSY and HR deficiency [107].

Table 3.
Mechanisms of pre-existing and acquired resistance to PARPi.
Another mechanism of PARPi resistance in BRCA1/2-deficient tumor cells is due to the downregulation of PARP1 expression, thereby resulting in PARylation-independent cell proliferation and/or the inefficiency of PARP1 trapping by PARPi. In fact, the expression level of PARP1 was remarkably reduced in colorectal carcinoma HCT116 clones that were refractory against PARPi and temozolomide [109]. Moreover, in patients with ovarian cancer, a missense mutation (1771C>T) in PARP1 was described to induce the de novo attenuation of sensitivity to PARPi [110]. Additionally, the overexpression of P-glycoprotein efflux pumps has been shown to result in resistance to PARPi olaparib in a long-term treatment in murine triple-negative mammary carcinomas [15].
A third mechanism related to the acquired PARPi resistance is associated with the alteration of DSB end resection. In this circumstance, via a complex of 53BP1-RIF1, the 53BP1-mediated suppression of DSB end resection leads to enhanced c-NHEJ activity, reducing the dependency on PARP1-dependent a-NHEJ and thus causing PARPi resistance [111,112]. Hence, the inhibition of 53BP1 in BRCA1-null murine embryonic stem cells promotes DSB end resection to restrict c-NHEJ, thus sensitizing cells for PARPi [132]. On the other hand, the activity of 53BP1 is also required to maintain the efficiency of PARPi olaparib in BRCA1-mutated breast cancer cells [115]. In these cells, the somatic loss of 53BP1 leads to olaparib resistance due to the partial restoration of HR repair activity, decreasing the response of BRCA1-deficient mouse mammary tumors to the inhibitor [113,114]. Additionally, the blockage of 53BP1 localization to DSBs also contributes to the inactivation of PARPi in BRCA1-deficient tumors, mediated by the increased expression of TIRR [116]. Moreover, the inactivation of SHLD1/2/3 complex and the decreased expression of other proteins in DSB end resection, including RIF1 and REV7, restore the functional activity of HR, diminishing the efficacy of PARPi [117,118,119]. In the same mechanistic manner, TRIP13 is a negative regulator of the SHLD1/2/3 complex by dissociating REV7-SHLD1/2/3 to promote HR [120,121]. Therefore, the amplification of TRIP13 induces resistance to PARPi in BRCA1/2-mutated cancers [122]. Furthermore, another DSB end resection-associated mechanism is acquired by the decrease in DNLL1, increasing the DSB end resection potential and recovering HR functionality in BRCA1-mutated cells [123]. In addition, it has been recently shown that the CHAMP1-POGZ heterochromatin complex counteracts the 53BP1 inhibitory effect against HR and binds directly to REV7 to repress the REV7-SHLD1/2/3 complex, elevating HR repair activity via promoting DSB end-resection [124,125]. Hence, the overexpression of CHAMP1 confers PARPi resistance; meanwhile, the depletion of CHAMP1 restores functional 53BP1 and the complex of REV7-SHLD1/2/3 to restrict DSB end resection, leading to PARPi sensitivity.
Besides functioning in the HR pathway, BRCA1 and BRCA2 have been reported to play a role in maintenance of the integrity of replication forks [126,133]. The acquired resistance to PARPi in BRCA1- and BRCA2-deficient cells can be caused by the decreased expression of the MRE11 and MUS81 nucleases via the loss of EZH2, leading to a protective response toward replication forks [126,127]. Furthermore, the deficient activities of PTIP and SLFN11 maintain the stability of replication forks by the prevention of fork degradation, decreasing the efficiency of PARPi [128,129].
In addition, going beyond mechanisms of acquired PARPi resistance based on the restoration of HR or replication fork protection, a recent study has reported that the resistance can be induced by a reduction/loss of DNA replication gaps via the restoration of Okazaki fragment processing (OFP) [53]. Basically, replication gap has been reported as a major determinant of PARPi response in BRCA1/2-deficient cells [53]. In general, deficiency in BRCA1/2 extends replication gaps, leading to the PARPi sensitivity of the cells. However, in a cell model with a simultaneous knock-out of BRCA1 and 53BP1, the OFP was recovered by the restoration of XRCC1-LIG3, leading to the decrease/depletion of replication gaps and, eventually, to the resistance to PARPi. On the contrary, a double knock-out of 53BP1 and LIG3 re-sensitizes BRCA1-mutated cells to PARPi [53].
Additionally, the arising of alternative factors in DSB repair pathways is an important mechanism of acquired resistance to PARPi. For example, RAD52-mediated DNA repair can play a role as a backup of HR when BRCA1/2-RAD51-RAD54 are not fully functional [134]. At a DSB, RAD52 stimulates DNA pairing (a pivotal step of D-loop formation) and single-strand DNA annealing to regions of homology (>30 bps) [135]. The annealed DNA is then processed by nucleases (e.g., ERCC1/XPF) to generate an error-prone repair [136,137]. Therefore, the overexpression of RAD52 can result in the inefficiency of PARPi in HRD cells. Our group has successfully demonstrated that the simultaneous targeting of PARP1 and RAD52 induces “dual synthetic lethality” in BRCA1/2-deficient cancer cells [33].
Besides RAD52, another alternative factor in DSB repair which is widely being studied is Polθ, encoded by POLQ. Polθ facilitates MMEJ, a branch of a-NHEJ [11], and Polθ inactivation causes synthetic lethality in HRD cells [138,139,140]. The over-activation of Polθ also confers resistance to radiation and chemotherapies including PARPi [138,140,141,142]. Therefore, the pharmacological inhibition of Polθ is being developed to overcome the resistance to PARPi in a cohort of HRD cancers [130,131].
7. Pre-Existing Resistance to PARPi: The Bone Marrow Microenvironment
Hematopoietic stem cells (HSCs) originate from a specific hematopoietic tissue: the bone marrow (BM). Anatomically, the bone marrow (BM) is established by various types of stromal cells, including mesenchymal stromal/stem cells and endothelial cells, together with sympathetic nerve-related cells, macrophages, osteoblasts, fibroblasts, megakaryocytes and others, all functioning under the hypoxic conditions. The term “HSC niche” was introduced in 1978 by Ray Schofield to initially elicit the importance of hematopoietic tissues, such as BM and the spleen, for HSCs’ biology [143]. This has led to numerous studies being conducted during the past four decades to validate the function of the bone marrow microenvironment (BMM) [144]. Overall, the BMM plays a significant role in the maintenance, self-renewal and differentiation of HSCs, while molecular interactions between HSCs and cellular components of the BMM are set to maintain the balance between the self-renewal and differentiation of HSCs [144].
While the BMM plays a key role in hematopoiesis, there is a reciprocal relationship between leukemia stem cells (LSCs) and the BMM. LSCs are responsible for the initiation of the disease, as well as the reprogramming of the BMM. Meanwhile, the BMM has been documented to provide essential factors for the maintenance and survival of LSCs [145,146]. Therefore, the protective feature of the BMM towards LSCs often renders malignant cells refractory against chemotherapeutic agents and TKi [147,148,149]. Moreover, in primary xenograft conditions, the CD34+ leukemia cells remodeled murine BM including bone marrow stromal cells, to promote an unfavorable microenvironment for normal HSCs but provide a protective and pro-survival milieu for LSCs [150,151,152]. Altogether, BMM can be a potential weakness of LSCs that can be therapeutically exploited to eliminate hematopoietic malignant cells in the BMM.
Recently, we discovered a pre-existing mechanism of resistance to PARPi in hematopoietic malignant cells residing in the BMM [35]. HR and/or c-NHEJ-deficient hematological malignancies, which are sensitive to PARPi-mediated synthetic lethality in conditions mimicking the peripheral blood microenvironment (PBM), become resistant to PARPi in conditions mimicking the BMM. We show that transforming growth factor beta 1 (TGF-β1) produced by bone marrow stromal cells activates a hypoxia-induced TGF-β receptor (TGFβR) kinase–SMAD2/3 canonical pathway in leukemic cells to promote DSBs repair (Figure 3). The TGFβR kinase inhibitor did not alter the sensitivity of leukemia cells to PARPi in the PBM, indicating that the regulation of the TGFβR-SMAD3 signaling in DSB repair activities is exclusive in the BMM. Mechanistically, TGFβR kinase signaling stimulates the repair of DSBs in leukemia cells in the BMM via the upregulation of BRCA1/2, ATM, DNA-PKcs and LIG4. This finding suggests potential clinical applications of TGFβR kinase inhibitors in PARPi-mediated interventions against hematopoietic malignancies. In fact, the inhibition of TGFβR kinase by galunisertib, which has been applied in clinical trials of several cancers [153,154,155,156,157,158], restored the PARPi sensitivity of leukemia cells in the BMM. Therefore, we postulate that the addition of galunisertib to PARPi treatment should improve the therapeutic outcome.
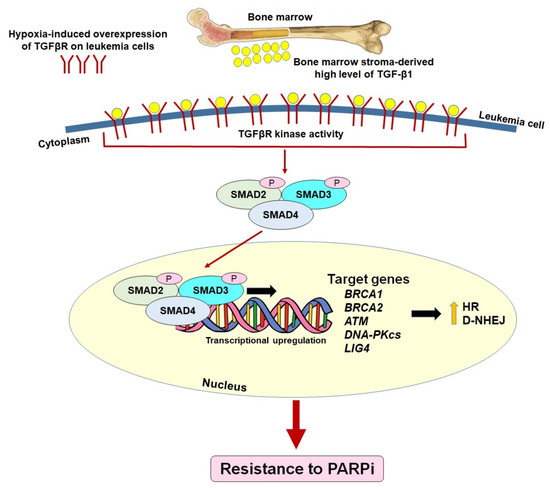
Figure 3.
Pre-existing mechanism of PARP inhibitor resistance by the restoration of HR/c-NHEJ in HR/c-NHEJ-deficient leukemias via the activation of TGF-β1—TGFβR—SMAD2/3 signaling in the hypoxic bone marrow microenvironment.
8. Pre-Existing Resistance to PARPi: Tumor-Inducing Mutations
Our group has shown that OTK c-KIT (N822K mutant in c-KIT receptor tyrosine kinase) induces PARPi resistance in BRCA1/2-deficient AML1-ETO-positive AML [18,29]. The inhibition of the oncogenic c-KIT kinase by TKi avapritinib re-sensitized leukemia cells to PARPi. Furthermore, somatic variants in DNMT3A are identified as mutations in hematological malignancies that affect the epigenetic regulation of DNA methylation. They often co-occur with activating mutations in OTKs such as FLT3ITD, BCR-ABL1, JAK2V617F and MPLW515L. We reported that DNMT3A-deficient cells favor HR/c-NHEJ, owing to the downregulation of PARP1 and the reduction of a-NHEJ [85]. In addition, a recent study has reported that DNMT3A-mutated leukemia cells exhibit impaired PARP1 recruitment, p53 activation and increased DNA damage after being challenged by replication stress-induced medications [108]. Consequently, DNMT3A-deficient leukemia cells are resistant to PARPi. However, the disruption of TET2 dioxygenase activity and/or the TET2–Wilms tumor 1 (WT1) binding ability are responsible for HR/c-NHEJ repair defects, the restoration of a-NHEJ activity and the sensitivity to PARPi. Therefore, TET2 dioxygenase inhibitors should be explored therapeutically to reverse PARPi resistance in DNMT3A-deficient leukemias. Besides the recent findings of our group, other researchers also discovered that the activation of tyrosine kinase activities of EGFR and IGF-1R stimulates the activity of HR by increasing BRCA1 and RAD51, respectively, causing resistance to PARPi [30,31]. Therefore, using the tyrosine kinase inhibition of EGFR and IGF-1R induces the “BRCAness” and HRD phenotypes in breast and ovarian cancers, leading to PARPi sensitivity in the cancer cells.
9. Conclusions
In light of the ever-increasing list of mechanistic pathways responsible for resistance to PARPi, the pharmacological inhibition of pre-existing and acquired PARPi resistance has to be achieved in order to improve therapeutic efficiency of PARPi-mediated synthetic lethality. For example, pre-existing PARPi resistance in leukemia cells could be addressed by combining inhibitors of PARP and TGFβR kinase (to re-sensitize leukemia cells in the BMM [35]), inhibitors of PARP and TET2 dioxygenase (to re-sensitize leukemias carrying DNMT3A mutations [85]) and inhibitors of PARP and OTK (FLT3(ITD), JAK2(V617F), c-KIT(N822K), EGFR, IGF-1R [27,28,29,30,31]). Acquired PARPi resistance could be prevented by the more aggressive “dual synthetic lethality” approach (simultaneously targeting PARP1 and RAD52 [33]), which eliminates more tumors cells in a shorter time, thus reducing the chances of the time-dependent emergence of resistant clones. Moreover, if PARPi resistance emerges, these clones could be eliminated by the inhibition of another DNA repair mechanism, e.g., Polθ-mediated TMEJ [130,131,159].
Author Contributions
Conceptualization, Writing—original and final draft preparation, B.V.L.; Review and editing, P.P.-B.; Review and editing, Supervision, T.S. and K.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by NIH R01CA186238, R01CA237286, R01CA244044, R01CA244179, and R01CA247707; and the Leukemia and Lymphoma Society Translational Research Program awards 6565-19 and 6628-21 (all to T.S.). B.V.L. has been supported by the European Union’s Horizon 2020 Research and Innovation Program under the Marie Sklodowska-Curie grant agreement no. 665735 and by the funding from Polish Ministry of Science and Higher Education funds for the implementation of international projects, 2016–2020 (to K.P.). P.P.-B. has been supported by the research grant 2018/30/M/NZ3/00274 from the Polish National Science Centre. The APC was funded by NIH grant R01CA244044.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kaelin, W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 2005, 5, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Dudas, A.; Chovanec, M. DNA double-strand break repair by homologous recombination. Mutat. Res. 2004, 566, 131–167. [Google Scholar] [CrossRef]
- Yang, H.; Zhong, Y.; Peng, C.; Chen, J.Q.; Tian, D. Important role of indels in somatic mutations of human cancer genes. BMC Med. Genet. 2010, 11, 128. [Google Scholar] [CrossRef]
- Featherstone, C.; Jackson, S.P. DNA double-strand break repair. Curr. Biol. CB 1999, 9, R759–R761. [Google Scholar] [CrossRef]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef]
- Humphryes, N.; Hochwagen, A. A non-sister act: Recombination template choice during meiosis. Exp. Cell Res. 2014, 329, 53–60. [Google Scholar] [CrossRef]
- Caldecott, K.W. Mammalian single-strand break repair: Mechanisms and links with chromatin. DNA Repair 2007, 6, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Iliakis, G. Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Transl. Cancer Res. 2013, 2, 163–177. [Google Scholar]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.-H.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef] [PubMed]
- Kent, T.; Chandramouly, G.; McDevitt, S.M.; Ozdemir, A.Y.; Pomerantz, R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015, 22, 230–237. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Yap, T.A.; Sandhu, S.K.; Carden, C.P.; de Bono, J.S. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J. Clin. 2011, 61, 31–49. [Google Scholar] [CrossRef]
- Rottenberg, S.; Jaspers, J.E.; Kersbergen, A.; van der Burg, E.; Nygren, A.O.; Zander, S.A.; Derksen, P.W.; de Bruin, M.; Zevenhoven, J.; Lau, A.; et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. USA 2008, 105, 17079–17084. [Google Scholar] [CrossRef] [PubMed]
- Fok, J.H.L.; Ramos-Montoya, A.; Vazquez-Chantada, M.; Wijnhoven, P.W.G.; Follia, V.; James, N.; Farrington, P.M.; Karmokar, A.; Willis, S.E.; Cairns, J.; et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 2019, 10, 5065. [Google Scholar] [CrossRef]
- Chan, C.Y.; Tan, K.V.; Cornelissen, B. PARP Inhibitors in Cancer Diagnosis and Therapy. Clin. Cancer Res. 2021, 27, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Nieborowska-Skorska, M.; Sullivan, K.; Dasgupta, Y.; Podszywalow-Bartnicka, P.; Hoser, G.; Maifrede, S.; Martinez, E.; Di Marcantonio, D.; Bolton-Gillespie, E.; Cramer-Morales, K.; et al. Gene expression and mutation-guided synthetic lethality eradicates proliferating and quiescent leukemia cells. J. Clin. Investig. 2017, 127, 2392–2406. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O'Connor, M.J.; et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Gunjur, A. Talazoparib for BRCA-mutated advanced breast cancer. Lancet Oncol. 2018, 19, e511. [Google Scholar] [CrossRef]
- Hurvitz, S.A.; Gonçalves, A.; Rugo, H.S.; Lee, K.H.; Fehrenbacher, L.; Mina, L.A.; Diab, S.; Blum, J.L.; Chakrabarti, J.; Elmeliegy, M.; et al. Talazoparib in Patients with a Germline BRCA-Mutated Advanced Breast Cancer: Detailed Safety Analyses from the Phase III EMBRACA Trial. Oncol. 2019, 25, e439–e450. [Google Scholar] [CrossRef] [PubMed]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Podszywalow-Bartnicka, P.; Wolczyk, M.; Kusio-Kobialka, M.; Wolanin, K.; Skowronek, K.; Nieborowska-Skorska, M.; Dasgupta, Y.; Skorski, T.; Piwocka, K. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle 2014, 13, 3727–3741. [Google Scholar] [CrossRef] [PubMed]
- Maifrede, S.; Martin, K.; Podszywalow-Bartnicka, P.; Sullivan-Reed, K.; Langer, S.K.; Nejati, R.; Dasgupta, Y.; Hulse, M.; Gritsyuk, D.; Nieborowska-Skorska, M.; et al. IGH/MYC Translocation Associates with BRCA2 Deficiency and Synthetic Lethality to PARP1 Inhibitors. Mol. Cancer Res. 2017, 15, 967–972. [Google Scholar] [CrossRef]
- Esposito, M.T.; Zhao, L.; Fung, T.K.; Rane, J.K.; Wilson, A.; Martin, N.; Gil, J.; Leung, A.Y.; Ashworth, A.; So, C.W. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat. Med. 2015, 21, 1481–1490. [Google Scholar] [CrossRef]
- Faraoni, I.; Giansanti, M.; Voso, M.T.; Lo-Coco, F.; Graziani, G. Targeting ADP-ribosylation by PARP inhibitors in acute myeloid leukaemia and related disorders. Biochem. Pharmacol. 2019, 167, 133–148. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Maifrede, S.; Dasgupta, Y.; Sullivan, K.; Flis, S.; Le, B.V.; Solecka, M.; Belyaeva, E.A.; Kubovcakova, L.; Nawrocki, M.; et al. Ruxolitinib-induced defects in DNA repair cause sensitivity to PARP inhibitors in myeloproliferative neoplasms. Blood 2017, 130, 2848–2859. [Google Scholar] [CrossRef]
- Maifrede, S.; Nieborowska-Skorska, M.; Sullivan-Reed, K.; Dasgupta, Y.; Podszywalow-Bartnicka, P.; Le, B.V.; Solecka, M.; Lian, Z.; Belyaeva, E.A.; Nersesyan, A.; et al. Tyrosine kinase inhibitor-induced defects in DNA repair sensitize FLT3(ITD)-positive leukemia cells to PARP1 inhibitors. Blood 2018, 132, 67–77. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Paietta, E.M.; Levine, R.L.; Fernandez, H.F.; Tallman, M.S.; Litzow, M.R.; Skorski, T. Inhibition of the mutated c-KIT kinase in AML1-ETO-positive leukemia cells restores sensitivity to PARP inhibitor. Blood Adv. 2019, 3, 4050–4054. [Google Scholar] [CrossRef]
- Nowsheen, S.; Cooper, T.; Stanley, J.A.; Yang, E.S. Synthetic Lethal Interactions between EGFR and PARP Inhibition in Human Triple Negative Breast Cancer Cells. PLoS ONE 2012, 7, e46614. [Google Scholar] [CrossRef] [PubMed]
- Amin, O.; Beauchamp, M.-C.; Nader, P.A.; Laskov, I.; Iqbal, S.; Philip, C.-A.; Yasmeen, A.; Gotlieb, W.H. Suppression of Homologous Recombination by insulin-like growth factor-1 inhibition sensitizes cancer cells to PARP inhibitors. BMC Cancer 2015, 15, 817. [Google Scholar] [CrossRef] [PubMed]
- D'Andrea, A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Sullivan-Reed, K.; Bolton-Gillespie, E.; Dasgupta, Y.; Langer, S.; Siciliano, M.; Nieborowska-Skorska, M.; Hanamshet, K.; Belyaeva, E.A.; Bernhardy, A.J.; Lee, J.; et al. Simultaneous Targeting of PARP1 and RAD52 Triggers Dual Synthetic Lethality in BRCA-Deficient Tumor Cells. Cell Rep. 2018, 23, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Sakai, W.; Swisher, E.M.; Karlan, B.Y.; Agarwal, M.K.; Higgins, J.; Friedman, C.; Villegas, E.; Jacquemont, C.; Farrugia, D.J.; Couch, F.J.; et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008, 451, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Le, B.V.; Podszywalow-Bartnicka, P.; Maifrede, S.; Sullivan-Reed, K.; Nieborowska-Skorska, M.; Golovine, K.; Yao, J.C.; Nejati, R.; Cai, K.Q.; Caruso, L.B.; et al. TGFβR-SMAD3 Signaling Induces Resistance to PARP Inhibitors in the Bone Marrow Microenvironment. Cell Rep. 2020, 33, 108221. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed]
- O'Neil, N.J.; Bailey, M.L.; Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Nijman, S.M. Synthetic lethality: General principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011, 585, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dobzhansky, T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics 1946, 31, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Grabarz, A.; Barascu, A.; Guirouilh-Barbat, J.; Lopez, B.S. Initiation of DNA double strand break repair: Signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am. J. Cancer Res. 2012, 2, 249–268. [Google Scholar] [PubMed]
- Mazin, A.V.; Bornarth, C.J.; Solinger, J.A.; Heyer, W.D.; Kowalczykowski, S.C. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 2000, 6, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Mazin, A.V.; Alexeev, A.A.; Kowalczykowski, S.C. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003, 278, 14029–14036. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.M.; Yanez, D.A.; Stark, J.M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015, 11, e1004943. [Google Scholar] [CrossRef] [PubMed]
- Boboila, C.; Jankovic, M.; Yan, C.T.; Wang, J.H.; Wesemann, D.R.; Zhang, T.; Fazeli, A.; Feldman, L.; Nussenzweig, A.; Nussenzweig, M.; et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA 2010, 107, 3034–3039. [Google Scholar] [CrossRef] [PubMed]
- Keijzers, G.; Maynard, S.; Shamanna, R.A.; Rasmussen, L.J.; Croteau, D.L.; Bohr, V.A. The role of RecQ helicases in non-homologous end-joining. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 463–472. [Google Scholar] [CrossRef]
- Parsons, J.L.; Dianova, II; Allinson, S. L.; Dianov, G.L. Poly(ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J. 2005, 272, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, V.J.; Rouleau, M.; Poirier, G.G. PARP-1, a determinant of cell survival in response to DNA damage. Exp. Hematol. 2003, 31, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Kuzminov, A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 2001, 98, 8241–8246. [Google Scholar] [CrossRef] [PubMed]
- Iglehart, J.D.; Silver, D.P. Synthetic lethality--a new direction in cancer-drug development. N. Engl. J. Med. 2009, 361, 189–191. [Google Scholar] [CrossRef] [PubMed]
- McCann, K.E.; Hurvitz, S.A. Advances in the use of PARP inhibitor therapy for breast cancer. Drugs Context 2018, 7, 212540. [Google Scholar] [CrossRef] [PubMed]
- Mittica, G.; Ghisoni, E.; Giannone, G.; Genta, S.; Aglietta, M.; Sapino, A.; Valabrega, G. PARP Inhibitors in Ovarian Cancer. Recent Pat. Anticancer Drug Discov. 2018, 13, 392–410. [Google Scholar] [CrossRef] [PubMed]
- Cong, K.; Peng, M.; Kousholt, A.N.; Lee, W.T.C.; Lee, S.; Nayak, S.; Krais, J.; VanderVere-Carozza, P.S.; Pawelczak, K.S.; Calvo, J.; et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell 2021, 81, 3128–3144.e3127. [Google Scholar] [CrossRef] [PubMed]
- Panzarino, N.J.; Krais, J.J.; Cong, K.; Peng, M.; Mosqueda, M.; Nayak, S.U.; Bond, S.M.; Calvo, J.A.; Doshi, M.B.; Bere, M.; et al. Replication Gaps Underlie BRCA Deficiency and Therapy Response. Cancer Res. 2021, 81, 1388–1397. [Google Scholar] [CrossRef]
- Czyz, M.; Toma, M.; Gajos-Michniewicz, A.; Majchrzak, K.; Hoser, G.; Szemraj, J.; Nieborowska-Skorska, M.; Cheng, P.; Gritsyuk, D.; Levesque, M.; et al. PARP1 inhibitor olaparib (Lynparza) exerts synthetic lethal effect against ligase 4-deficient melanomas. Oncotarget 2016, 7, 75551–75560. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J.; Szabo, C. Therapeutic applications of PARP inhibitors: Anticancer therapy and beyond. Mol. Aspects Med. 2013, 34, 1217–1256. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Huang, S.Y.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, T.A.; Ainsworth, W.B.; Ellis, P.A.; Donawho, C.K.; DiGiammarino, E.L.; Panchal, S.C.; Abraham, V.C.; Algire, M.A.; Shi, Y.; Olson, A.M.; et al. PARP1 Trapping by PARP Inhibitors Drives Cytotoxicity in Both Cancer Cells and Healthy Bone Marrow. Mol. Cancer Res. 2019, 17, 409–419. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Renaud, A.; Zhang, Y.; Ji, J.; Takeda, S.; Morris, J.; Teicher, B.; Doroshow, J.H.; Pommier, Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014, 13, 433–443. [Google Scholar] [CrossRef]
- Meehan, R.S.; Chen, A.P. New treatment option for ovarian cancer: PARP inhibitors. Gynecol. Oncol. Res. Pract. 2016, 3, 3. [Google Scholar] [CrossRef]
- Kim, G.; Ison, G.; McKee, A.E.; Zhang, H.; Tang, S.; Gwise, T.; Sridhara, R.; Lee, E.; Tzou, A.; Philip, R.; et al. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin. Cancer Res. 2015, 21, 4257–4261. [Google Scholar] [CrossRef] [PubMed]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Friedlander, M.; Matulonis, U.; Gourley, C.; du Bois, A.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Long-term efficacy, tolerability and overall survival in patients with platinum-sensitive, recurrent high-grade serous ovarian cancer treated with maintenance olaparib capsules following response to chemotherapy. Br. J. Cancer 2018, 119, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.O.; Hochhauser, D.; Arnold, D.; Oh, D.Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- De Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Oza, A.M.; Tinker, A.V.; Oaknin, A.; Shapira-Frommer, R.; McNeish, I.A.; Swisher, E.M.; Ray-Coquard, I.; Bell-McGuinn, K.; Coleman, R.L.; O'Malley, D.M.; et al. Antitumor activity and safety of the PARP inhibitor rucaparib in patients with high-grade ovarian carcinoma and a germline or somatic BRCA1 or BRCA2 mutation: Integrated analysis of data from Study 10 and ARIEL2. Gynecol. Oncol. 2017, 147, 267–275. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Abida, W.; Campbell, D.; Patnaik, A.; Sautois, B.; Shapiro, J.; Vogelzang, N.J.; Bryce, A.H.; McDermott, R.; Ricci, F.; Rowe, J.; et al. 846PD - Preliminary results from the TRITON2 study of rucaparib in patients (pts) with DNA damage repair (DDR)-deficient metastatic castration-resistant prostate cancer (mCRPC): Updated analyses. Ann. Oncol. 2019, 30, v327–v328. [Google Scholar] [CrossRef]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.M.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef]
- Moore, K.N.; Secord, A.A.; Geller, M.A.; Miller, D.S.; Cloven, N.; Fleming, G.F.; Wahner Hendrickson, A.E.; Azodi, M.; DiSilvestro, P.; Oza, A.M.; et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): A multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019, 20, 636–648. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- De Bono, J.; Ramanathan, R.K.; Mina, L.; Chugh, R.; Glaspy, J.; Rafii, S.; Kaye, S.; Sachdev, J.; Heymach, J.; Smith, D.C.; et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov. 2017, 7, 620–629. [Google Scholar] [CrossRef]
- Exman, P.; Barroso-Sousa, R.; Tolaney, S.M. Evidence to date: Talazoparib in the treatment of breast cancer. Onco Targets Ther. 2019, 12, 5177–5187. [Google Scholar] [CrossRef] [PubMed]
- Ettl, J.; Quek, R.G.W.; Lee, K.H.; Rugo, H.S.; Hurvitz, S.; Gonçalves, A.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Quality of life with talazoparib versus physician's choice of chemotherapy in patients with advanced breast cancer and germline BRCA1/2 mutation: Patient-reported outcomes from the EMBRACA phase III trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1939–1947. [Google Scholar] [CrossRef]
- Coleman, R.L.; Fleming, G.F.; Brady, M.F.; Swisher, E.M.; Steffensen, K.D.; Friedlander, M.; Okamoto, A.; Moore, K.N.; Efrat Ben-Baruch, N.; Werner, T.L.; et al. Veliparib with First-Line Chemotherapy and as Maintenance Therapy in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2403–2415. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Pommier, Y. Classification of PARP Inhibitors Based on PARP Trapping and Catalytic Inhibition, and Rationale for Combinations with Topoisomerase I Inhibitors and Alkylating Agents. In PARP Inhibitors for Cancer Therapy; Curtin, N.J., Sharma, R.A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 261–274. [Google Scholar]
- Maifrede, S.; Martinez, E.; Nieborowska-Skorska, M.; Di Marcantonio, D.; Hulse, M.; Le, B.V.; Zhao, H.; Piwocka, K.; Tempera, I.; Sykes, S.M.; et al. MLL-AF9 leukemias are sensitive to PARP1 inhibitors combined with cytotoxic drugs. Blood Adv. 2017, 1, 1467–1472. [Google Scholar] [CrossRef] [PubMed]
- Piao, J.; Takai, S.; Kamiya, T.; Inukai, T.; Sugita, K.; Ohyashiki, K.; Delia, D.; Masutani, M.; Mizutani, S.; Takagi, M. Poly (ADP-ribose) polymerase inhibitors selectively induce cytotoxicity in TCF3-HLF-positive leukemic cells. Cancer Lett. 2017, 386, 131–140. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Radivoyevitch, T.; Nagata, Y.; Khurshed, M.; Przychodzen, B.; Makishima, H.; Xu, M.; Bleeker, F.E.; Wilmink, J.W.; Carraway, H.E.; et al. IDH1/2 Mutations Sensitize Acute Myeloid Leukemia to PARP Inhibition and This Is Reversed by IDH1/2-Mutant Inhibitors. Clin. Cancer Res. 2018, 24, 1705–1715. [Google Scholar] [CrossRef]
- Deutsch, E.; Jarrousse, S.; Buet, D.; Dugray, A.; Bonnet, M.L.; Vozenin-Brotons, M.C.; Guilhot, F.; Turhan, A.G.; Feunteun, J.; Bourhis, J. Down-regulation of BRCA1 in BCR-ABL-expressing hematopoietic cells. Blood 2003, 101, 4583–4588. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, L.; Dolgalev, I.; Wang, Y.; Yoshimi, A.; Martin, G.H.; Wang, J.; Ng, V.; Xia, B.; Witkowski, M.T.; Mitchell-Flack, M.; et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 2017, 170, 1079–1095.e1020. [Google Scholar] [CrossRef]
- Inoue, S.; Li, W.Y.; Tseng, A.; Beerman, I.; Elia, A.J.; Bendall, S.C.; Lemonnier, F.; Kron, K.J.; Cescon, D.W.; Hao, Z.; et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell 2016, 30, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Maifrede, S.; Le, B.V.; Nieborowska-Skorska, M.; Golovine, K.; Sullivan-Reed, K.; Dunuwille, W.M.B.; Nacson, J.; Hulse, M.; Keith, K.; Madzo, J.; et al. TET2 and DNMT3A Mutations Exert Divergent Effects on DNA Repair and Sensitivity of Leukemia Cells to PARP Inhibitors. Cancer Res. 2021, 81, 5089–5101. [Google Scholar] [CrossRef] [PubMed]
- Dkhissi, F.; Aggoune, D.; Pontis, J.; Sorel, N.; Piccirilli, N.; LeCorf, A.; Guilhot, F.; Chomel, J.C.; Ait-Si-Ali, S.; Turhan, A.G. The downregulation of BAP1 expression by BCR-ABL reduces the stability of BRCA1 in chronic myeloid leukemia. Exp. Hematol. 2015, 43, 775–780. [Google Scholar] [CrossRef]
- Alcalay, M.; Meani, N.; Gelmetti, V.; Fantozzi, A.; Fagioli, M.; Orleth, A.; Riganelli, D.; Sebastiani, C.; Cappelli, E.; Casciari, C.; et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 2003, 112, 1751–1761. [Google Scholar] [CrossRef] [PubMed]
- Sulkowski Parker, L.; Corso Christopher, D.; Robinson Nathaniel, D.; Scanlon Susan, E.; Purshouse Karin, R.; Bai, H.; Liu, Y.; Sundaram Ranjini, K.; Hegan Denise, C.; Fons Nathan, R.; et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 2017, 9, eaal2463. [Google Scholar] [CrossRef]
- Sulkowski, P.L.; Oeck, S.; Dow, J.; Economos, N.G.; Mirfakhraie, L.; Liu, Y.; Noronha, K.; Bao, X.; Li, J.; Shuch, B.M.; et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 2020, 582, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Lin, H.P.; Zhou, W.J.; He, C.X.; Zhang, Z.Y.; Cheng, Z.L.; Song, J.B.; Liu, P.; Chen, X.Y.; Xia, Y.K.; et al. SNIP1 Recruits TET2 to Regulate c-MYC Target Genes and Cellular DNA Damage Response. Cell Rep. 2018, 25, 1485–1500.e1484. [Google Scholar] [CrossRef]
- Deutsch, E.; Dugray, A.; AbdulKarim, B.; Marangoni, E.; Maggiorella, L.; Vaganay, S.; M'Kacher, R.; Rasy, S.D.; Eschwege, F.; Vainchenker, W.; et al. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood 2001, 97, 2084–2090. [Google Scholar] [CrossRef]
- Tothova, Z.; Valton, A.-L.; Gorelov, R.A.; Vallurupalli, M.; Krill-Burger, J.M.; Holmes, A.; Landers, C.C.; Haydu, J.E.; Malolepsza, E.; Hartigan, C.; et al. Cohesin mutations alter DNA damage repair and chromatin structure and create therapeutic vulnerabilities in MDS/AML. JCI Insight 2021, 6, e142149. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, M.O.; Falinski, R.; Koptyra, M.; Slupianek, A.; Stoklosa, T.; Gloc, E.; Nieborowska-Skorska, M.; Blasiak, J.; Skorski, T. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood 2004, 104, 3746–3753. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Kopinski, P.K.; Ray, R.; Hoser, G.; Ngaba, D.; Flis, S.; Cramer, K.; Reddy, M.M.; Koptyra, M.; Penserga, T.; et al. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood 2012, 119, 4253–4263. [Google Scholar] [CrossRef] [PubMed]
- Marty, C.; Lacout, C.; Droin, N.; Le Couédic, J.P.; Ribrag, V.; Solary, E.; Vainchenker, W.; Villeval, J.L.; Plo, I. A role for reactive oxygen species in JAK2 V617F myeloproliferative neoplasm progression. Leukemia 2013, 27, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Mufti, G.; Estey, E.; Popat, R.; Mattison, R.; Menne, T.; Azar, J.; Bloor, A.; Gaymes, T.; Khwaja, A.; Juckett, M. Results of a phase 1 study of BMN 673, a potent and specific PARP-1/2 inhibitor, in patients with advanced hematological malignancies. In Haematologica, 2014; Ferrata Storti Foundation: Pavia, Italy, 2014; pp. 33–34. [Google Scholar]
- Gojo, I.; Beumer, J.H.; Pratz, K.W.; McDevitt, M.A.; Baer, M.R.; Blackford, A.L.; Smith, B.D.; Gore, S.D.; Carraway, H.E.; Showel, M.M.; et al. A Phase 1 Study of the PARP Inhibitor Veliparib in Combination with Temozolomide in Acute Myeloid Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Pratz, K.W.; Rudek, M.A.; Gojo, I.; Litzow, M.R.; McDevitt, M.A.; Ji, J.; Karnitz, L.M.; Herman, J.G.; Kinders, R.J.; Smith, B.D.; et al. A Phase I Study of Topotecan, Carboplatin and the PARP Inhibitor Veliparib in Acute Leukemias, Aggressive Myeloproliferative Neoplasms, and Chronic Myelomonocytic Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Pratt, G.; Yap, C.; Oldreive, C.; Slade, D.; Bishop, R.; Griffiths, M.; Dyer, M.J.S.; Fegan, C.; Oscier, D.; Pettitt, A.; et al. A multi-centre phase I trial of the PARP inhibitor olaparib in patients with relapsed chronic lymphocytic leukaemia, T-prolymphocytic leukaemia or mantle cell lymphoma. Br. J. Haematol. 2018, 182, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Morice, P.M.; Leary, A.; Dolladille, C.; Chrétien, B.; Poulain, L.; González-Martín, A.; Moore, K.; O'Reilly, E.M.; Ray-Coquard, I.; Alexandre, J. Myelodysplastic syndrome and acute myeloid leukaemia in patients treated with PARP inhibitors: A safety meta-analysis of randomised controlled trials and a retrospective study of the WHO pharmacovigilance database. Lancet Haematol. 2021, 8, e122–e134. [Google Scholar] [CrossRef]
- Baer, M.R.; Kogan, A.A.; Bentzen, S.M.; Mi, T.; Lapidus, R.G.; Duong, V.H.; Emadi, A.; Niyongere, S.; O'Connell, C.L.; Youngblood, B.A.; et al. Phase I Clinical Trial of DNA Methyltransferase Inhibitor Decitabine and PARP Inhibitor Talazoparib Combination Therapy in Relapsed/Refractory Acute Myeloid Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 1313–1322. [Google Scholar] [CrossRef]
- Quigley, D.; Alumkal, J.J.; Wyatt, A.W.; Kothari, V.; Foye, A.; Lloyd, P.; Aggarwal, R.; Kim, W.; Lu, E.; Schwartzman, J.; et al. Analysis of Circulating Cell-Free DNA Identifies Multiclonal Heterogeneity of BRCA2 Reversion Mutations Associated with Resistance to PARP Inhibitors. Cancer Discov. 2017, 7, 999–1005. [Google Scholar] [CrossRef]
- Edwards, S.L.; Brough, R.; Lord, C.J.; Natrajan, R.; Vatcheva, R.; Levine, D.A.; Boyd, J.; Reis-Filho, J.S.; Ashworth, A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 2008, 451, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Pettitt, S.J.; Frankum, J.R.; Punta, M.; Lise, S.; Alexander, J.; Chen, Y.; Yap, T.A.; Haider, S.; Tutt, A.N.J.; Lord, C.J. Clinical BRCA1/2 Reversion Analysis Identifies Hotspot Mutations and Predicted Neoantigens Associated with Therapy Resistance. Cancer Discov. 2020, 10, 1475–1488. [Google Scholar] [CrossRef] [PubMed]
- Jacot, W.; Thezenas, S.; Senal, R.; Viglianti, C.; Laberenne, A.C.; Lopez-Crapez, E.; Bibeau, F.; Bleuse, J.P.; Romieu, G.; Lamy, P.J. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 2013, 13, 523. [Google Scholar] [CrossRef] [PubMed]
- Marzio, A.; Puccini, J.; Kwon, Y.; Maverakis, N.K.; Arbini, A.; Sung, P.; Bar-Sagi, D.; Pagano, M. The F-Box Domain-Dependent Activity of EMI1 Regulates PARPi Sensitivity in Triple-Negative Breast Cancers. Mol. Cell 2019, 73, 224–237.e226. [Google Scholar] [CrossRef] [PubMed]
- Marzio, A.; Kurz, E.; Sahni, J.M.; Di Feo, G.; Puccini, J.; Jiang, S.; Hirsch, C.A.; Arbini, A.A.; Wu, W.L.; Pass, H.I.; et al. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion. Cell 2022, 185, 169–183.e119. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, K.; Feng, Y.; Nowialis, P.; Xu, H.; Shabashvili, D.E.; Berntsen, C.M.; Kaur, P.; Krajcik, K.I.; Taragjini, C.; Zaroogian, Z.; et al. DNMT3A Harboring Leukemia-Associated Mutations Directs Sensitivity to DNA Damage at Replication Forks. Clin. Cancer Res. 2022, 28, 756–769. [Google Scholar] [CrossRef]
- Liu, X.; Han, E.K.; Anderson, M.; Shi, Y.; Semizarov, D.; Wang, G.; McGonigal, T.; Roberts, L.; Lasko, L.; Palma, J.; et al. Acquired resistance to combination treatment with temozolomide and ABT-888 is mediated by both base excision repair and homologous recombination DNA repair pathways. Mol. Cancer Res. 2009, 7, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Pettitt, S.J.; Krastev, D.B.; Brandsma, I.; Drean, A.; Song, F.; Aleksandrov, R.; Harrell, M.I.; Menon, M.; Brough, R.; Campbell, J.; et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 2018, 9, 1849. [Google Scholar] [CrossRef]
- Mirman, Z.; Lottersberger, F.; Takai, H.; Kibe, T.; Gong, Y.; Takai, K.; Bianchi, A.; Zimmermann, M.; Durocher, D.; de Lange, T. 53BP1–RIF1–shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 2018, 560, 112–116. [Google Scholar] [CrossRef]
- Ghezraoui, H.; Oliveira, C.; Becker, J.R.; Bilham, K.; Moralli, D.; Anzilotti, C.; Fischer, R.; Deobagkar-Lele, M.; Sanchiz-Calvo, M.; Fueyo-Marcos, E.; et al. 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature 2018, 560, 122–127. [Google Scholar] [CrossRef]
- Jaspers, J.E.; Kersbergen, A.; Boon, U.; Sol, W.; van Deemter, L.; Zander, S.A.; Drost, R.; Wientjens, E.; Ji, J.; Aly, A.; et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov. 2013, 3, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-m.; Liao, X.-m.; Chen, Y.; Shen, Y.-y.; Yang, X.-y.; Su, Y.; Sun, Y.-m.; Gao, Y.-l.; Ding, J.; Zhang, A.; et al. Combining 53BP1 with BRCA1 as a biomarker to predict the sensitivity of poly(ADP-ribose) polymerase (PARP) inhibitors. Acta Pharmacol. Sin. 2017, 38, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Nacson, J.; Krais, J.J.; Bernhardy, A.J.; Clausen, E.; Feng, W.; Wang, Y.; Nicolas, E.; Cai, K.Q.; Tricarico, R.; Hua, X.; et al. BRCA1 Mutation-Specific Responses to 53BP1 Loss-Induced Homologous Recombination and PARP Inhibitor Resistance. Cell Rep. 2018, 24, 3513–3527.e3517. [Google Scholar] [CrossRef] [PubMed]
- Drané, P.; Brault, M.-E.; Cui, G.; Meghani, K.; Chaubey, S.; Detappe, A.; Parnandi, N.; He, Y.; Zheng, X.-F.; Botuyan, M.V.; et al. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature 2017, 543, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Dev, H.; Chiang, T.-W.W.; Lescale, C.; de Krijger, I.; Martin, A.G.; Pilger, D.; Coates, J.; Sczaniecka-Clift, M.; Wei, W.; Ostermaier, M.; et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 2018, 20, 954–965. [Google Scholar] [CrossRef]
- Xu, G.; Chapman, J.R.; Brandsma, I.; Yuan, J.; Mistrik, M.; Bouwman, P.; Bartkova, J.; Gogola, E.; Warmerdam, D.; Barazas, M.; et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 2015, 521, 541–544. [Google Scholar] [CrossRef]
- Tomida, J.; Takata, K.-i.; Bhetawal, S.; Person, M.D.; Chao, H.-P.; Tang, D.G.; Wood, R.D. FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J. 2018, 37, e99543. [Google Scholar] [CrossRef]
- Sarangi, P.; Clairmont, C.S.; D'Andrea, A.D. Disassembly of the Shieldin Complex by TRIP13. Cell Cycle 2020, 19, 1565–1575. [Google Scholar] [CrossRef]
- Xie, W.; Wang, S.; Wang, J.; de la Cruz, M.J.; Xu, G.; Scaltriti, M.; Patel, D.J. Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2024512118. [Google Scholar] [CrossRef]
- Clairmont, C.S.; Sarangi, P.; Ponnienselvan, K.; Galli, L.D.; Csete, I.; Moreau, L.; Adelmant, G.; Chowdhury, D.; Marto, J.A.; D’Andrea, A.D. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol. 2020, 22, 87–96. [Google Scholar] [CrossRef]
- He, Y.J.; Meghani, K.; Caron, M.-C.; Yang, C.; Ronato, D.A.; Bian, J.; Sharma, A.; Moore, J.; Niraj, J.; Detappe, A.; et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 2018, 563, 522–526. [Google Scholar] [CrossRef]
- Fujita, H.; Ikeda, M.; Ui, A.; Ouchi, Y.; Mikami, Y.; Kanno, S.-i.; Yasui, A.; Tanaka, K. CHAMP1-POGZ counteracts the inhibitory effect of 53BP1 on homologous recombination and affects PARP inhibitor resistance. Oncogene 2022, 41, 2706–2718. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Sarangi, P.; Iyer, D.R.; Feng, H.; Moreau, L.; Nguyen, H.; Clairmont, C.; D’Andrea, A.D. CHAMP1 binds to REV7/FANCV and promotes homologous recombination repair. Cell Rep. 2022, 40, 111297. [Google Scholar] [CrossRef] [PubMed]
- Lomonosov, M.; Anand, S.; Sangrithi, M.; Davies, R.; Venkitaraman, A.R. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003, 17, 3017–3022. [Google Scholar] [CrossRef] [PubMed]
- Rondinelli, B.; Gogola, E.; Yucel, H.; Duarte, A.A.; van de Ven, M.; van der Sluijs, R.; Konstantinopoulos, P.A.; Jonkers, J.; Ceccaldi, R.; Rottenberg, S.; et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017, 19, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Ray Chaudhuri, A.; Callen, E.; Ding, X.; Gogola, E.; Duarte, A.A.; Lee, J.-E.; Wong, N.; Lafarga, V.; Calvo, J.A.; Panzarino, N.J.; et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016, 535, 382–387. [Google Scholar] [CrossRef]
- Lok, B.H.; Gardner, E.E.; Schneeberger, V.E.; Ni, A.; Desmeules, P.; Rekhtman, N.; de Stanchina, E.; Teicher, B.A.; Riaz, N.; Powell, S.N.; et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 523–535. [Google Scholar] [CrossRef]
- Zatreanu, D.; Robinson, H.M.R.; Alkhatib, O.; Boursier, M.; Finch, H.; Geo, L.; Grande, D.; Grinkevich, V.; Heald, R.A.; Langdon, S.; et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 2021, 12, 3636. [Google Scholar] [CrossRef]
- Zatreanu, D.; Robinson, H.; Alkhatib, O.; Boursier, M.; Finch, H.; Geo, L.; Grande, D.; Grinkevich, V.; Heald, R.; Langdon, S.; et al. Abstract 5697: Targeting PARP inhibitor resistance with Polθ inhibitors. Cancer Res. 2022, 82, 5697. [Google Scholar] [CrossRef]
- Chapman, J.R.; Barral, P.; Vannier, J.B.; Borel, V.; Steger, M.; Tomas-Loba, A.; Sartori, A.A.; Adams, I.R.; Batista, F.D.; Boulton, S.J. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 2013, 49, 858–871. [Google Scholar] [CrossRef]
- Schlacher, K.; Wu, H.; Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef]
- Sung, P.; Klein, H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 739–750. [Google Scholar] [CrossRef]
- Rossi, M.J.; DiDomenico, S.F.; Patel, M.; Mazin, A.V. RAD52: Paradigm of Synthetic Lethality and New Developments. Front. Genet. 2021, 12, 780293. [Google Scholar] [CrossRef] [PubMed]
- Motycka, T.A.; Bessho, T.; Post, S.M.; Sung, P.; Tomkinson, A.E. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J. Biol. Chem. 2004, 279, 13634–13639. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet. TIG 2016, 32, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.; O'Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Mateos-Gomez, P.A.; Gong, F.; Nair, N.; Miller, K.M.; Lazzerini-Denchi, E.; Sfeir, A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 2015, 518, 254–257. [Google Scholar] [CrossRef]
- Higgins, G.S.; Boulton, S.J. Beyond PARP-POLtheta as an anticancer target. Science 2018, 359, 1217–1218. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Wyatt, D.W.; Takata, K.; Mu, Y.; Hensley, S.C.; Tomida, J.; Bylund, G.O.; Doublié, S.; Johansson, E.; Ramsden, D.A.; et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014, 10, e1004654. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.S.; Prevo, R.; Lee, Y.F.; Helleday, T.; Muschel, R.J.; Taylor, S.; Yoshimura, M.; Hickson, I.D.; Bernhard, E.J.; McKenna, W.G. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010, 70, 2984–2993. [Google Scholar] [CrossRef]
- Schofield, R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978, 4, 7–25. [Google Scholar] [PubMed]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef]
- Konopleva, M.Y.; Jordan, C.T. Leukemia stem cells and microenvironment: Biology and therapeutic targeting. J. Clin. Oncol. 2011, 29, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Hanoun, M.; Frenette, P.S. This niche is a maze; an amazing niche. Cell Stem Cell 2013, 12, 391–392. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Konopleva, M.; Konoplev, S.; Hu, W.; Zaritskey, A.Y.; Afanasiev, B.V.; Andreeff, M. Stromal cells prevent apoptosis of AML cells by up-regulation of anti-apoptotic proteins. Leukemia 2002, 16, 1713–1724. [Google Scholar] [CrossRef]
- Tabe, Y.; Konopleva, M. Advances in understanding the leukaemia microenvironment. Br. J. Haematol. 2014, 164, 767–778. [Google Scholar] [CrossRef]
- Colmone, A.; Amorim, M.; Pontier, A.L.; Wang, S.; Jablonski, E.; Sipkins, D.A. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science 2008, 322, 1861–1865. [Google Scholar] [CrossRef]
- Kode, A.; Manavalan, J.S.; Mosialou, I.; Bhagat, G.; Rathinam, C.V.; Luo, N.; Khiabanian, H.; Lee, A.; Murty, V.V.; Friedman, R.; et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature 2014, 506, 240–244. [Google Scholar] [CrossRef]
- Kode, A.; Mosialou, I.; Manavalan, S.J.; Rathinam, C.V.; Friedman, R.A.; Teruya-Feldstein, J.; Bhagat, G.; Berman, E.; Kousteni, S. FoxO1-dependent induction of acute myeloid leukemia by osteoblasts in mice. Leukemia 2016, 30, 1–13. [Google Scholar] [CrossRef]
- Herbertz, S.; Sawyer, J.S.; Stauber, A.J.; Gueorguieva, I.; Driscoll, K.E.; Estrem, S.T.; Cleverly, A.L.; Desaiah, D.; Guba, S.C.; Benhadji, K.A.; et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway. Drug Des. Devel. Ther. 2015, 9, 4479–4499. [Google Scholar] [CrossRef]
- Azaro, A.; Rodon, J.; Carducci, M.; Sepulveda-Sanchez, J.M.; Gueorguieva, I.; Cleverly, A.L.; Desaiah, D.; Namaseevayam, S.P.; Holdhoff, M.; Lahn, M.M. Case Series of Cancer Patients Treated with Galunisertib, a Transforming Growth Factor-Beta Receptor I Kinase Inhibitor in a First-in-Human Dose Study. J. Med. Cases 2014, 5, 603–609. [Google Scholar] [CrossRef][Green Version]
- Wick, A.; Desjardins, A.; Suarez, C.; Forsyth, P.; Gueorguieva, I.; Burkholder, T.; Cleverly, A.L.; Estrem, S.T.; Wang, S.; Lahn, M.M.; et al. Phase 1b/2a study of galunisertib, a small molecule inhibitor of transforming growth factor-beta receptor I, in combination with standard temozolomide-based radiochemotherapy in patients with newly diagnosed malignant glioma. Investig. New Drugs 2020, 38, 1570–1579. [Google Scholar] [CrossRef] [PubMed]
- Melisi, D.; Garcia-Carbonero, R.; Macarulla, T.; Pezet, D.; Deplanque, G.; Fuchs, M.; Trojan, J.; Oettle, H.; Kozloff, M.; Cleverly, A.; et al. Galunisertib plus gemcitabine vs. gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br. J. Cancer 2018, 119, 1208–1214. [Google Scholar] [CrossRef]
- Santini, V.; Valcarcel, D.; Platzbecker, U.; Komrokji, R.S.; Cleverly, A.L.; Lahn, M.M.; Janssen, J.; Zhao, Y.; Chiang, A.; Giagounidis, A.; et al. Phase II Study of the ALK5 Inhibitor Galunisertib in Very Low-, Low-, and Intermediate-Risk Myelodysplastic Syndromes. Clin. Cancer Res. 2019, 25, 6976–6985. [Google Scholar] [CrossRef]
- Kelley, R.K.; Gane, E.; Assenat, E.; Siebler, J.; Galle, P.R.; Merle, P.; Hourmand, I.O.; Cleverly, A.; Zhao, Y.; Gueorguieva, I.; et al. A Phase 2 Study of Galunisertib (TGF-beta1 Receptor Type I Inhibitor) and Sorafenib in Patients With Advanced Hepatocellular Carcinoma. Clin. Transl. Gastroenterol. 2019, 10, e00056. [Google Scholar] [CrossRef]
- Zhou, J.; Gelot, C.; Pantelidou, C.; Li, A.; Yücel, H.; Davis, R.E.; Färkkilä, A.; Kochupurakkal, B.; Syed, A.; Shapiro, G.I.; et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2021, 2, 598–610. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).