Simple Summary
Pain is experienced by ~55% of patients who undergo anti-cancer treatment. However, many genetic factors that are involved in the efficacy of opioid analgesics in the treatment of cancer pain remain unidentified. The aim of our genome-wide association study (GWAS) was to comprehensively explore genetic variations that are associated with opioid analgesic requirements in the treatment of cancer pain. We identified several single-nucleotide polymorphisms (SNPs) that are associated with opioid analgesic requirements, two of the most potent of which were the rs1283671 and rs1283720 SNPs in the ANGPT1 gene region, and SNPs in the SLC2A14 gene were also associated with the phenotype. These results indicate that these SNPs in the ANGPT1 and SLC2A14 genes could serve as markers that predict the efficacy of opioid analgesics in the treatment of cancer pain.
Abstract
Considerable individual differences have been widely observed in the sensitivity to opioids. We conducted a genome-wide association study (GWAS) in patients with cancer pain to identify potential candidate single-nucleotide polymorphisms (SNPs) that contribute to individual differences in opioid analgesic requirements in pain treatment by utilizing whole-genome genotyping arrays with more than 650,000 markers. The subjects in the GWAS were 428 patients who provided written informed consent and underwent treatment for pain with opioid analgesics in a palliative care unit at Higashi-Sapporo Hospital. The GWAS showed two intronic SNPs, rs1283671 and rs1283720, in the ANGPT1 gene that encodes a secreted glycoprotein that belongs to the angiopoietin family. These two SNPs were strongly associated with average daily opioid requirements for the treatment of pain in both the additive and recessive models (p < 5.0000 × 10−8). Several other SNPs were also significantly associated with the phenotype. In the gene-based analysis, the association was significant for the SLC2A14 gene in the additive model. These results indicate that these SNPs could serve as markers that predict the efficacy of opioid analgesics in cancer pain treatment. Our findings may provide valuable information for achieving satisfactory pain control and open new avenues for personalized pain treatment.
1. Introduction
Pain is defined by the International Association for the Study of Pain as “An unpleasant sensory and emotional experience associated with, or resembling that associated with, actual or potential tissue damage” [1]. Although pain is reportedly experienced by 55% of patients who undergo anti-cancer treatment and by 66% of patients who have advanced, metastatic, or terminal disease, many more cancer patients may potentially experience pain according to a recent systematic review and meta-analysis [2]. The World Health Organization recommends that any opioid may be considered for the maintenance of pain relief (alone or combined with nonsteroidal anti-inflammatory drugs [NSAIDs] and/or paracetamol) in adults (including older persons) and adolescents with cancer-related pain to achieve sustained, effective, and safe pain control, depending on clinical assessment and pain severity [3]. However, their effects are not uniform across all patients. Patient responses to opioid medications vary by patient, specific medication, and other factors [3,4,5]. Such significant variations in opioid responsiveness or sensitivity may influence analgesic effects that are necessary to achieve sufficient pain relief, which may hinder the clinical management of pain. Although twin studies by Angst et al. indicated that genetic effects on the opioid-induced elevation of heat or cold-pressor pain thresholds accounted for 12–60% of the observed response variance [6,7], many genetic factors remain to be elucidated.
To identify genetic variations, mostly single-nucleotide polymorphisms (SNPs), that are associated with human opioid sensitivity or responsiveness and related phenotypes, many candidate gene association studies have been conducted [8,9,10]. The most extensively investigated genetic variation is the functional A118G (rs1799971) SNP in the µ-opioid receptor gene, OPRM1. A recent meta-analysis indicated that G-allele carriers (AG+GG) of this SNP required higher opioid doses for cancer pain management than AA homozygotes [11]. Genetic polymorphisms in the COMT gene, which encodes catechol-O-methyltransferase, have also been often investigated, and SNPs in this gene were found to be associated with responsiveness to opioids even in advanced cancer patients [12,13]. Moreover, several other SNPs in the ABCB1, IL6, GCH1, and TAOK3 gene have also been found to be associated with the response to opioids in patients with cancer pain using a candidate gene approach [14].
Genome-wide association studies (GWASs) focus on genetic variations in all regions of the human genome without regard to preexisting theories about the relationship between variations and the phenotypes of interest [15]. GWASs can also be used to investigate genetic factors that are related to individual differences in analgesic effects of opioids. However, only a few studies have examined human opioid responsiveness and sensitivity based on GWASs. One of these studies investigated associations between SNPs and total morphine requirements as a quantitative trait locus in a retrospective population of opioid-naive children (4–18 years old) who had undergone day surgery tonsillectomy and adenoidectomy. This study identified an association between the rs795484 and rs1277441 SNPs at the TAOK3 gene locus and morphine dose [16]. Additionally, we performed a GWAS of traits that are linked to opioid sensitivity. We chose participants who were scheduled to undergo aesthetic orthognathic surgery for mandibular prognathism [17] and found that the best candidate was the rs2952768 SNP, located near the METTL21A (FAM119A) and CREB1 gene regions [18]. A GWAS was also conducted in patients with cancer from 11 European countries using a prospective cross-sectional multinational, multicenter design [19]. The rs12948783 SNP, which corresponds to chromosome 17 upstream of the RHBDF2 gene, had the strongest association with responsiveness to opioids in this study’s patients who received opioid treatment for moderate or severe pain [20]. However, no GWAS has yet been conducted in populations other than of European origin, such as Asian populations.
In the present study, we conducted a GWAS in patients with cancer pain to identify potential genetic variants that contribute to the efficacy of opioid analgesics that are used to treat pain, reflected by analgesic requirements.
2. Materials and Methods
2.1. Patients
2.1.1. Patients Who Underwent Cancer Pain Treatment with Opioid Analgesics
We enrolled 428 adult patients (20–94 years old, 213 males and 215 females) who suffered from various types of cancer and were hospitalized at Higashi-Sapporo Hospital (Hokkaido, Japan) for the treatment of cancer pain in 2017–2019 and were apparently Japanese. Higashi-Sapporo Hospital was established in 1983 as a hospital that specializes in cancer care, particularly palliative care. Characteristics of the hospital and palliative cancer care were detailed in previous reports [21,22]. All of the patients who were recruited in the present study were treated with opioid analgesics, and many were also appropriately treated with NSAIDs and/or other supplementary analgesics for the treatment of pain. We excluded patients who were considered to be unsuitable by their primary care doctors. Detailed demographic and clinical data of the subjects are provided in Table 1. Peripheral blood samples were collected from these subjects for gene analysis.

Table 1.
Demographic and clinical data of patient subjects.
The study was conducted according to guidelines of the Declaration of Helsinki and approved by the Institutional Review Board or Ethics Committee of Higashi-Sapporo Hospital and Tokyo Metropolitan Institute of Medical Science (Tokyo, Japan). Written informed consent was obtained from all of the patients.
2.1.2. Patient Characteristics and Clinical Data
In the patient subjects, we obtained data on surgical history, treatment history, pain status (e.g., presence/absence of somatic pain, visceral pain, and neuropathic pain), drug treatments, and disease status (e.g., lung cancer, breast cancer, stomach cancer, etc.; Table 1). Some of the patients were affected by multiple diseases.
The treatment of pain was mainly conducted by administering opioid analgesics (e.g., morphine and oxycodone; Table 1). Various types of drugs, such as NSAIDs (e.g., loxoprofen and diclofenac) and/or other supplementary analgesics (e.g., pregabalin and dexamethasone), were also administered at the discretion of primary care doctors if required. To allow intersubject comparisons of opioid analgesic doses that were required for cancer pain treatment, the opioid doses were converted to equivalent doses of oral morphine as described in Supplementary Table S1, based on previous reports with slight modification [23,24,25]. The total dose of converted opioid analgesics that were administered was calculated daily, and the total dose of analgesics was calculated as a daily average based on the amount of 5 days of administration, 3–7 days before blood collection. This average total dose was used as the endpoint of opioid requirements for the genetic association analysis in the present study. Doses of analgesics that were administered were normalized to body weight. The detailed clinical data of the subjects are detailed in Table 1.
2.2. Whole-Genome Genotyping, Quality Control, and Gene-Based and Gene-Set Analyses
2.2.1. Whole-Genome Genotyping and Quality Control
A total of 428 DNA samples from the patients were used for genotyping. Total genomic DNA was extracted from whole-blood samples using standard procedures. The extracted DNA was dissolved in TE buffer (10 mM Tris-HCl and 1 mM ethylenediaminetetraacetic acid [EDTA], pH 8.0). The DNA concentration was adjusted to 50 ng/μL for whole-genome genotyping using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
According to the manufacturer’s recommendations, whole-genome genotyping was performed by utilizing the Infinium Assay II with an iScan system (Illumina, San Diego, CA, USA). Infinium Asian Screening Array-24 v. 1.0 BeadChips (one kind) were utilized to genotype 428 patient samples (total markers: 659,184). Numerous copy number variation markers were included in the BeadChips, but the majority of the BeadChips were for SNP markers on the human autosome or sex chromosome.
GenomeStudio with the Genotyping v. 2.0.4 module (Illumina) was used to examine data for samples that had their entire genomes genotyped to assess quality of the findings. Following data cleaning, samples with genotype call rates less than 0.95 were not included in the remaining studies. No patient samples were consequently disregarded for subsequent investigations (i.e., all of the 428 samples met the 0.95 quality control cutoff, and the entire samples were considered for further association analyses). In the subsequent association analyses, markers with genotype call frequencies less than 0.95 and “Cluster sep” (a measure of genotype cluster separation) values less than 0.1 were not included. The patient samples retained a total of 650,504 SNP markers after this screening step. Markers were further filtered based on a test of Hardy–Weinberg equilibrium (HWE). Markers with p values (df = 1) less than 0.001 were considered to be deviated in the HWE tests and thus were excluded. Finally, the analysis only included the 648,817 SNPs that passed the full filtration procedure.
A log quantile-quantile (QQ) p-value plot as a result of the GWAS for the entire sample was subsequently drawn to check the pattern of the generated p-value distribution in the association studies between the SNPs and phenotype, in which the observed p values against the values that were expected from the null hypothesis of a uniform distribution, calculated as –log10 (p value), were plotted for each model. All of the plots were mostly concordant with the expected line (y = x), especially over the range of 0 < –log10 (p value) < 2 for each model, indicating no apparent population stratification of the samples that were used in the study (Supplementary Figure S1). Additionally, results of the GWAS are presented as Manhattan plots, in which the –log10 (p value) for each SNP on each chromosome was plotted simultaneously.
2.2.2. Gene-Based and Gene-Set Analyses
Gene-based and gene-set approaches were adopted with Multi-marker Analysis of GenoMic Annotation (MAGMA) v. 1.06 [26], which is also available on the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA GWAS) v. 1.3.9 platform [27], to better understand genetic backgrounds and molecular mechanisms that underlie complex traits, such as opioid requirements in cancer pain treatment. To examine the combined relationship between all genetic markers in the gene and the phenotype, genetic marker data were aggregated to the level of full genes in the gene-based analysis. Similarly, individual genes were compiled into groupings of genes with similar biological, functional, or other properties for the gene-set analysis. Gene-set analysis can thus shed light on the role that particular biological pathways or cellular processes may play in the genetic basis of a trait [26]. In these analyses, associations were explored for genes on autosomes 1–22 and the X chromosome, and the window of the genes to assign SNPs was set to 20 kb, thereby assigning SNPs within the 20 kb window of the gene (both sides) to that gene. For the reference panel, the 1000 Genome Phase3 EAS population was selected (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ accessed on 26 July 2022). In the gene-set analysis, gene sets were defined using the Molecular Signatures Database (MSigDB) v. 7.5.1 (https://www.gsea-msigdb.org/gsea/msigdb accessed on 26 September 2022) [28]. A total of 10,678 gene sets (curated gene sets: 4761, GO terms: 5917) from MsigDB were tested. In both analyses, Bonferroni correction for multiple testing was performed for all tested genes and gene sets. Adjusted values of p < 0.05 in the results were considered significant.
2.3. Statistical Analysis
In the association studies of patients who underwent cancer pain treatment with opioids, the total dose of analgesics as a daily average that was calculated based on the amount of 5 days of administration, 3–7 days before blood collection, was used as an index of opioid sensitivity. Before the analyses, the quantitative values of total analgesic requirements that were equivalent to oral morphine (mg/kg) were natural log-transformed for approximation to the normal distribution according to the following formula: Value for analyses = Ln (1 + total analgesic requirements on average per day [mg/kg]). To explore associations between the SNPs and phenotypes, linear regression analyses were conducted, in which total analgesic requirements as a daily average (mg/kg; log-transformed) and genotype data for each SNP were incorporated as dependent and independent variables, respectively. Additionally, weight and binary data on the presence/absence of neuropathic pain were incorporated as covariables because these parameters were strongly associated with the dependent variable in a preliminary study (data not shown). Additive, dominant, and recessive genetic models were used for the analyses because of previously insufficient knowledge about genetic factors that are associated with opioid sensitivity. Male genotypes were not included in the analysis of X chromosome markers, whereas both male and female individuals were included in the association study for autosomal markers. PLINK v. 1.07 (https://zzz.bwh.harvard.edu/plink/index.shtml accessed on 28 July 2022) [29], gPLINK v. 2.050 [29], and Haploview v. 4.2 [30] were used to perform the statistical analyses. The criterion for significance in the GWAS was set at p < 5 × 10−8, which is widely known to be a conventional criterion for the level of significance in GWASs [31,32]. Additionally, Hardy–Weinberg equilibrium was tested using Exact Tests for genotypic distributions of SNPs that were significantly associated with the phenotype.
2.4. Additional in Silico Analysis
2.4.1. Power Analysis
G*Power 3.0.5 software (https://www.psychologie.hhu.de/arbeitsgruppen/allgemeine-psychologie-und-arbeitspsychologie/gpower accessed on 26 September 2022) was used to perform preliminary statistical power analyses [33]. When the type I error probability was set at 1 × 10−7 (less than 5 × 10−8) and the sample size was 428, power assessments for the linear regression analysis showed that the expected power (1 minus type II error probability) was over 99.4% for a Cohen’s conventional “middle” effect size of 0.15 [34]. When Cohen’s conventional “small” effect size was 0.02, however, for the same type I error probability and sample sizes of 428, the estimated power decreased to 0.7%. In contrast, assuming the same type I error probability and sample sizes of 428 to obtain 80% power, the predicted effect size was 0.0919. For effect sizes from large to moderately small but not too small, a single analysis in the preceding study was anticipated to detect true associations with the phenotype with 80% statistical power, although the precise effect size has not been well understood in cases of SNPs that significantly influence opioid sensitivity.
2.4.2. Linkage Disequilibrium Analysis
Linkage disequilibrium (LD) analysis was performed using Haploview v. 4.2 [30] for a total of 428 samples from patients who underwent cancer pain treatment with opioids for the locus of the ~243 kbp region that was annotated as the ANGPT1 gene and its flanking region on chromosome 8 based on an annotation file that was provided by Illumina (San Diego, CA, USA) to identify relationships between SNPs in the ANGPT1 gene region. The commonly used D′ and r2 values were pairwise calculated using the genotype dataset for each SNP to estimate the strength of LD between SNPs. Linkage disequilibrium blocks were defined as in a previous study [35].
2.4.3. Reference of Databases
Several databases and bioinformatic tools were referenced to more thoroughly examine the candidate SNP that may be related to human opioid analgesic sensitivity, including the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov accessed on 28 July 2022), Genotype-Tissue Expression (GTEx) portal (https://www.gtexportal.org/home/; accessed on 28 July 2022) [36], HaploReg v. 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php; accessed on 28 July 2022) [37], and SNPinfo Web Server (https://snpinfo.niehs.nih.gov/; accessed on 28 July 2022) [38]. The GTEx project, an ongoing effort to create a comprehensive public resource to research tissue-specific gene expression and regulation [36], is the basis for the GTEx portal, which offers open access to such data as gene expression, quantitative trait loci, and histology images. HaploReg is a tool for investigating non-coding genomic annotations at variations in haplotype blocks, such as potential regulatory SNPs at disease-associated sites [37]. The SNPinfo Web Server is a set of web-based SNP selection tools (freely available at https://snpinfo.niehs.nih.gov/; accessed on 28 July 2022) where investigators can specify genes or linkage regions and select SNPs based on GWAS results, LD, and predicted functional characteristics of both coding and non-coding SNPs [38].
3. Results
3.1. Identification of Genetic Polymorphisms Associated with Opioid Analgesic Requirements in the Treatment of Cancer Pain by GWAS
We comprehensively explored genetic variations that were associated with opioid analgesic requirements per day in the treatment of cancer pain in a total of 428 patients. A total of 648,817 SNPs that met the quality control standards in the GWAS of all patients were examined for relationships with the phenotype in the additive, dominant, and recessive models. The top 20 potential SNPs for the additive, dominant, and recessive models are shown in Table 2, Table 3 and Table 4, respectively. Significant associations were found for the rs1283671 and rs1283720 SNPs on chromosome 8 and the GSA-rs72752701 (rs72752701) SNP on chromosome 9 in the additive model (p = 3.876 × 10−11 for rs1283671, p = 4.007 × 10−11 for rs1283720, p = 3.879 × 10−8 for GSA-rs72752701; Table 2, Figure 1a). Significant associations were also found for the kgp11947181 (rs79524268) SNP on chromosome 6, 9:133013640 (rs2417163) SNP on chromosome 9, and rs763315292 SNP on chromosome 10 in the dominant model (p = 1.592 × 10−8 for kgp11947181, p = 1.730 × 10−8 for 9:133013640, p = 3.807 × 10−8 for rs763315292; Table 3, Figure 1b). Significant associations were also found in the recessive model for the same three SNPs as in the additive model (p = 7.698 × 10−11 for rs1283671, p = 7.698 × 10−11 for rs1283720, p = 3.814 × 10−8 for GSA-rs72752701; Table 4, Figure 1c). Many of the examined SNPs’ computed -log10 p values (observed p values), which are based on the null hypothesis of a uniform distribution in the QQ plot, differed from the predicted values (Supplementary Figure S1). The values for SNPs with significant relationships in Table 2, Table 3 and Table 4 (rs1283671 and rs1283720) and other SNPs were obviously higher than predicted values (Supplementary Figure S1). The gene that encodes angiopoietin-1, ANGPT1, was in the vicinity of the rs1283671 and rs1283720 SNPs (i.e., the best candidate SNPs in the additive and recessive models). Meanwhile, the kgp11947181 SNP, the best candidate SNP in the dominant model, was located in an intergenic region on chromosome 6 (Supplementary Figure S2b). The rs1283671 and rs1283720 SNPs were in strong LD with each other (r2 = 0.97), although these two SNPs were not in strong LD (r2 < 0.80) with other neighboring SNPs within and around the ANGPT1 gene (Supplementary Figure S3). As shown in Table 2 and Table 4, an increase in the minor allele carry in the rs1283671 and rs1283720 SNPs was associated with greater opioid analgesic requirements in cancer pain treatment.

Table 2.
Top 30 candidate SNPs selected from the GWAS (additive model).

Table 3.
Top 30 candidate SNPs selected from the GWAS (dominant model).

Table 4.
Top 30 candidate SNPs selected from the GWAS (recessive model).
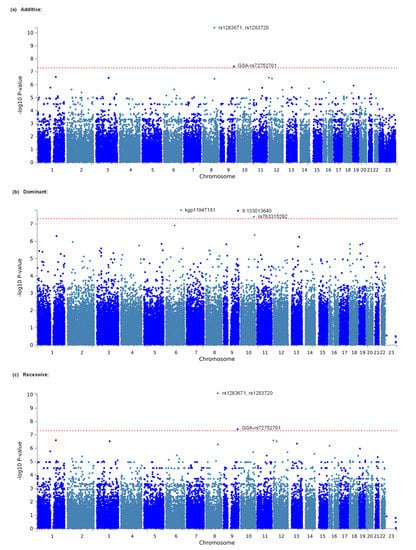
Figure 1.
Manhattan plot of GWAS outcomes. (a) Analytical plot in the additive model. (b) Analytical plot in the dominant model. (c) Analytical plot in the recessive model. The dashed red line shows the significant association threshold.
3.2. Identification of Genes and Gene Sets Associated with Opioid Analgesic Requirements in the Treatment of Cancer Pain by Gene-Based and Gene-Set Analyses
Considering that the effects of individual markers tend to be too weak to be detected by comprehensive analyses, such as GWASs, that target only single polymorphisms, we conducted gene-based and gene-set analyses, which are statistical methods that are used to analyze multiple genetic markers simultaneously to determine their joint effect. In both analyses, using MAGMA software [26], which was made accessible in the FUMA GWAS platform [27], we investigated genes and gene sets that were related to the amount of opioid analgesics that were required daily for the management of cancer pain in a total of 428 individuals. As a result, 648,817 SNPs from the selected candidate genes and gene sets in the additive, dominant, and recessive models were included in the analyses of all patients. The top 20 candidate genes that were found in each genetic model by the gene-based analysis are listed in Table 5. In the additive model, SLC2A14, the top candidate gene, was significantly associated with the phenotype (adjusted p = 0.03722; Table 5, Figure 2a). However, in both the dominant and recessive models, none of the genes were significantly associated with the phenotype (Table 5, Figure 2b,c), although the association with the SLC2A14 gene was marginally significant in the recessive model (p = 0.07384; Table 5). The top 10 candidate gene sets that were found in each genetic model using the gene-set analysis are listed in Supplementary Table S1. However, in none of the genetic models were any of the gene sets significantly related to the phenotype (Supplementary Table S1).

Table 5.
Top 20 candidate genes selected from gene-based analysis.
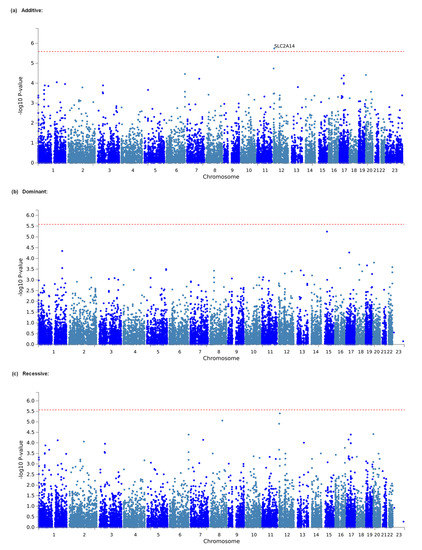
Figure 2.
Manhattan plot of results of gene-based analysis. (a) Analytical plot in the additive model. (b) Analytical plot in the dominant model. (c) Analytical plot in the recessive model. The dashed red line shows the significant association threshold.
4. Discussion
To identify potential genetic variants that contribute to the requirements of opioid analgesics that were used to treat cancer pain, we conducted a GWAS in patient subjects who were diagnosed with various types of cancers (Table 1). The GWAS results suggested that carriers of minor alleles of the rs1283671 and rs1283720 SNPs within the ANGPT1 gene region were less sensitive to opioid analgesics and thus required greater amounts of daily opioid analgesics in cancer pain treatment (Table 2 and Table 4). Our GWAS also identified other potent SNPs that are possibly associated with opioid sensitivity, including GSA-rs72752701 (rs72752701) in the additive and recessive models (Table 2 and Table 4) and kgp11947181 (rs79524268), 9:133013640 (rs2417163), and rs763315292 in the dominant model (Table 3). Additionally, numerous SNPs in or near the SLC2A14 gene region collectively increased the amount of analgesic that was required for cancer pain management (Table 5). No relevant gene sets that may have been related to the trait were identified (Supplementary Table S1). These findings show that in the genetic etiology of the trait, individual genes did not properly aggregate to groups of genes that shared specific biological, functional, or other features. Future studies with larger sample sizes are required to find candidate gene sets that are associated with the phenotype and corroborate the significant results that were identified in our GWAS of single SNPs and gene-set analysis.
Previous studies identified several genetic variations that are associated with phenotypes that are related to opioid responsiveness in the treatment of cancer pain [11,12,13,14,20]. Included in those candidate genetic variations are the rs1799971, rs1319339, and rs34427887 SNPs in the OPRM1 gene, rs4680, rs4818, rs4633, rs4646316, and rs35478083 SNPs in the COMT gene, rs1045642 SNP in the ABCB1 gene, −174G>C (rs1800795) SNP in the IL6 gene, rs8007267, rs3783641, and rs10483639 SNPs in the GCH1 gene, rs12948783 SNP in the RHBDF2 gene, and rs1277441 and rs795484 SNPs in the TAOK3 gene [11,12,13,14,20]. Among these SNPs, rs1799971, rs4680, rs4633, rs4646316, rs1045642, rs1800795, rs8007267, rs3783641, rs10483639, rs12948783, and rs795484 SNPs were included in the SNP array that was used in the present study. However, no SNPs except rs8007267 (p = 0.007411) and rs3783641 (p = 0.006942) in the GCH1 gene were even nominally significantly associated with opioid analgesic requirements (p > 0.05) in our association analysis results (details not shown). The GCH1 gene encodes GTP cyclohydrolase 1, which is the first and rate-limiting enzyme in tetrahydrobiopterin (BH4) biosynthesis. Although these two SNPs, with the other rs10483639 SNP, were reported to be associated with opioid therapy initiation in cancer pain patients [39], these three SNPs were not shown to be associated with the responsiveness to opioids itself. These results might indicate the general difficulty of replicating results of human genetic association studies, probably because of the heterogeneity of study designs, diagnoses of different types of cancer, pain severity, types of opioid analgesics used, and genetic background of the subjects, among other factors.
The best candidate SNPs in the present study were rs1283671 and rs1283720, which are found in the intronic region of the ANGPT1 gene on chromosome 8. These SNPs had the lowest p values among the candidate SNPs. Angiopoietin-1, a secreted glycoprotein that is a member of the angiopoietin family, is encoded by the ANGPT1 gene. Angiopoietin-1 is an angiogenic growth factor with antipermeability and anti-inflammatory properties [40]. Chronic or uncontrolled inflammation plays an important role in many diseases [41], including stroke [42]. When the angiopoetin-1 receptor, Tie2, becomes inactivated, important molecular brakes are released in the endothelium, which in turn potentiate inflammation and vascular leakage [43]. ANGPT1 mRNA is known to be broadly expressed in fat, the lungs, the prostate, and several other tissues in humans, according to the NCBI database. Mice that were engineered to lack angiopoietin-1 exhibited angiogenic deficits [44]. A missense mutation (807G>T, A119S) that was identified in a family was associated with hereditary angioedema in humans [45]. Meanwhile, angiogenesis may be protected against by angiotensin-converting enzyme (ACE) inhibitors [46], and the inhibition of ACE activity might prevent or even reverse hypoglycemia-associated autonomic failure (HAAF) [47]. β-endorphin has also been reported to influence the autonomic response to hypoglycemia via opioid receptor activation [48]. The opioid receptor antagonist naloxone improved counterregulatory responses [49] and prevented HAAF [48] in humans. Therefore, both the inhibition of ACE activity and antagonism of opioid receptors may protect against angiogenesis and HAAF in humans, suggesting that angiogenesis, in which angiopoietin-1 is involved, could also be modulated by actions of opioids. Therefore, a possible explanation for the present results may be that genetic variations in the ANGPT1 gene could influence individual differences in angiogenic effects, which in turn may lead to individual differences in opioid analgesic efficacy. However, to date, no genetic variations of the ANGPT1 gene have been reported to be related to functions of the opioid system. The rs1283671 and rs1283720 SNPs (i.e., the best candidate SNPs in the present study) have not been previously reported to be associated with any phenotypes to date. Although the rs1283671 and rs1283720 SNPs were in strong LD with each other (r2 = 0.97), these two SNPs were not in strong LD (r2 < 0.80) with other neighboring SNPs in our data (Supplementary Figure S3). When these SNPs were referenced in HaploReg v. 4.1 [37] (accessed on 2 August 2022), they were in strong LD (r2 ≥ 0.80) with 20 neighboring SNPs (data not shown). HaploReg v. 4.1 also showed that the rs1283671 and rs1283720 SNPs could change eight and three motifs, respectively, for DNA-binding proteins. Nevertheless, none of these SNPs were significantly associated with mRNA expression levels of any genes in any tissues, according to the GTEx portal (accessed on 2 August 2022), suggesting that these SNPs unlikely influence variations in opioid sensitivity among individuals by influencing the mRNA expression of some genes.
Our GWAS identified several other potent SNPs beyond rs1283671 and rs1283720 that were significantly associated with opioid requirements in cancer pain patients, including GSA-rs72752701 (rs72752701) in the additive and recessive models (Table 2 and Table 4) and kgp11947181 (rs79524268), 9:133013640 (rs2417163), and rs763315292 in the dominant model (Table 3). Among these, the GSA-rs72752701, kgp11947181, and 9:133013640 SNPs are located in intergenic regions, whereas rs763315292 is located in the SEC23 interacting protein gene (SEC23IP) region. Although no significant expression quantitative trait loci (eQTLs) were found for kgp11947181 and rs763315292, the GSA-rs72752701 SNP is associated with mRNA expression of the heat shock protein family A (Hsp70) member 5 (HSPA5) gene, and the 9:133013640 SNP is associated with mRNA expression of the hemicentin 2 (HMCN2) and long intergenic non-protein coding RNA 963 (LINC00963) genes, according to the GTEx portal [36]. The SEC23IP, HMCN2, and LINC00963 genes have not been previously reported to be related to the opioid system or its receptors. In a previous quantitative mass spectrometry proteomics study of purified synaptosomes that were isolated from the caudate in two groups of rhesus macaques that were chronically infected with simian immunodeficiency virus (SIV) and differed with regard to morphine treatment regimens, the upregulation of HSPA5 was found in the SIV+morphine group, indicating an increase in cellular stress as a result of the SIV/morphine interaction, thereby leading to central nervous system dysfunction [50]. This previous study suggests that the expression and function of HSPA5 could interact with the opioid system, and alterations of HSPA5 mRNA expression through the GSA-rs72752701 SNP might influence the function of opioids.
Although none of the top 30 candidate SNPs in the additive model in the present study included SNPs that were annotated as this gene (Table 2), the SLC2A14 gene was significantly associated with the trait in the gene-based analysis of all patients (Table 5, Figure 2a). The SLC2A14 gene encodes solute carrier family 2 member 14 (SLC2A14), which is a member of the glucose transporter (GLUT) family. Members of the GLUT family, including SLC2A14, are highly conserved integral membrane proteins that transport hexoses, such as glucose and fructose, into all mammalian cells. However, the functional relationship between this protein and the opioid system is unknown. SLC2A14 mRNA is known to be highly expressed in the testis [51], followed by bone marrow, the urinary bladder, the placenta, the gall bladder, the appendix, and the brain, according to the NCBI database. In human genetic studies, SNPs in the SLC2A14 gene region were reported to be associated with some diseases, including Alzheimer’s disease and inflammatory bowel disease [52,53]. Notably, several SNPs in the SLC2A14 gene region were also reported to be associated with alcohol dependence at the p ≤ 10−4 level of significance [54], which is known to be at least partly pertinent to the opioid system in its etiology [55]. Since alcohol dependence could be caused by the rewarding effects of alcohol, in which the opioid system is also involved, an antagonist of opioid receptors, naltrexone, is widely used for the treatment of alcohol dependence [56]. One of the associated SNPs, rs11612319, was also included in our GWAS and not strongly but rather modestly associated with the phenotype (p = 4.325 × 10−4). Although the impact of this SNP on SLC2A14 function and the opioid system remains unknown, this SNP may have some influence on both the risk for alcohol dependence and opioid sensitivity.
One limitation of the present study was the relatively small sample size, which might have led to an inability to detect significant associations in the gene-set analysis. Other limitations include heterogeneity of the primary disease and the level of cancer pain. In the present study, patients with various types of cancer were recruited, and a GWAS was conducted in all samples. With much larger sample sizes, GWASs could be performed in a particular subset of patients, such as only in patients with lung cancer (Table 1), which was actually difficult in the present study because only a small number of such patients (<100) would have been included in the GWAS. The lack of data on the level of cancer pain in each patient in the present study also hampered the ability to conduct a GWAS only in patients with cancer pain of relatively homogeneous severity. Notwithstanding these limitations, the present study identified several SNPs and the SLC2A14 gene that were significantly associated with the phenotype. Regardless of the type of cancer or severity of cancer pain, these SNPs and this gene might commonly impact opioid sensitivity, and they were associated with the need for opioids to treat cancer pain in analyses of whole samples.
The results of the present study suggest that the rs1283671 and rs1283720 SNPs of the ANGPT1 gene could be clinical markers that predict analgesic requirements, in which the minor alleles of these SNPs are possibly associated with lower opioid sensitivity and thus greater requirements for opioid analgesics in the treatment of cancer pain. Before clinically applying these findings, further studies should be designed to better predict analgesic requirements, such as studies of the construction and validation of prediction formulas for analgesic requirements, similar to previous studies [57,58], by considering the influence of other possible candidate SNPs and non-genetic factors.
5. Conclusions
In conclusion, our GWASs discovered SNPs and a gene that were associated with opioid analgesic requirements in the treatment of cancer pain, including the ANGPT1 rs1283671 and rs1283720 SNPs and SLC2A14 gene. Although the present results need to be corroborated by more research with larger sample sizes, these findings indicate that these SNPs in the ANGPT1 and SLC2A14 genes could serve as markers that predict the efficacy of opioid analgesics in the treatment of cancer pain.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cancers14194692/s1, Figure S1: Log quantile-quantile (QQ) p-value plot as a result of the GWAS for all patient samples; Figure S2: Regional plots of potent loci that were associated with daily opioid requirements in cancer pain treatment; Figure S3: State of linkage disequilibrium (LD) between SNPs in the genomic region of the ANGPT1 gene; Table S1: Estimated systemic dose equipotent to 90 mg oral morphine (mg); Table S2: Top 10 candidate gene sets selected from gene-set analysis.
Author Contributions
Conceptualization, T.T. and K.I. (Kazutaka Ikeda); methodology, D.N., J.H., K.N. and Y.E.; software, D.N.; validation, D.N., J.H., K.N. and Y.E.; formal analysis, D.N.; investigation, D.N. and S.K.; resources, T.T. and K.I. (Kunihiko Ishitani); data curation, D.N., J.H., K.N. and Y.E.; writing—original draft preparation, D.N.; writing—review and editing, D.N., T.T. and K.I. (Kazutaka Ikeda); visualization, D.N.; supervision, T.T. and K.I. (Kazutaka Ikeda); project administration, K.I. (Kazutaka Ikeda); funding acquisition, D.N. and K.I. (Kazutaka Ikeda). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. 22790518, 23390377, 24790544, 26293347, JP22H04922 [AdAMS], 17H04324, 17K08970, 18K08829, 20K09259, and 21H03028), Ministry of Health, Labour, and Welfare (MHLW) of Japan (no. H26-Kakushintekigan-ippan-060), Japan Agency for Medical Research and Development (AMED; no. JP19ek0610011 and JP19dk0307071), Smoking Research Foundation (Tokyo, Japan), and Japan Research Foundation for Clinical Pharmacology (JRFCP). The article processing charge (APC) was funded by the Japan Society for the Promotion of Science (JSPS) KAKENHI.
Institutional Review Board Statement
The study was conducted according to guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Higashi-Sapporo Hospital (protocol code: none, date of approval: November 20, 2016) and Tokyo Metropolitan Institute of Medical Science (protocol code: 17-1, date of approval: 31 March 2017).
Informed Consent Statement
Informed consent was obtained from all subjects who were involved in the study.
Data Availability Statement
Data that are presented in this study are available upon request from the corresponding author.
Acknowledgments
We thank Michael Arends for editing the manuscript. We are grateful to the volunteers for their participation in the study and anesthesiologists and surgeons for collecting the clinical data.
Conflicts of Interest
Kazutaka Ikeda has received support from Asahi Kasei Pharma Corporation for a project that is unrelated to this research and speaker’s and consultant’s fees from MSD K.K., VistaGen Therapeutics, Inc., Atheneum Partners Otsuka Pharmaceutical Co. Ltd., Taisho Pharmaceutical Co. Ltd., Eisai, Daiichi-Sankyo, Inc., Sumitomo Pharma, Japan Tobacco, Inc., EA Pharma Co. Ltd., and Nippon Chemiphar. The authors declare no other conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in writing the manuscript; or in the decision to publish the results.
References
- Raja, S.N.; Carr, D.B.; Cohen, M.; Finnerup, N.B.; Flor, H.; Gibson, S.; Keefe, F.J.; Mogil, J.S.; Ringkamp, M.; Sluka, K.A.; et al. The revised International Association for the Study of Pain definition of pain: Concepts, challenges, and compromises. Pain 2020, 161, 1976–1982. [Google Scholar] [CrossRef]
- Van den Beuken-van Everdingen, M.H.; Hochstenbach, L.M.; Joosten, E.A.; Tjan-Heijnen, V.C.; Janssen, D.J. Update on prevalence of pain in patients with cancer: Systematic review and meta-analysis. J. Pain Symptom Manag. 2016, 51, 1070–1090. [Google Scholar] [CrossRef]
- WHO Guidelines for the Pharmacological and Radiotherapeutic Management of Cancer Pain in Adults and Adolescents. Available online: https://www.who.int/publications/i/item/9789241550390 (accessed on 9 August 2022).
- Ikeda, K.; Ide, S.; Han, W.; Hayashida, M.; Uhl, G.R.; Sora, I. How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol. Sci. 2005, 26, 311–317. [Google Scholar] [CrossRef]
- Nishizawa, D.; Ikeda, K. Genome-wide association studies and human opioid sensitivity. In Neuropathology of Drug Addictions and Substance Misuse Volume 1: Foundations of Understanding, Tobacco, Alcohol, Cannabinoids and Opioids; Preedy, V.R., Ed.; Academic Press: London, UK, 2016; pp. 909–921. [Google Scholar]
- Angst, M.S.; Phillips, N.G.; Drover, D.R.; Tingle, M.; Galinkin, J.L.; Christians, U.; Swan, G.E.; Lazzeroni, L.C.; Clark, J.D. Opioid pharmacogenomics using a twin study paradigm: Methods and procedures for determining familial aggregation and heritability. Twin Res. Hum. Genet. 2010, 13, 412–425. [Google Scholar] [CrossRef]
- Angst, M.S.; Phillips, N.G.; Drover, D.R.; Tingle, M.; Ray, A.; Swan, G.E.; Lazzeroni, L.C.; Clark, J.D. Pain sensitivity and opioid analgesia: A pharmacogenomic twin study. Pain 2012, 153, 1397–1409. [Google Scholar] [CrossRef]
- Branford, R.; Droney, J.; Ross, J.R. Opioid genetics: The key to personalized pain control? Clin. Genet. 2012, 82, 301–310. [Google Scholar] [CrossRef]
- Droney, J.; Riley, J.; Ross, J. Opioid genetics in the context of opioid switching. Curr. Opin. Supportive Palliat. Care 2012, 6, 10–16. [Google Scholar] [CrossRef]
- Vuilleumier, P.H.; Stamer, U.M.; Landau, R. Pharmacogenomic considerations in opioid analgesia. Pharm. Pers. Med. 2012, 5, 73–87. [Google Scholar] [CrossRef][Green Version]
- Yu, Z.; Wen, L.; Shen, X.; Zhang, H. Effects of the OPRM1 A118G polymorphism (rs1799971) on opioid analgesia in cancer pain: A systematic review and meta-analysis. Clin. J. Pain 2019, 35, 77–86. [Google Scholar] [CrossRef]
- Matic, M.; Jongen, J.L.; Elens, L.; de Wildt, S.N.; Tibboel, D.; Sillevis Smitt, P.A.; van Schaik, R.H. Advanced cancer pain: The search for genetic factors correlated with interindividual variability in opioid requirement. Pharmacogenomics 2017, 18, 1133–1142. [Google Scholar] [CrossRef]
- Yennurajalingam, S.; Astolfi, A.; Indio, V.; Beccaro, M.; Schipani, A.; Yu, R.; Shete, S.; Reyes-Gibby, C.; Lu, Z.; Williams, J.L.; et al. Genetic factors associated with pain severity, daily opioid dose requirement, and pain response among advanced cancer patients receiving supportive care. J. Pain Symptom Manag. 2021, 62, 785–795. [Google Scholar] [CrossRef]
- Yang, G.S.; Barnes, N.M.; Lyon, D.E.; Dorsey, S.G. Genetic variants associated with cancer pain and response to opioid analgesics: Implications for precision pain management. Semin. Oncol. Nurs. 2019, 35, 291–299. [Google Scholar] [CrossRef]
- Daly, A.K. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 2010, 11, 241–246. [Google Scholar] [CrossRef]
- Cook-Sather, S.D.; Li, J.; Goebel, T.K.; Sussman, E.M.; Rehman, M.A.; Hakonarson, H. TAOK3, a novel genome-wide association study locus associated with morphine requirement and postoperative pain in a retrospective pediatric day surgery population. Pain 2014, 155, 1773–1783. [Google Scholar] [CrossRef]
- Fukuda, K.; Hayashida, M.; Ide, S.; Saita, N.; Kokita, Y.; Kasai, S.; Nishizawa, D.; Ogai, Y.; Hasegawa, J.; Nagashima, M.; et al. Association between OPRM1 gene polymorphisms and fentanyl sensitivity in patients undergoing painful cosmetic surgery. Pain 2009, 147, 194–201. [Google Scholar] [CrossRef]
- Nishizawa, D.; Fukuda, K.; Kasai, S.; Hasegawa, J.; Aoki, Y.; Nishi, A.; Saita, N.; Koukita, Y.; Nagashima, M.; Katoh, R.; et al. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol. Psychiatry 2014, 19, 55–62. [Google Scholar] [CrossRef]
- Kurita, G.P.; Sjogren, P.; Ekholm, O.; Kaasa, S.; Loge, J.H.; Poviloniene, I.; Klepstad, P. Prevalence and predictors of cognitive dysfunction in opioid-treated patients with cancer: A multinational study. J. Clin. Oncol. 2011, 29, 1297–1303. [Google Scholar] [CrossRef]
- Galvan, A.; Skorpen, F.; Klepstad, P.; Knudsen, A.K.; Fladvad, T.; Falvella, F.S.; Pigni, A.; Brunelli, C.; Caraceni, A.; Kaasa, S.; et al. Multiple loci modulate opioid therapy response for cancer pain. Clin. Cancer Res. 2011, 17, 4581–4587. [Google Scholar] [CrossRef]
- Terui, T.; Koike, K.; Hirayama, Y.; Kusakabe, T.; Ono, K.; Mihara, H.; Kobayashi, K.; Takahashi, Y.; Nakajima, N.; Kato, J.; et al. Recent advances in palliative cancer care at a regional hospital in Japan. Am. J. Hosp. Palliat. Med. 2014, 31, 717–722. [Google Scholar] [CrossRef]
- Hirayama, Y.; Terui, T.; Kusakabe, T.; Koike, K.; Ono, K.; Kato, J.; Ishitani, K. A survey of patients who were referred to our palliative care division from other hospitals and appeared to have obvious indications for cancer chemotherapies. Am. J. Hosp. Palliat. Care 2014, 31, 804–807. [Google Scholar] [CrossRef]
- McPherson, M.L.; Walker, K.A.; Davis, M.P.; Bruera, E.; Reddy, A.; Paice, J.; Malotte, K.; Lockman, D.K.; Wellman, C.; Salpeter, S.; et al. Safe and appropriate use of methadone in hospice and palliative care: Expert consensus white paper. J. Pain Symptom Manag. 2019, 57, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Quigley, C.; Wiffen, P. A systematic review of hydromorphone in acute and chronic pain. J. Pain Symptom Manag. 2003, 25, 169–178. [Google Scholar] [CrossRef]
- Cherny, N.I.; Fallon, M.T.; Kaasa, S.; Portenoy, R.K.; Currow, D.C. Oxford Textbook of Palliative Medicine, 6th ed.; Oxford University Press: Oxford, UK, 2021. [Google Scholar]
- De Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- De Bakker, P.I.; Ferreira, M.A.; Jia, X.; Neale, B.M.; Raychaudhuri, S.; Voight, B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008, 17, R122–R128. [Google Scholar] [CrossRef]
- Barsh, G.S.; Copenhaver, G.P.; Gibson, G.; Williams, S.M. Guidelines for genome-wide association studies. PLoS Genet. 2012, 8, e1002812. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences, revised ed.; Academic Press: New York, NY, USA, 1977. [Google Scholar]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic. Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic. Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef] [PubMed]
- Lotsch, J.; Klepstad, P.; Doehring, A.; Dale, O. A GTP cyclohydrolase 1 genetic variant delays cancer pain. Pain 2010, 148, 103–106. [Google Scholar] [CrossRef]
- Chen, J.; Yang, T.; Yu, H.; Sun, K.; Shi, Y.; Song, W.; Bai, Y.; Wang, X.; Lou, K.; Song, Y.; et al. A functional variant in the 3’-UTR of angiopoietin-1 might reduce stroke risk by interfering with the binding efficiency of microRNA 211. Hum. Mol. Genet. 2010, 19, 2524–2533. [Google Scholar] [CrossRef]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef]
- Elkind, M.S. Inflammation, atherosclerosis, and stroke. Neurologist 2006, 12, 140–148. [Google Scholar] [CrossRef]
- Parikh, S.M. Angiopoietins and Tie2 in vascular inflammation. Curr. Opin. Hematol. 2017, 24, 432–438. [Google Scholar] [CrossRef]
- Suri, C.; Jones, P.F.; Patan, S.; Bartunkova, S.; Maisonpierre, P.C.; Davis, S.; Sato, T.N.; Yancopoulos, G.D. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell 1996, 87, 1171–1180. [Google Scholar] [CrossRef]
- Bafunno, V.; Firinu, D.; D’Apolito, M.; Cordisco, G.; Loffredo, S.; Leccese, A.; Bova, M.; Barca, M.P.; Santacroce, R.; Cicardi, M.; et al. Mutation of the angiopoietin-1 gene (ANGPT1) associates with a new type of hereditary angioedema. J. Allergy Clin. Immunol. 2018, 141, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Converse, M.P.; Sobhanian, M.; Taber, D.J.; Houston, B.A.; Meadows, H.B.; Uber, W.E. Effect of angiotensin II inhibitors on gastrointestinal bleeding in patients with left ventricular assist devices. J. Am. Coll. Cardiol. 2019, 73, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Poplawski, M.M.; Mastaitis, J.W.; Mobbs, C.V. Naloxone, but not valsartan, preserves responses to hypoglycemia after antecedent hypoglycemia: Role of metabolic reprogramming in counterregulatory failure. Diabetes 2011, 60, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.; Cui, M.H.; Shamoon, H.; Gabriely, I. Hypoglycemia-associated autonomic failure is prevented by opioid receptor blockade. J. Clin. Endocrinol. Metab. 2009, 94, 3372–3380. [Google Scholar] [CrossRef] [PubMed]
- Caprio, S.; Gerety, G.; Tamborlane, W.V.; Jones, T.; Diamond, M.; Jacob, R.; Sherwin, R.S. Opiate blockade enhances hypoglycemic counterregulation in normal and insulin-dependent diabetic subjects. Am. J. Physiol. 1991, 260, E852–E858. [Google Scholar] [CrossRef] [PubMed]
- Pendyala, G.; Periyasamy, P.; Callen, S.; Fox, H.S.; Lisco, S.J.; Buch, S.J. Chronic SIV and morphine treatment increases heat shock protein 5 expression at the synapse. J. Neurovirol. 2015, 21, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Freeze, H.H. GLUT14, a duplicon of GLUT3, is specifically expressed in testis as alternative splice forms. Genomics 2002, 80, 553–557. [Google Scholar] [CrossRef]
- Wang, W.; Yu, J.T.; Zhang, W.; Cui, W.Z.; Wu, Z.C.; Zhang, Q.; Tan, L. Genetic association of SLC2A14 polymorphism with Alzheimer’s disease in a Han Chinese population. J. Mol. Neurosci. 2012, 47, 481–484. [Google Scholar] [CrossRef]
- Amir Shaghaghi, M.; Zhouyao, H.; Tu, H.; El-Gabalawy, H.; Crow, G.H.; Levine, M.; Bernstein, C.N.; Eck, P. The SLC2A14 gene, encoding the novel glucose/dehydroascorbate transporter GLUT14, is associated with inflammatory bowel disease. Am. J. Clin. Nutr. 2017, 106, 1508–1513. [Google Scholar] [CrossRef]
- Edenberg, H.J.; Koller, D.L.; Xuei, X.; Wetherill, L.; McClintick, J.N.; Almasy, L.; Bierut, L.J.; Bucholz, K.K.; Goate, A.; Aliev, F.; et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 2010, 34, 840–852. [Google Scholar] [CrossRef]
- Nutt, D.J. The role of the opioid system in alcohol dependence. J. Psychopharmacol. 2014, 28, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Michalak, A.; Biala, G. Alcohol dependence: Neurobiology and treatment. Acta Pol. Pharm. 2016, 73, 3–12. [Google Scholar] [PubMed]
- Yoshida, K.; Nishizawa, D.; Ichinomiya, T.; Ichinohe, T.; Hayashida, M.; Fukuda, K.; Ikeda, K. Prediction formulas for individual opioid analgesic requirements based on genetic polymorphism analyses. PLoS ONE 2015, 10, e0116885. [Google Scholar] [CrossRef]
- Cheng, H.; Zeng, R.; Kong, L.; Ding, C.; He, Y.; Zhuang, W.; Sun, Y. Establishment of predicting equation for individual sufentanil dosage postoperatively based on gene polymorphisms. Pain Pract. 2022, 22, 39–46. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).